Abstract
Telomeres are nucleoprotein complexes that cap the ends of all linear chromosomes and function to prevent aberrant repair and end-to-end chromosome fusions. In somatic cells, telomere shortening is a natural part of the aging process as it occurs with each round of cell division. In germ and stem cells, however, the enzyme telomerase synthesizes telomere DNA to counter-balance telomere shortening and help maintain cellular proliferation. Of the primary telomere end-binding proteins, TPP1 has recently emerged as a primary contributor in protecting telomere DNA and in recruiting telomerase to the telomere ends. In this review, we summarize the current knowledge regarding the role of TPP1 in telomere maintenance.
1. Introduction
All linear chromosomes end in specialized nucleoprotein moieties called telomeres [1–5]. The DNA component of telomeres includes long double-stranded stretches, which extend approximately eight thousand bases in embryonic human cells, decreasing in length with age. The double-stranded telomere DNA is then followed by a single-stranded DNA (ssDNA) overhang that is typically 50–200 nucleotides in length [6–8]. The DNA sequence in both the double and single-stranded stretches is composed of multiple tandem repeats that vary among species, but typically consist of short G/T-rich segments. In vertebrates this sequence is TTAGGG [6, 9].
One role of the repetitive telomere DNA sequence is to protect against the loss of genetic information during chromosome shortening that occurs during the natural aging process. Since DNA polymerases are unable to fully replicate the extreme ends of chromosomes [10, 11], a gradual attrition occurs at the telomere at each round of cell division [12, 13]. Eventually, the telomeres of adult somatic cells reach a critically short length that induces a state of replicative senescence, thereby halting cell division and, thus, limiting proliferative capacity. In some rapidly dividing and/or immortalized cells (e.g. during embryogenesis, in the germline, stem cells and in cancer), a specialized ribonucleoprotein complex called telomerase is upregulated. Telomerase is responsible for synthesizing telomere DNA at the ends of chromosomes, thereby helping to maintain an adequate telomere length for cell division to proceed indefinitely [14–18].
Telomere DNA is bound and shielded by specialized telomere-end binding proteins to mask it from appearing as damaged DNA and to avoid induction of the DNA damage response [19, 20]. In humans six specialized proteins, coined shelterin [20, 21], coordinately bind and protect both the double-stranded and single-stranded telomeric DNA. Telomeric Repeat binding factors (TRF1 and TRF2) bind the double-stranded telomeric DNA repeats, whereas the Protection of Telomere 1 (POT1) protein specifically binds the ssDNA overhang [20, 22–29]. Repressor-activator protein (RAP1) is an associated factor of TRF2 [30, 31] and TRF interacting protein (TIN2) connects TRF1 with TRF2 and facilitates interactions between the double- and single-stranded telomere end-binding proteins [32–34]. The sixth protein of the shelterin complex is called TPP1. The term “TPP1” is the result of combining the first letter of each name, TINT1 [35]; PTOP [33]; PIP1 [36], that it was given by the three groups that initially characterized the human protein. To muddle the nomenclature, the accepted gene symbol for TPP1 is ACD after the adrenocortical dysplasia phenotype exhibited in mice lacking the gene [37].
While TPP1 is not known to interact directly with telomere DNA, it plays a vital role in telomere maintenance. In addition to interacting with other shelterin proteins to properly protect DNA, TPP1 regulates the recruitment of telomerase to the telomere [38–43]. Although the cellular functions of TPP1 have been elucidated over the past decade or so, the molecular mechanisms accounting for the biological processes have only begun to emerge. In this review, we discuss the overall activities, functions, and roles of TPP1 in telomere maintenance. Though there are TPP1 homologs from other organisms, this review focuses primarily on the human protein unless otherwise stated.
2. Structure of TPP1
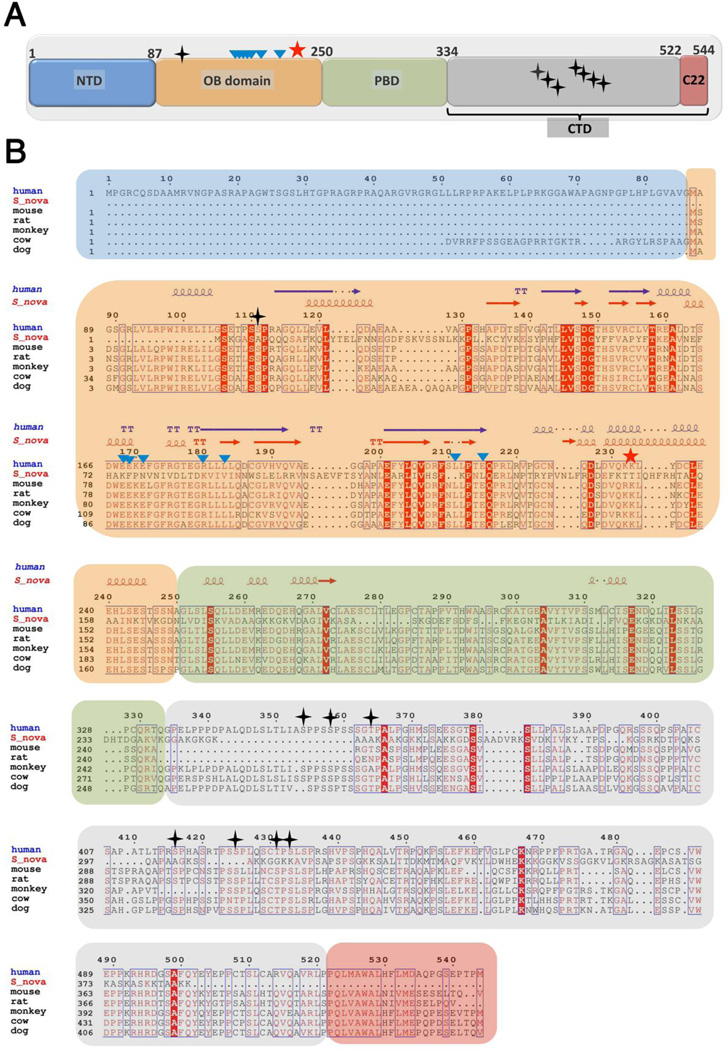
TPP1 is a multi-domain protein (544 amino acids in length) with defined regions that serve as interacting sites for other shelterin proteins and telomerase. The N-terminus (residues 1–86) is not conserved among species (Figure 1) and exhibits no sequence homology to known structural motifs. Most TPP1 orthologs, including that of mouse, dog, monkey and rat, lack this region entirely (Figure 1). Furthermore, the first 86 residues are dispensable for all known functions of TPP1 [39, 40, 44, 45]. Based on these observations, it might be argued that the TPP1 open-reading frame truly starts at Met87 with the first 86 residues assigned to the protein arising from a potential error in gene annotation. The next domain (residues 87–250 aa) comprises an oligosaccharide/oligonucleotide-binding (OB)-fold domain (referred to as TPP1-OBD). The crystal structure of the human TPP1-OBD reveals a typical OB-fold architecture that is common among many DNA-binding proteins [39]. Despite its name, the OB-fold of TPP1 is not known to facilitate direct interactions with oligonucleotides alone. However OB-fold domains have also been implicated in coordinating protein-protein interactions within multi-component complexes [46, 47]. In this regard, TPP1-OBD helps coordinate interactions with telomerase to facilitate its recruitment to the telomere [40–43]. These telomere-associated functions of TPP1 are covered in more detail later in this review, as well as elsewhere (for example [20, 25, 48–50]).
