Abstract
Oxo-lipids, a large family of oxidized human lipoxygenase (hLOX) products, are of increasing interest to researchers due to their involvement in different inflammatory responses in the cell. Oxo-lipids are unique because they contain electrophilic sites that can potentially form covalent bonds through a Michael addition mechanism with nucleophilic residues in protein active sites and thus increase inhibitor potency. Due to the resemblance of oxo-lipids to LOX substrates, the inhibitor potency of 4 different oxo-lipids; 5-oxo-6,8,11,14-(E,Z,Z,Z)-eicosatetraenoic acid (5-oxo-ETE), 15-oxo-5,8,11,13-(Z,Z,Z,E)-eicosatetraenoic acid (15-oxo-ETE), 12-oxo-5,8,10,14-(Z,Z,E,Z)-eicosatetraenoic acid (12-oxo-ETE), and 13-oxo-9,11-(Z,E)-octadecadienoic acid (13-oxo-ODE) were determined against a library of LOX isozymes; leukocyte 5-lipoxygenase (h5-LOX), human reticulocyte 15-lipoxygenase-1 (h15-LOX-1), human platelet 12-lipoxygenase (h12-LOX), human epithelial 15-lipoxygenase-2 (h15-LOX-2), soybean 15-lipoxygenase-1 (s15-LOX-1), and rabbit reticulocyte 15-LOX (r15-LOX). 15-oxo-ETE exhibited the highest potency against h12-LOX, with an IC50 = 1 ± 0.1 μM and was highly selective. Steady state inhibition kinetic experiments determined 15-oxo-ETE to be a mixed inhibitor against h12-LOX, with a Kic value of 0.087 ± 0.008 μM and a Kiu value of 2.10 ± 0.8 μM. Time-dependent studies demonstrated irreversible inhibition with 12-oxo-ETE and h15-LOX-1, however, the concentration of 12-oxo-ETE required (Ki = 36.8 ± 13.2 μM) and the time frame (k2 = 0.0019 ± .00032 s−1) were not biologically relevant. These data are the first observations that oxo-lipids can inhibit LOX isozymes and may be another mechanism in which LOX products regulate LOX activity.
Keywords: lipoxygenase, oxo-lipids, inhibitor, mechanism
1.1 Introduction
Human lipoxygenases (hLOX) generate hydroperoxyeicosatetraenoic acid (HpETE) and hydroperoxyoctadecadienoic acid (HpODE) as their primary products from polyunsaturated fatty acids, such as arachidonic acid (AA) [1] and linoleic acid (LA), respectively [2]. These hydroperoxide products are in turn reduced by cellular glutathione peroxidase to the secondary alcohol product, hydroxyeicosatetraenoic acid (HETE) and hydroxyoctadecadienoic acid (HODE) respectively [3,4]. The electrophilic oxo-lipids, such as 5-oxo-ETE, 15-oxo-ETE, 12-oxo-ETE, and 13-oxo-ODE, are derived either from HpETE and HpODE, such as in the macrophage [5] or from the corresponding HETE and HODE, by a dehydrogenase-mediated oxidation [6, 7]. These oxo-lipids are of interest because they are important biological molecules, whose interaction with LOX isozymes has not been fully explored.
The role of the various oxo-lipids in biology is significant and expanding. 5-oxo-6,8,11,14-(E,Z,Z,Z)-eicosatetraenoic acid (5-oxo-ETE) is a multi-functional oxo-lipid that has been found to stimulate proliferation in cancer cell lines through a specific Gi-coupled receptor [8, 9,10]. 5-oxo-ETE also plays an important role in the asthmatic inflammatory response [11], gastrointestinal diseases [12] and activation of peroxisome proliferator-activated receptor γ (PPARγ) transcriptional activity [13]. Another oxo-lipid which plays a role in the cell is 12-oxo-5,8,10,14-(Z,Z,E,Z)-eicosatetraenoic acid (12-oxo-ETE). Powell et al. [14,15] observed that 12-oxo-ETE had effects on cytosolic calcium levels at concentrations of 10 μM, but Naccache et al. [16] reported calcium effects as low as 10 nM. The third oxo-lipid generated from AA is 15-oxo-5,8,11,13-(Z,Z,Z,E)-eicosatetraenoic acid (15-oxo-ETE). For this oxo-lipid, an esterified, 15-oxo-ETE phospholipid has been detected in patients with cystic fibrosis (CF) [13] and shown to activate transcriptional activity in PPARγ [12]. Activation of PPARγ expression in CF mice ameliorates disease severity, suggesting that 15-oxo-ETE might potentially act to lower inflammation in CF [13]. Finally, there is an oxo-lipid generated from LA, 13-oxo-9,11-(Z,E)-octadecadienoic acid (13-oxo-ODE), which was found to be an endogenous ligand to PPARγ in intestinal epithelial cells (IEC). 13-oxo-ODE mediated the activation of PPARγ to reduce mucosal damage and down-regulate inflammation in several mouse models of intestinal colitis [17], implicating it as a possible therapeutic target for the treatment of inflammatory bowel disease [17].