Figure 1.
Domain organization and sequence alignment of TPP1 proteins. (A) Cartoon schematic depicting the individual domains of TPP1. The first 86 residues of human TPP1 (NTD; blue) are not conserved. The 87–250 region contains the OB-fold domain (TPP1-OBD; orange). Residues 250–334 comprise the POT1-binding domain (PBD; green). There are multiple Ser/Thr phosphorylation sites within the C-terminal domain (CTD; 334–544aa; grey), while the last 22 residues (C22; red) are most critical for TIN2 interactions. Phosphorylation sites (S111 in OBD and seven in CTD) are highlighted with black-stars. The TEL patch residues involved in telomerase interaction are marked as teal-triangles. Lys233 has been shown to be ubiquitylated and is labeled as a red star. (B) Multiple sequence alignment of TPP1 with its homologs. The multiple sequence alignment was performed using Clustal-omega [101] and further formatted using Espript [102] to highlight sequence similarities and identities among the proteins. Symbols and protein domain colors are the same as described in panel (A). The secondary structural elements of human TPP1(87–250) and S.nova (1–222) are depicted above the sequence alignment.
OB-folds are found in all three kingdoms of life and function as independent proteins, as tandem domains within a single polypeptide, and within multiple protein complexes [46, 47]. An individual OB-fold domain presents a conserved architecture that is comprised of five antiparallel β-strands that are highly curved and arranged to form a cylindrical β-barrel [51] (Figure 2). The ends of the β-barrel are usually capped by a conserved α-helix residing between the β3 and β4 strands and an N-terminal helix or loop at the opposite end [51, 52] (Figures 2 & 3). The residues forming the inner side of the β-barrel are typically hydrophobic in nature and the end-helices/loop act as protective gates, limiting access to the interior of the β-barrel. A cleft along one side of the β-barrel generally provides a platform rich in aromatic residues that facilitates base stacking with the substrate [53]. DNA typically binds directionally across the surface of the OB-fold, with its 5’ and 3’ ends proximal to strands 2 and 3, respectively, within the β-barrel [54, 55].
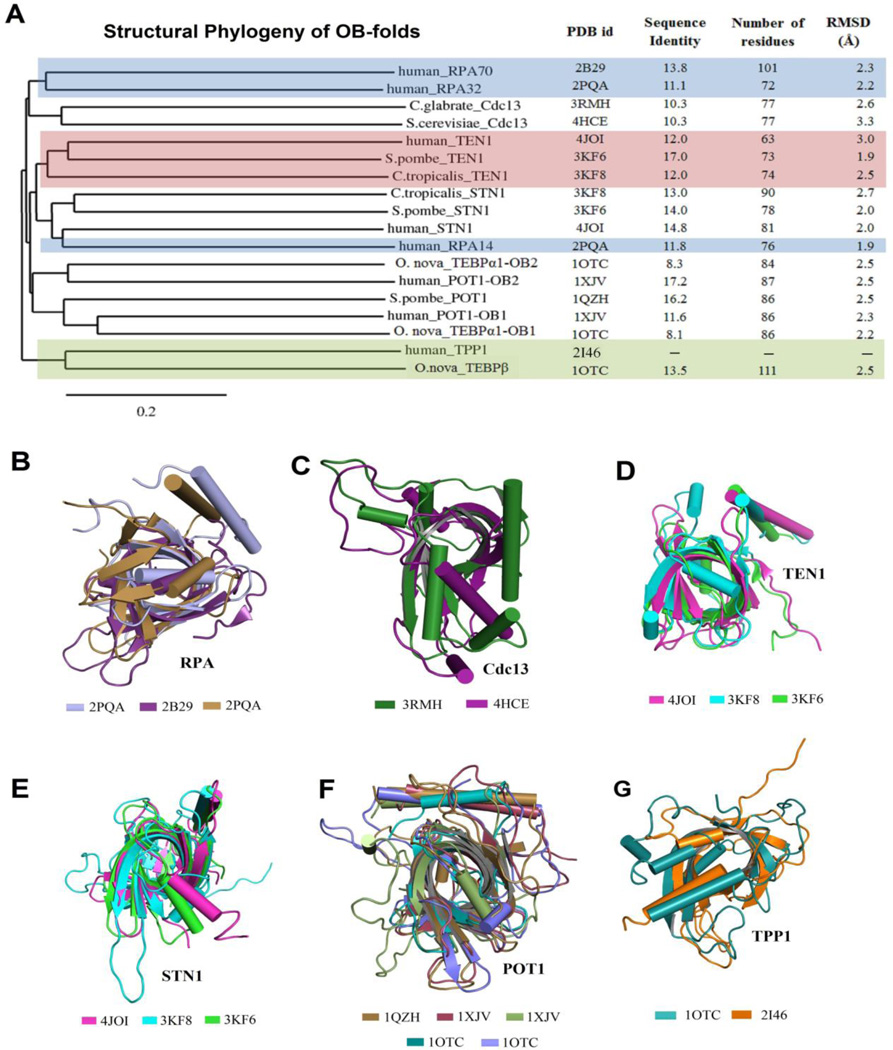
Figure 2.