Chemically, the oxo-lipids, like 5-oxo-ETE, 15-oxo-ETE, 12-oxo-ETE and 13-oxo-ODE, are unique in that they contain an α, β unsaturated carbonyl that can readily react with nucleophiles, such as proteins and glutathione (GSH), via Michael addition reaction, resulting in covalent modifications. The reversible conjugation of 13-oxo-ODE by GSH [18,19] has been shown to occur by both enzymatic and non-enzymatic pathways, with the conjugate being exported from the cell via an energy-dependent process [20]. Similar synthetic molecules that form covalent linkages to their targets have been considered as therapeutics, but have traditionally been disfavored due to concerns for their off-target reactivity, either through direct tissue damage or through haptenization of proteins, which could elicit an immune response [21]. However, as selectivity and drug resistance remain a serious issue for reversible inhibitors, a resurgence of interest in this class of therapeutics has emerged. For example, Taunton and coworkers, [22] developed a fluoromethylketone-substituted ligand (fmk), which irreversibly inactivates p90 ribosomal protein S6 kinase (RSK1/2) in human cells at nanomolar concentrations by modifying an active site cysteine, without inhibiting over 130 other kinases [22]. A similar covalent inhibitor, JNK-IN-8, was discovered as a specific, irreversible intracellular inhibitor against the mitogen-activated kinase JNK [23]. JNK-IN-8 inhibits phosphorylation of c-Jun, a direct substrate of JNK, by covalent modification of a conserved cysteine residue in the ATP-binding motif [23]. Both of these studies argue against the widely held view that electrophilic inhibitors are inherently nonselective [22] and therefore it is possible that the oxo-lipids target nonconserved, non-catalytic cysteines in many proteins in the cell, such as lipoxygenase.
Due to the fact that oxo-lipids have interesting biological properties, that they are potential covalent modifiers and that they have similar structures to LOX substrates, we hypothesized that oxo-lipids could potentially inhibit LOX isozymes at concentrations that are biologically relevant. This hypothesis is reinforced by the fact that certain LOX isozymes have non-catalytic cysteines in their active sites [24], which could serve as nucleophiles to oxo-lipids. In the current work, we present inhibitory data of a variety of oxo-lipids (5-oxo-ETE, 15-oxo-ETE, 12-oxo-ETE, and 13-oxo-ODE) against LOX isozymes (h5-LOX, h15-LOX-1, human platelet 12-lipoxygenase (h12-LOX), human epithelial 15-lipoxygenase-2 (h15-LOX-2), soybean 15-lipoxygenase-1 (s15-LOX-1), and rabbit reticulocyte 15-LOX (r15-LOX)) and demonstrate that certain oxo-lipids are LOX inhibitors.
1.2. Materials and Methods
1.2.1 Materials
All commercial fatty acids (Sigma-Aldrich Chemical Company) were re-purified using a Higgins HAIsil Semi-Preparative (5 mM, 250 × 10 mm) C-18 column. Solution A was 99.9% MeOH and 0.1% acetic acid; solution B was 99.9% H2O and 0.1% acetic acid. An isocratic elution of 85% A: 15% B was used to purify all fatty acids, which were stored at −80 °C for a maximum of 6 months. HPLC grade solvents were used for both semi-preparative HPLC purification and analytical HPLC analysis of LOX products. Large scale product purification was achieved by using a C18HAIsil 250 × 10 mm semi-preparative column, whereas a C18HAIsil 250 × 4.6 mm analytical column was used for product separation in tandem with MS/MS analysis. Both columns were purchased from Higgins Analytical (Mountain View, CA). All other chemicals were reagent grade or better and were used without further purification.