The structure-based phylogenetic analysis of a representative list of OB-fold containing proteins that are involved in telomere maintenance. Proteins with OB-folds were selected based on telomere-related function and on their structural availability. (A) A three-dimensional structure based comparison was performed using STRAP (STRuctural Alignments of Proteins) to construct a phylogenetic tree based on structural conservation. The tree was constructed using ‘neighbor-joining’ clustering methods in ClustalW. The branches of the tree are labeled with the organism and protein name. The last column is a structural analysis (in terms of RMSD) comparing the structure of individual OB-domains with human TPP1-OBD. Phylogenetic analysis clustered the various proteins into the following families:(B) RPA, (C) Cdc13, (D) Ten1 domain, (E) Stn1 domain, (F) POT1, and (G) TPP1-like. The similarities in OB-fold architecture, as well as the helix orientation among members, are apparent in the superpositions of members in each family. Note that variations between families are restricted primarily to the loop and helix orientations on the perimeter of the OB-fold architecture. Protein database (PDB) accession numbers are listed below the appropriate structures.
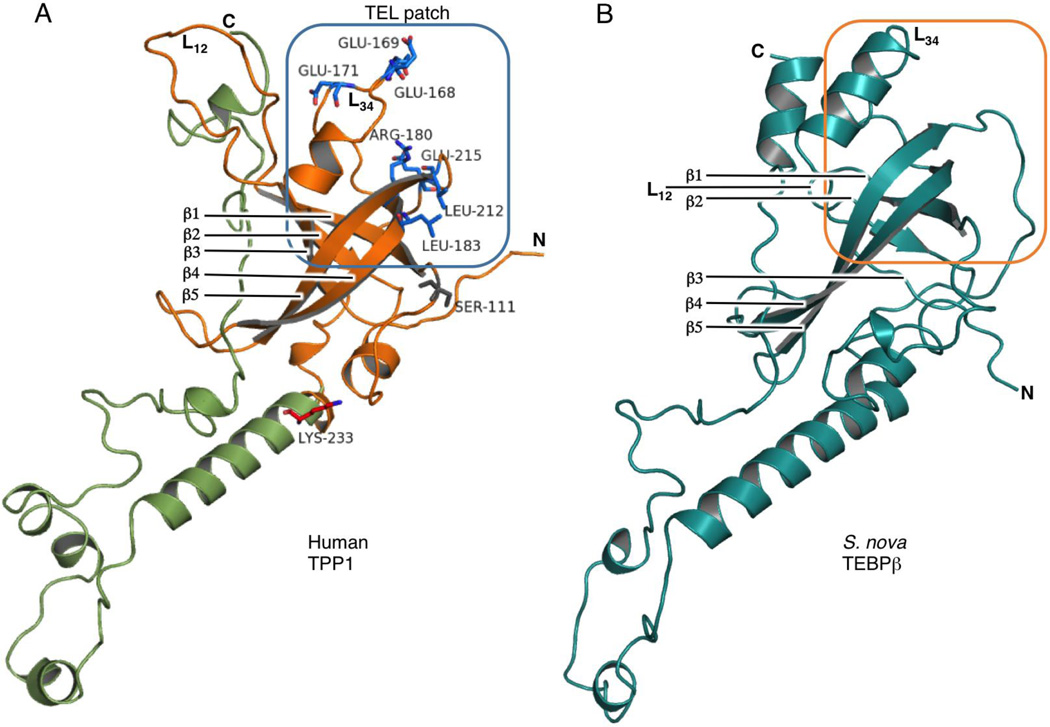
Figure 3.
Structural comparison of OBD and putative PBD of human TPP1 with S. nova TEBPβ. (A) The x-ray crystal structure of human TPP1-OBD (orange) [39], modeled with its predicted POT1-binding domain (green). Ser111, which is recognized as a phosphorylation site implicated in regulating telomerase recruitment, is labeled and represented as a grey stick model. The telomerase-interacting TEL-patch is in the upper box and labeled as such, with important residues represented as blue stick models. The putative PBD of TPP1, shown in green, is a homology model constructed from the analogous region of TEBPβ. (B) The x-ray crystal structure of S. nova TEBPβ within the context of the TEBPα-TEBPβ-ssDNA ternary complex [66]. Despite little sequence identity, human TPP1 and S. nova TEBPβ display significant structural similarities. Based on structural similarities with human TPP1, an analogous TEL patch for TEBPβ is predicted to reside inside the orange box area.
In spite of sharing a common scaffold, there are variations in the overall size of OB-folds, which ranges from about 70 – 150 amino acids in length. The difference in sequence length among OB-fold domains is generally accounted for by variations in the loops connecting the individual β-strands. These loop regions are involved in forming specific hydrogen-bonding interactions with the substrate and therefore augment the OB-fold binding platform to enhance recognition and specificity (e.g. DNA/RNA binding or protein-protein interactions) [51, 54]. The disparity in loop regions, along with a general lack of a specific sequence pattern, has made it difficult to predict putative OB-fold motifs in nucleic acid-binding proteins. This observation is punctuated by the low sequence identity among OB-fold proteins that associate with telomere DNA and have structural models presently available (Figure 2A).
To further evaluate a subset of OB-fold domains classified under telomere maintenance from different organisms, a secondary structure based sequence alignment was used to create a phylogenetic analysis to cluster the proteins into families based on structural conservation [56–59] (Figure 2 B–G). The comparison was performed to include the structures of human TPP1, POT1, RPA (Replication Protein A), Saccharomyces cerevisiae Stn1, Ten1, and Cdc13, Schizosaccharomyces pombe Pot1, Sterkiella nova TEBP (Telomere end-binding proteins) α and β, Candida tropicalis Stn1 and Ten1, Candida glabrata Cdc13 [60–68]. When those structures are superimposed with TPP1-OBD, the modest root-mean-square (r.m.s.) deviations (1.9–3.3 Å) display only minor structural variations, at least globally, within the OB-fold scaffolds between the subset of telomere-associated proteins (Figure 2A). In addition to structural homology, proteins belonging to the same family cluster based on similar functions. For example, phylogenetic analysis clustered all Stn1 proteins from different organisms into a single phylogenetic family. Similar results were obtained for Ten1, POT1, Cdc13, and RPA proteins (Figure 2A).
Superposition of the various sub-classes revealed that the phylogenetic families share similarities in the unique features of the OB-fold domain, namely the position and orientation of the loops and helices connecting the β-strands of the OB-fold (Figure 2B–G). The observed greater conformational changes in these regions support their role in recognition and selective binding of diverse substrate sequences. This phylogenetic analysis, along with prior reports, insinuates the OB-fold design as a favorable scaffold for global structural stability while allowing substantial variations at the sequence level to accommodate diverse recognition patterns.
The phylogenetic analysis clusters human TPP1 with S. nova TEBPβ, suggesting that the two proteins have co-evolved to maintain structural similarities. A more thorough comparison between the crystal structures of human TPP1-OBD and that of S. nova TEBPβ supports this assumption (Figure 3). In both proteins, a helix (158–163 aa in TPP1; 69–74 aa in TEBPβ) and the adjacent L34-loop that connects the β3–β4 strands (164–180 aa in TPP1; 75–93 in TEBPβ) contain multiple hydrophobic residues that form a lid to the OBD in both TPP1 and TEBPβ (Figures 1 & 3). Also, there is a striking conservation in the presence and location of specific regions that are known to play key roles in function. For example, a serine-rich motif resides in the N-terminal region of the two proteins (S90-X(15)-S106-X(3)-S110S111 in TPP1-OBD and S12-X(16)-S29-X(2)-S32S33 in TEBPβ). As discussed in more detail below, many of the serine residues, and particularly Ser111 of TPP1, have been identified as phosphorylation sites and are important for regulating protein-protein interactions with telomerase.
One surface of the OBD of TPP1, formed by the L34-loop (167–172 aa in TPP1) along with the adjacent β4-strand (201–212 aa in TPP1), is rich in glutamate and leucine residues that are conserved among mammalian species [36, 40, 41]. These residues (comprehensively called the TEL-patch) are responsible for telomerase interactions and its recruitment to the telomere. Like TPP1, TEBPβ is responsible for telomerase recruitment [69]. Based on the conservation in structure and function between these two proteins, it is likely that TEBPβ interacts with telomerase using a region of the protein that is analogous to the TEL patch of human TPP1 (Figure 3).
The POT1-Binding Domain (PBD; 250–334 aa) resides just C-terminal to the TPP1-OBD. Although there is currently no structural information for the PBD in human TPP1, this domain is likely to be structurally analogous to the corresponding region of the S. nova TEBPβ protein in the context of the TEBPα-TEBPβ-DNA ternary complex [66]. Support for this analogy stems from genetic mutations and deletions of the predicted PBD of human TPP1, which results in the failure to recognize and bind POT1 [33, 41]. Following the PBD, there is a C-Terminal Domain (CTD) that is responsible for TIN2 recognition, with the last twenty-two residues critically important for TIN2 binding (C22) [35, 70]. Within the CTD of TPP1 there are additional serine residues that have been implicated in regulating telomerase recruitment through phosphorylation [35, 71].
3. TPP1-POT1-ssDNA interaction
Early studies aimed at determining the cellular function of POT1 and TPP1 were done by overexpression or knockdown of either protein in cells. Through these studies it became clear that both proteins play important roles in telomere end protection through prevention of DNA damage responses and chromosomal fusions [61, 72, 73]. Interestingly, knockdown of mouse TPP1 elicits a p53-dependent growth arrest and ATM-dependent DNA damage response (DDR), while POT1 depletion results in an ATR-dependent DDR. In the absence of a functional DDR, reduction of mouse TPP1 levels can promote chromosomal instability, cellular transformation, and tumorigenesis [61]. Additionally, mouse cells completely lacking TPP1 display an increase in chromosomal fusions with non-homologous chromosomes [74]. It is important to note that the aforementioned studies focused on the effects of manipulating cellular levels of either POT1 or TPP1 independently. However, it cannot be ruled out that POT1-TPP1 functions explicitly as a heterodimer in vivo, and the adverse effects are due to disrupting these interactions and/or the POT1:TPP1 stoichiometry that is normally maintained equivalently at ~50–100 per telomere [75].