1.2.2 Protein Expression
All the LOX isozymes used in this publication were expressed and purified as previously published (h5-LOX [28], h12-LOX [25], h15-LOX-1 [25] and s15-LOX-1 [26], h15-LOX-2 [27] and r15-LOX [29]).
1.2.3 General Procedure for the Synthesis of Oxo-Lipids
The synthesis of all oxo-lipids consists of two steps, the first step is enzymatic while the second step is synthetic. In the synthesis of 13-oxo-ODE, s15-LOX-1 is reacted with linoleic acid (LA) in 100 mL of 100 mM Borate (pH 9.2) generating 13-HpODE. 15-oxo-ETE is generated by reaction between h15-LOX-2 and 40 μM arachidonic acid (AA) in 100 mL of 25 mM HEPES (pH 7.5), generating 15-HpETE. 12-oxo-ETE is generated by reaction between h12-LOX and 40 μM AA in 100 mL 25 mM HEPES (pH 8.0), generating 12-HpETE. 5-oxo-ETE is generated by reaction between 5-LOX and 40 μM AA in 100 mL 25 mM HEPES (pH 7.3), 0.3 mM CaCl2, 0.1 mM EDTA, 0.2 mM ATP, generating 5-HpETE. The reactions are quenched with 1–2% acetic acid and extracted using dichloromethane (DCM). The formation of 13-HpODE, 15-HpETE, 12-HpETE and 5-HpETE are monitored at 234 nm with a Perkin Elmer Lambda 40 UV/Vis spectrophotometer. The second step is an overnight synthetic reaction in which the hydroperoxy products are reacted with acetic anhydride and pyridine at 4°C in a 1:1 ratio to generate 13-oxo-ODE, 15-oxo-ETE, 12-oxo-ETE and 5-oxo-ETE respectively. The reactions are quenched with cold Milli-Q water for 2 hours. The oxo-lipids are purified via high performance liquid chromatography (HPLC) using a Higgins HAIsiL Semi-Preparative C-18 column. Solution A was 99.9% ACN and 0.1% acetic acid; solution B was 99.9% H2O and 0.1% acetic acid. An isocratic elution of 55% A: 45% B was used to purify each oxo-compound. The retention times for each oxo-lipid at 280 nm are as follows: 13-oxo-ODE (30 min), 15-oxo-ETE (33 min), 12-oxo-ETE (90 min) and 5-oxo-ETE (70 min). Analytical analysis was performed by liquid chromatography-mass spectrometry (LC-MS/MS). Solution A was 99.9% H2O and 0.1% formic acid; solution B was 99.9% ACN and 0.1% formic acid. Oxo-lipids were injected onto a Phenomenex Synergi (4 μM, 150 mm × 4.6 mm) C-18 column attached to a Thermo LTQ LC-MS/MS. The elution protocol consisted of 200 μL/min, with an isocratic mobile phase of 45% solution A and 55% solution B. Negative ion MS/MS was utilized (collision energy of 35 eV) to determine the fragmentation patterns of all the oxo-lipids. 13-oxo-ODE, parent m/z = 293, fragments m/z = 113, 249, 293; 15-oxo-ETE, parent m/z = 317, fragments m/z = 113, 273, 299; 12-oxo-ETE, parent m/z = 317, fragments m/z = 153, 179, 273; 5-oxo-ETE, parent m/z = 317, fragments m/z = 129, 203, 273.
The concentrations of the purified oxo-lipids are quantified using a Perkin Elmer Lambda 40 UV/Vis spectrophotometer based on the ε280 value of 13-oxo-ODE’s (28,000 M−1cm−1). The extinction coefficient for 13-oxo-ODE was determined by weighing the compound on an analytical balance, dissolving it with a known mass of HPLC grade methanol and measuring the absorbance (280 nm) for various concentrations of 13-oxo-ODE (Perkin-Elmer Lambda 40 UV/Vis spectrophotometer). A standard curve plot was used to extract the extinction coefficient for 13-oxo-ODE at 280 nm.