The most complete structural information to-date of single-stranded telomeric DNA bound by the specialized telomere end-binding proteins is the S. nova TEBPα-TEBPβ-DNA complex [66] The structure of the ternary complex reveals that TEBPβ interacts with TEBPα through a domain consisting of four helices intermingled within an extended peptide loop [66]. The long, N-terminal helix is comprised of multiple charged and polar residues, which form a protein-protein interaction with TEBPα through a network of hydrogen bonds. The C-terminal region of this helix interacts with TEBPα through three hydrophobic residues (L156, I160, and V164) that appear on consecutive turns of the α-helix. A sequence comparison demonstrates that all of these residues are conserved, at least in physicochemical properties, between S. nova TEBPβ and human TPP1 (Figure 1). Furthermore, two regions of this TEBPβ peptide loop cluster into short helices that are composed of hydrophobic residues (I172, V175, I189, and V190) which interact with hydrophobic residues in TEBPα to further facilitate protein-protein interactions [66]. The analogous residues in human TPP1 (L254, L257, L271, and V272) are also hydrophobic, suggesting a similar protein-protein interaction is formed between these residues and POT1. Based on these distinct features, a homology model of the POT1-interacting domain of TPP1 was constructed using the structure of S. nova TEBPβ as a template (Figure 3).
The S. nova ternary structure shows that two OB-folds in the N-terminus of the POT1 homolog, TEBPα, are primarily responsible for binding the G-rich ssDNA, with a single OB-fold in the TPP1 homolog, TEBPβ, participating to kink the DNA into a deep cleft [66]. This interaction results in the nucleobases of the ssDNA being buried within the complex and the phosphate backbone residing closer to the surface. Early conjecture based upon the TEBPα-TEBPβ-DNA structure noted that this bound configuration would likely shield the DNA from enzyme accessibility, thereby potentially protecting it from nuclease digestion and possibly limiting interactions with telomerase. This notion was supported by biochemical evidence showing that TEBPα bound to the terminal DNA end prevented telomerase accessibility [76]. By extension, it was hypothesized that the primary functions of POT1-TPP1 were to protect the DNA ends from insults and negatively regulate telomere extension by telomerase in humans as well.
While POT1 is capable of binding a minimal nine or ten nucleotide-binding site alone, association with TPP1 increases its DNA binding affinity by ten-fold [38, 39, 67, 77]. TPP1 also significantly enhances DNA-RNA discrimination and sequence selectivity for the POT1-TPP1 heterodimer compared to POT1 alone [78]. Currently, it is not known whether the increase in affinity and selectivity is due to conformational changes in POT1 that occur upon TPP1 binding or by a direct interaction between the ssDNA and TPP1 that occurs in the ternary complex, but not between TPP1 and ssDNA alone.
One unique aspect of G-rich single-stranded telomeric DNA lies in its ability to form highly stable structures called G-quadruplexes [79, 80]. Even though POT1 alone is capable of coating G-quadruplex DNA [45, 81], FRET (Förster/fluorescence resonance energy transfer) probes monitored in real-time revealed that TPP1 confers the ability for the complex to dynamically slide along bound DNA templates [82]. A separate study deduced that POT1 is capable of unfolding G-quadruplexes with an antiparallel topology, but not those folded in the more stable, parallel conformation [83]. Meanwhile, POT1-TPP1 together, but not POT1 alone, inhibits binding of RPA to both parallel and antiparallel G-quadruplex DNA templates. Additionally, the binding profile of POT1-TPP1 to telomeric DNA sequences lacking G-quadruplex structure indicates multiple binding events may involve cooperativity [81]. Loading of POT1-TPP1 onto telomeric DNA makes it more susceptible to nuclease digestion compared to uncoated, or POT1-coated, DNA [81]. Together, these properties indicate that POT1-TPP1 may facilitate telomere accessibility for telomerase-mediated extension by promoting subsequent binding events and unfolding of the extended G-quadruplex DNA.
The first study to demonstrate that end-binding proteins helped to regulate telomere DNA length was in conjunction with Pot1 discovery in S. pombe. The deletion of Pot1 in fission yeast led to telomere loss combined with chromosome circularization [84]. In human cells, overexpression of POT1 in a telomerase-positive cell line led to an increase in telomere length [85]. A decrease in either POT1 or TPP1 correlated with increased telomere length and chromosomal instability [33, 35, 36, 86, 87]. Individual contributions of each protein toward regulating telomere length were difficult to decipher, however, as TPP1 is indispensable for POT1 nuclear import and, perhaps, telomere localization [33, 72, 88, 89]. These seemingly contradictory results, in which both increased and decreased POT1 concentrations correlate with elongated telomeres, may have more to do with the functions of TPP1 linking the double-stranded and single-stranded telomeric segments [70, 90]. When the telomeric ends are sufficiently long, TIN2-TPP1-POT1 may normally link the two segments into a telomerase inaccessible closed conformation. However, when the telomeres shorten the TIN2-TPP1 link may become unstable and reveal an open conformation suitable for telomerase extension [21]. Disruption of the telomere end-binding protein stoichiometry may inhibit the ability of these proteins to form the closed conformation. With the single-stranded overhang more accessible, telomerase may elongate the telomeres through processive addition. Recent data supports this hypothesis, at least in fission yeast, where Tpz1 (the ortholog of TPP1) was shown to establish the telomerase non-extendible state by directly linking regions of telomeric double- and single-stranded DNA through shelterin interactions [91].
4. TPP1-Telomerase interaction
In recent years, specific protein-protein interactions have been identified as integral to regulating telomerase activity, with TPP1 emerging as the principal participant [38, 92, 93]. In mouse embryonic fibroblasts, deletion of the Tpp1 gene results in increased sister telomere fusions, a decreased binding of telomerase at telomeres, and perinatal death [94]. The selective depletion of TPP1 in HeLa cells also abolishes telomerase colocalization at telomeres [42]. Telomerase recruitment relies on a physical interaction with TPP1, which has been shown to be coordinated through the TPP1-OBD [42, 95]. A direct interaction has been demonstrated in cells by successfully localizing telomerase to TPP1-OBD tethered to non-telomeric, chromatin loci [43].
Co-immunopurification experiments, along with mutational analysis, have pinpointed the molecular interactions that reside between telomerase and TPP1. These studies have identified G100 in the N-terminus of telomerase to be a primary contributor to coordinated interactions with TPP1 [93]. On TPP1, a series of mutations of conserved residues within the L34-loop (167–172 aa) disrupted its association with the telomerase enzyme [41]. Other mutational studies confined the points of interaction between telomerase and a specific patch of the TPP1-OBD [40]. Of the mutations interrogated (12 single point mutations and 2 double mutants) many resulted in partial recruitment, while one double mutant (E169A/E171A) completely eliminated telomerase recruitment and telomere elongation. The region of TPP1 responsible for telomerase recruitment has been dubbed the “TEL” patch due to the high conservation of glutamate and leucine residues (i.e. TPP1 glutamate (E) and leucine (L)-rich) in this region [40] (Figures 1 & 3).
Independent of recruitment, the POT1-TPP1 heterodimer increases telomerase processivity, or the ability to add multiple repeats to an oligonucleotide following a single binding event [39]. TPP1, along with POT1, contributes to telomerase processivity by reducing dissociation of telomerase from a DNA primer [44]. The increased stability of the telomerase-DNA complex due to the presence of POT1-TPP1 stabilizes telomerase translocation, the critical step of telomerase processivity in which telomerase translocates and rebinds its RNA template to synthesize subsequent hexameric repeats.
5. TPP1-TIN2-Shelterin interaction
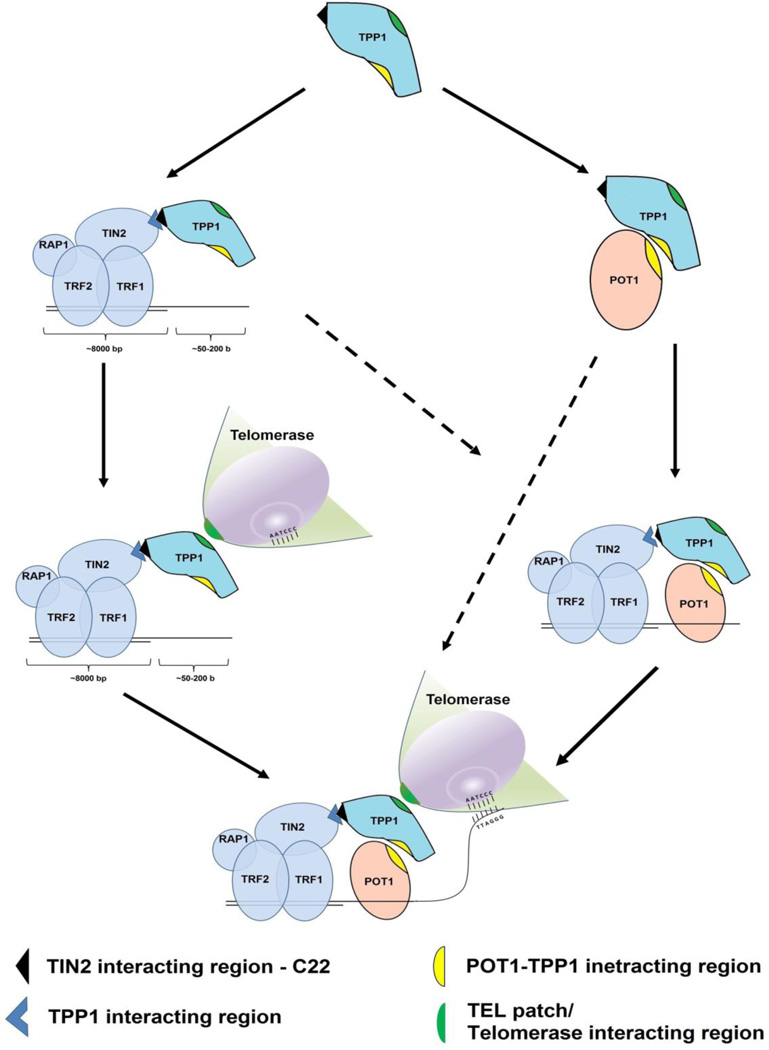
The properties and interactions of POT1 and TPP1 might present a model in which POT1 recognizes and binds the telomere DNA overhang while TPP1 recruits and, through its interaction with POT1, delivers telomerase to the telomere. Other data present an alternative scenario in which the localization of POT1 at the telomere is dependent on the presence of TPP1 [33, 42, 61, 72, 88, 90]. POT1 constructs with intact OB-folds necessary for recognizing and binding telomere DNA, but with deleted TPP1-interacting domains are defective in telomere localization [33, 38]. Conversely, POT1 constructs with OB-fold deletions, but with an intact TPP1-interacting domain, continue to localize at telomeres even though telomere length is increased [86]. Similarly, knockdown of TPP1 inhibits the recruitment of both POT1 [33, 36] and telomerase [42] to telomeres. Together, these data indicate that TPP1 might be responsible for POT1 recruitment to the telomere (Figure 4). However, it is important to note that the POT1 localization results are challenging to interpret in the context of the telomere. Since the double-stranded region (~10 kb) is substantially longer than the ssDNA overhang (50–200 nts), the consequences of abrogating the interactions between POT1 and TRF1 and TRF2 (through TIN2-TPP1) are significantly easier to detect in comparison to the relatively few direct interactions between POT1 and the shorter ssDNA section. As such, it is possible that POT1 is recruited to the ssDNA through its DNA affinity and independently to the shelterin complex through its interactions with TPP1.
Figure 4.
Schematic representation of TPP1 and its role in telomere maintenance. TPP1 interacts with various members of the shelterin complex. In addition to shelterin proteins, TPP1 interacts with and recruits telomerase to the telomere. Derived pathways for TPP1 localization at the telomere and telomerase recruitment are shown.