1.2.4 Lipoxygenase UV-Vis-based IC50 Assay
The initial one-point inhibition percentages were determined by following the formation of the conjugated diene product at 234 nm (ε = 25,000 M−1cm−1) with a Perkin-Elmer Lambda 40 UV/Vis spectrophotometer at one inhibitor concentration. The full IC50 experiments were done with at least five different inhibitor concentrations. All reactions were 2 mL in volume and constantly stirred using a magnetic stir bar at room temperature (23°C) with the appropriate amount of LOX isozyme (h5-LOX (~ 200 nM); h12-LOX (~ 100 nM); h15-LOX-1 (~ 60 nM); r15-LOX (~50 nM); h15-LOX-2 (~ 200 nM); s15-LOX-1 (~ 2 nM)). The protein concentrations are the total protein concentration, however active protein concentration will be significantly less due to incomplete metallation. Incomplete metallation of the enzymes will not affect inhibitor potency, due to the relative nature of the IC50 calculation. Reactions with h12-LOX were carried out in 25 mM HEPES (pH 8.0) 0.01% Triton X-100 and 10 μM AA. Reactions with the crude, ammonium sulfate precipitated h5-LOX were carried out in 25 mM HEPES (pH 7.3), 0.3 mM CaCl2, 0.1 mM EDTA, 0.2 mM ATP, 0.01% Triton X100 and 10 μM AA. Reactions with h15-LOX-1, r15-LOX and h15-LOX-2 were carried out in 25 mM HEPES buffer (pH 7.5), 0.01% Triton X-100 and 10 μM AA. Reactions with s15-LOX-1 were carried out in 100 mM Borate (pH 9.2) 0.01% Triton X-100 and 10 μM AA. The concentration of AA was quantitated by allowing the enzymatic reaction to proceed to completion. IC50 values were obtained by determining the enzymatic rate at various inhibitor concentrations and plotted against inhibitor concentration, followed by a hyperbolic saturation curve fit. The data used for the saturation curves were performed in duplicate or triplicate, depending on the quality of the data.
1.2.5 Incubation Activity Assay with oxo-lipids and LOX
h15-LOX-1 and s15-LOX-1 rates and buffer conditions were utilized as described above, with the following modifications. A specific volume and concentration of h15-LOX-1 (or s15-LOX-1) was added to either the 12-oxo-ETE or 13-oxo-ODE oil (no solvent) and incubated on ice to ensure that the isozymes did not lose activity. It should be emphasized that the oxo-lipid was added as the oil, so as not to introduce solvent, which could inhibit the LOX isozyme. Aliquots of approximately 20 μL of the incubated mixture were then added at designated time periods (intervals of 2 minutes, upwards to 30 minutes total) to a constantly stirring 2 mL cuvette, containing 10 μM AA. The control to this reaction was the same as above, but with no oxo-lipid oil added. This procedure was repeated for at least five different concentrations of 12-oxo-ETE or 13-oxo-ODE. The ln (% Activity) was plotted vs. time (sec) to generate a slope = ka. A second plot of ka against [I]incubation allowed us to obtain Ki and k2.
1.2.6 Steady-State Inhibition Kinetics
h12-LOX rates were determined by monitoring the formation of the conjugated product, 12-HpETE, at 234 nm (ε = 25,000 M−1cm−1) with a Perkin Elmer Lambda 40 UV/Vis spectrophotometer. Reactions were initiated by adding h12-LOX to a constantly stirring 2 mL cuvette containing 0.4 μM – 10 μM AA in 25 mM HEPES buffer (pH 8.0), in the presence of 0.01% Triton X-100. The substrate concentration was quantitated by allowing the enzymatic reaction to proceed to completion. Kinetic data were obtained by recording initial enzymatic rates, at varied inhibitor concentrations (15-oxo-ETE), and subsequently fitted to the Henri-Michaelis-Menten equation, using KaleidaGraph (Synergy) to determine the microscopic rate constants, Vmax (μmol/min/mg) and Vmax/Km (μmol/min/mg/μM). These rate constants were subsequently re-plotted 1/Vmax and Km/Vmax versus inhibitor concentration, to yield Kiu and Kic, which are defined as the equilibrium constant of dissociation from the secondary and catalytic sites, respectively.