As TPP1 is important for POT1 localization, the presence of TPP1 at the telomere appears to rely on its interactions with TIN2. TIN2 is associated with the double-stranded region of the telomere, through interactions with TRF1 and TRF2 [19, 34, 35, 88]. Thus, on the double-stranded region of the telomere, TIN2 contributes to the stabilization of TRF1 and TRF2 in binding DNA [19, 34, 35, 96]. Localization of POT1-TPP1 to the single-stranded region of the telomere is facilitated through interactions with TIN2. Knock-down of TIN2 in mice diminishes the amount of POT1-TPP1 detected at the telomere and results in accumulation of replication protein A (RPA) instead, thereby triggering DNA damage response via ATR signaling [90]. Depletion of TIN2 also abrogates TPP1-dependent telomerase recruitment to the telomere [42].
The interaction between TPP1 and TIN2 has been elucidated using several reconstitution studies. A yeast two-hybrid screen identified the C-terminus (residues 425–544) of TPP1 to be responsible for TIN2 interactions [35, 70]. Subsequent experiments using truncation mutants in mammalian cells identified the last 22 residues of TPP1 to facilitate interactions with TIN2 [70]. In addition to providing a physical link between the two proteins, the TIN2-TPP1 interaction is also important in stabilizing TRF1-TRF2 subcomplexes. For example, TRF1-TIN2-TRF2 co-immunoprecipitate as a complex only in the presence of TPP1, and overexpression of TPP1 enhances TIN2-TRF2 association [70]. These findings suggest that the double-stranded DNA binding proteins (TRF1-TIN2-TRF2) and the ssDNA binding protein POT1 are linked via TPP1, an indispensable member in shelterin complex assembly and stabilization.
6. Post-translational modifications of TPP1
It is clear that TPP1 uses a series of conserved residues on the surface of its OBD to recruit telomerase to the telomere, and recent data indicates that this interaction may be enhanced by phosphorylation of TPP1 in a cell-cycle dependent manner. Studies of telomere end binding proteins from yeast and ciliates have demonstrated that phosphorylation of certain components can switch from telomere end-protective roles to telomerase recruitment. Specifically, the phosphorylation of Cdc13 in S. cerevisiae by CDK1 kinase favors an interaction with the Est1 subunit of telomerase (telomerase recruitment) over interaction with Stn1/Ten1 proteins (end protection) [97, 98]. Similarly, in the ciliate, S. lemnae, the phosphorylation of TEBPβ plays an integral part in regulating G-quadruplex formation and unfolding of telomere DNA in vivo [99]. Additional experiments revealed that phosphorylation of TEBPβ in S. lemnae enhanced telomerase recruitment at the telomere, which may contribute to improved G-quadruplex unfolding [69].
On human TPP1, at least eight phosphorylation sites have been identified, all of which are phosphorylated during cell cycle progression [38, 39, 71]. One of these phosphorylation sites is located in the TPP1-OBD (Ser111) and the others reside in the CTD (Figures 1 & 3). In particular, the phosphorylation (presumably by a cyclin-dependent kinase) of Ser111 in the TPP1-OBD during S to G2/M phases has been associated with telomere length maintenance through enhanced telomerase recruitment [71]. These findings indicate that Ser111 of TPP1 might function as a cell-cycle dependent molecular switch, whereupon phosphorylation enhances telomerase recruitment and activity. Alternatively, enhanced end-protective functions have been associated with increased TPP1 homodimerization induced through phosphorylation by the Akt kinase [71]. In this report, the TPP1-TPP1 interacting domain was localized to its OB-fold domain and Akt was essential for promoting this association. However, the particular site of phosphorylation that induces the homodimerization of TPP1 has not been explicitly identified. Nonetheless, taken together, these data suggest that cell-cycle dependent phosphorylation of TPP1 plays a role in determining whether telomerase recruitment or end-protective functions are promoted.
In addition to putative regulation by phosphorylation, ubiquitylation of TPP1 has been shown to enhance its telomere localization [100]. In this report, it was shown in mouse cells that the E3-ubiquitin protein ligase, Rnf8, interacts with Tpp1 to generate Lys63 polyubiquitin chains that stabilize Tpp1 at the telomeres. The ubiquitylation event was dependent upon on the conserved Lys233 in Tpp1. Tpp1-Rnf8 interaction improves Tpp1 nuclear retention and telomere localization to help prevent rapid telomere shortening and chromosome fusions. These recent reports demonstrate that post-translational modifications are important mechanisms in the modulation of TPP1 function, and provide a glimpse into areas for further investigation.
7. Conclusions
Based on these collated reports, it is clear that TPP1 plays a major role in diverse functions, which are important for proper telomere maintenance. First, it is a primary component of the shelterin complex and contributes to protecting the telomere from deleterious events. TPP1 serves as a molecular fulcrum in the shelterin complex by governing the interactions with TIN2 and POT1, thus bridging the double- and single-stranded binding regions of the telomere. Secondly, because of its direct interaction with telomerase, TPP1 is a fundamental component in telomerase recruitment and regulator of telomere length. For telomerase recruitment, it is now established that the TPP1-OBD, which harbors multiple sites for post-translational modification and therefore opportunities for functional regulation, is responsible for TPP1 interactions with telomerase. In a simple on-off model, post-translation modifications of TPP1 may function as molecular switches between telomere association/protection and stimulating telomerase activity for telomere length extension. The coming years will undoubtedly shed even more light on the molecular interactions that govern telomere length and how the telomere end-binding proteins regulate important cellular functions ranging from the DNA damage response to telomerase-mediated telomere extension and perhaps other, yet to be discovered, roles.
Acknowledgements
We thank all members of the Taylor lab for the helpful discussions of this manuscript. Research in the Taylor lab is supported by the National Institutes of Health (DP2 CA186571 and R21 CA169611), the American Cancer Society (RSG-13-211-01-DMC), and the American Heart Association (11SDB5580010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Telomeres: no end in sight. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 3.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 4.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 6.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 8.Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3'-overhang. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 9.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 11.Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides. Doklady Akademii nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 12.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 13.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 14.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 15.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 17.Bacchetti S, Counter C. Telomeres and telomerase in human cancer (review) Int J Oncol. 1995;7:423–432. [PubMed] [Google Scholar]
- 18.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 20.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 21.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 22.Karlseder J. Telomere repeat binding factors: keeping the ends in check. Cancer Lett. 2003;194:189–197. doi: 10.1016/s0304-3835(02)00706-1. [DOI] [PubMed] [Google Scholar]
- 23.Martinez P, Blasco MA. Role of shelterin in cancer and aging. Aging cell. 2010;9:653–666. doi: 10.1111/j.1474-9726.2010.00596.x. [DOI] [PubMed] [Google Scholar]
- 24.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRFdeficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songyang Z, Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Critical reviews in eukaryotic gene expression. 2006;16:103–118. doi: 10.1615/critreveukargeneexpr.v16.i2.10. [DOI] [PubMed] [Google Scholar]
- 26.Parsons CA, Baumann P, Van Dyck E, West SC. Precise binding of single-stranded DNA termini by human RAD52 protein. Embo J. 2000;19:4175–4181. doi: 10.1093/emboj/19.15.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 28.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. Embo J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 34.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 35.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 36.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keegan CE, Hutz JE, Else T, Adamska M, Shah SP, Kent AE, Howes JM, Beamer WG, Hammer GD. Urogenital and caudal dysgenesis in adrenocortical dysplasia (acd) mice is caused by a splicing mutation in a novel telomeric regulator. Hum Mol Genet. 2005;14:113–123. doi: 10.1093/hmg/ddi011. [DOI] [PubMed] [Google Scholar]
- 38.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 40.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. Embo J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor DJ, Podell ER, Taatjes DJ, Cech TR. Multiple POT1-TPP1 Proteins Coat and Compact Long Telomeric Single-Stranded DNA. J Mol Biol. 2011;410:10–17. doi: 10.1016/j.jmb.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 47.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. Embo J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donate LE, Blasco MA. Telomeres in cancer and ageing, Philosophical transactions of the Royal Society of London. Series B. Biological sciences. 2011;366:76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristofari G, Sikora K, Lingner J. Telomerase unplugged. ACS chemical biology. 2007;2:155–158. doi: 10.1021/cb700037c. [DOI] [PubMed] [Google Scholar]
- 51.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 53.Ashton NW, Bolderson E, Cubeddu L, O'Byrne KJ, Richard DJ. Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BMC molecular biology. 2013;14:9. doi: 10.1186/1471-2199-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickey TH, Altschuler SE, Wuttke DS. Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure. 2013;21:1074–1084. doi: 10.1016/j.str.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gille C, Frommel C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- 60.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garber DA, O'Mara LA, Gangadhara S, McQuoid M, Zhang X, Zheng R, Gill K, Verma M, Yu T, Johnson B, Li B, Derdeyn CA, Ibegbu C, Altman JD, Hunter E, Feinberg MB. Deletion of specific immune-modulatory genes from modified vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in rhesus macaques. Journal of virology. 2012;86:12605–12615. doi: 10.1128/JVI.00246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mason M, Wanat JJ, Harper S, Schultz DC, Speicher DW, Johnson FB, Skordalakes E. Cdc1OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Structure. 2013;21:109–120. doi: 10.1016/j.str.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. Structure of the human telomeric Stn1-Ten1 capping complex. PloS one. 2013;8:e66756. doi: 10.1371/journal.pone.0066756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 67.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 68.Rath BK, Hegerl R, Leith A, Shaikh TR, Wagenknecht T, Frank J. Fast 3D motif search of EM density maps using a locally normalized cross-correlation function. J Struct Biol. 2003;144:95–103. doi: 10.1016/j.jsb.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 69.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 70.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Chen LY, Han X, Xie W, Kim H, Yang D, Liu D, Songyang Z. Phosphorylation of TPPregulates cell cycle-dependent telomerase recruitment. Proc Natl Acad Sci U S A. 2013;110:5457–5462. doi: 10.1073/pnas.1217733110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 73.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Else T, Theisen BK, Wu Y, Hutz JE, Keegan CE, Hammer GD, Ferguson DO. Tpp1/Acd maintains genomic stability through a complex role in telomere protection. Chromosome Res. 2007;15:1001–1013. doi: 10.1007/s10577-007-1175-5. [DOI] [PubMed] [Google Scholar]
- 75.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5'-TAGGGTTAG-3' minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 78.Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 80.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corriveau M, Mullins MR, Baus D, Harris ME, Taylor DJ. Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA. J Biol Chem. 2013;288:16361–16370. doi: 10.1074/jbc.M113.471896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, Johnson S, Ivanova E, Kost-Alimova M, Protopopov A, Wang YA, Shirihai OS, Chin L, DePinho RA. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–663. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc Natl Acad Sci U S A. 2014;111:2990–2995. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 85.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 86.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 87.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 88.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 89.Chen LY, Liu D, Songyang Z. Telomere maintenance through spatial control of telomeric proteins. Mol Cell Biol. 2007;27:5898–5909. doi: 10.1128/MCB.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POTrequires tethering to TIN2. Mol Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jun HI, Liu J, Jeong H, Kim JK, Qiao F. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes Dev. 2013;27:1917–1931. doi: 10.1101/gad.219485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.T M, Martinez P, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:733–735. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 97.Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tseng SF, Shen ZJ, Tsai HJ, Lin YH, Teng SC. Rapid Cdc13 turnover and telomere length homeostasis are controlled by Cdk1-mediated phosphorylation of Cdc13. Nucleic Acids Res. 2009;37:3602–3611. doi: 10.1093/nar/gkp235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 100.Rai R, Li JM, Zheng H, Lok GT, Deng Y, Huen MS, Chen J, Jin J, Chang S. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nat Struct Mol Biol. 2011;18:1400–1407. doi: 10.1038/nsmb.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]