1.2.7 Determination of Oxo-lipids Substrate Activity for LOX
The oxo-lipids were reacted with LOX to determine if they act as substrates due to their resemblance to LOX substrates. Two different experimental conditions were investigated. The first were the UV-Vis-based IC50 assay conditions (see section 1.2.4), while the second were the incubation activity assay conditions (see section 1.2.5). For the IC50 assay conditions, the following oxo-lipid and LOX combinations were followed by UV-Vis: h15-LOX-1, h12-LOX, and h5-LOX were each reacted with 10 μM 5-oxo-ETE; h5-LOX and h15-LOX-1 were reacted with 10 μM 12-oxo-ETE; and h12-LOX was reacted with 10 μM 15-oxo-ETE. All reactions were in 2 mL of the enzyme’s specific buffer and no change of absorbance at 234 nm or 280 nm was observed for any of the reactions. One pair, h12-LOX and 10 μM 15-oxo-ETE, was also quenched with 2% acetic acid, extracted with DCM and shown with LC-MS that no oxidation product formation, confirming that 15-oxo-ETE is not a substrate to h12-LOX under the IC50 conditions. However, the pair that did result in a change of absorbance at 234 nm was h5-LOX with 10 μM 15-oxo-ETE. This reaction was also quenched with 2% acetic acid, extracted with DCM and shown with LC-MS that multiple oxidation products were formed. Therefore, no inhibition data was gathered for this pair.
In the second set of conditions (Incubation conditions), h12-LOX and h15-LOX-1 were each incubated on ice with 10 μM 15-oxo-ETE for 15 minutes. Once incubation was complete, each reaction was diluted with 2 mL of the corresponding enzyme buffer, quenched with 2% acetic acid, extracted with DCM, evaporated to dryness and brought up in 50 μL of methanol. HPLC (205 nm, 234 nm, 280 nm) and LC-MS analysis did not show significant degradation, confirming that 15-oxo-ETE is not a substrate for either h12-LOX or h15-LOX-1, under the time-dependence incubation conditions.
1.3 Results and Discussion
1.3.1 Evaluation of Oxo-Lipids As Inhibitors of LOXs
To determine the inhibitory potency and selectivity of 5-oxo-ETE, 12-oxo-ETE, 13-oxo-ODE and 15-oxo-ETE, each oxo-lipid was screened as an inhibitor against h5-LOX, h12-LOX, h15-LOX-1, h15-LOX-2, r15-LOX and s15-LOX-1 (Table 1). To summarize Table 1, 15-oxo-ETE exhibited the highest potency against h12-LOX, with an IC50 value of 1.0 ± 0.1 μM and was also found to be the most selective oxo-lipid, with low potency against the other LOX isozymes (IC50 values greater than 25 μM). 12-oxo-ETE inhibited both h12-LOX and h15-LOX-1, but with an almost three-fold difference, revealing a higher potency against h15-LOX-1 (IC50 = 3.0 ± 0.1 μM) than h12-LOX (IC50 = 8.0 ± 1 μM). On the contrary, 5-oxo-ETE did not inhibit any of the LOXs significantly, with only a slight activity against h12-LOX (IC50 = 15 ± 2 μM). The oxo-lipid found to be the least potent against all the LOXs studied was 13-oxo-ODE with IC50 values greater than 100 μM for all LOXs.
Table 1.
Inhibition Potencies of Various Oxo-Lipids Against LOX Isozymes.a
| h5-LOX | h12-LOX | h15-LOX-1 | h15-LOX-2 | r15-LOX | s15-LOX-1 | |
|---|---|---|---|---|---|---|
| 5-oxo-ETE |
-----b | 15 ± 2 | > 25c | > 100 | > 25 | > 100 |
12-oxo-ETE
|
> 50 | 8.0 ± 1 | 3.0 ± 0.1 | > 100 | > 50 | > 100 |
13-oxo-ODE
|
> 100 | > 100 | > 100 | > 100 | > 100 | > 100 |
| 15-oxo-ETE |
> 100 | 1 ± 0.1 | > 25 | > 100 | > 50 | > 100 |
All IC50 values (μM) were conducted with 10 μM AA, 0.01% Triton X100 and LOX isozyme specific buffers (See Methods Section). Each experiment was done in triplicate, with the average and SD presented.
Exhibited substrate activity.
The IC50 value of weak inhibitors were approximated from one point inhibitor screens (in triplicate), at 25 μM inhibitor. Inhibitors with less than 5% inhibition at 25 μM were designated > 100 μM. Inhibitors with less than 25% inhibition but greater than 5% inhibition were designated > 50 μM, and inhibitors with less than 50% inhibition but greater than 25% inhibition were designated > 25 μM. Inhibitors with greater than 50% inhibition at 25 μM were studied further and full IC50 values determined.
1.3.2 Probing the Mechanism of Inhibition Via Time Dependence Experiments
To further examine the mode of inhibitory action of oxo-lipids against LOX, time incubation experiments were performed. Due to these oxo-lipids’ capability of behaving as general Michael acceptors, it was thought that these molecules could react with a nucleophilic residue in the active site, inactivating the enzyme. Such an action between a target-specific covalent inhibitor and an enzyme can be described by the general mechanism in Figure 1 [21].
Figure 1.
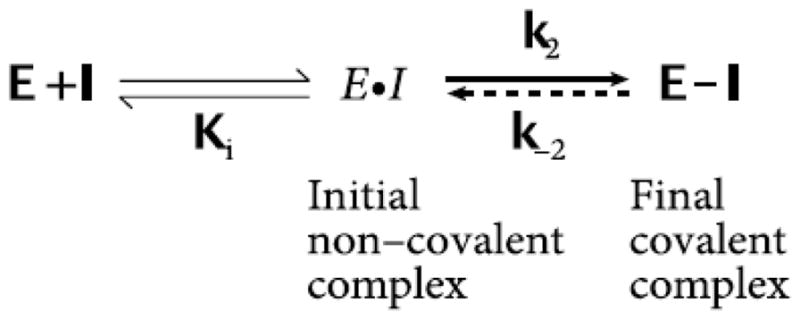
General Mechanism of a Covalent Inhibitor
The first event is the non-covalent binding of the oxo-lipid to the LOX active site, placing it close to an active site nucleophilic residue. At which point the nucleophile attacks the Michael acceptor of the oxo-lipid. If these oxo-lipids behave as pure irreversible covalent inhibitors then k−2 will be essentially zero. Inhibitors that behave in this manner can be described by their equilibrium inhibitor constant (Ki) and their rate of covalent modification (k2) [21]. To test if such a mechanism occurred with LOX inhibition, an oxo-lipid was incubated with a LOX isozyme where the concentration of the inhibitor was kept constant, while the incubation time was varied. The extent of inhibition was measured at several time points, and the observed rate of inhibition, ka, extracted from the slope of a plot of the ln (% Activity) vs. time (Data not shown). These graphs were repeated at various oxo-lipid concentrations and the ka values were re-plotted against the concentration of oxo-lipids and then fitted to the hyperbolic equation, kobs = k2[I]/(Ki+[I]), which yields Ki and k2. For 12-oxo-ETE and h15-LOX-1, the data revealed a fit to the hyperbolic equation above, with a Ki of 36.8 ± 13.2 μM and a k2 of 0.0019 ± .00032 s−1. These data indicate that 12-oxo-ETE is a poor irreversible inhibitor to h15-LOX-1. Considering that the time dependence is in minutes and only occurred at high concentrations of 12-oxo-ETE, we infer that this irreversible mechanism is not biologically relevant in the cell. This experiment was also performed with h15-LOX-1 vs. 15-oxo-ETE and s15-LOX-1 vs. 13-oxo-ODE, both of which had similarly large Ki values and long k2 rates. Therefore, it appears that the irreversible Michael addition mechanism is not a biologically relevant mode of action by which these oxo-lipids inhibit LOX.
1.3.3 Steady State Inhibition Kinetics
The mode of inhibition of 15-oxo-ETE against h12-LOX was probed further with steady state inhibition kinetics. The formation of 12-HpETE was monitored as a function of substrate and inhibitor concentration in the presence of 0.01% Triton-X-100. A Dixon plot (Figure 2A) and replot (Figure 2B) of the kinetic data yeilded an Kic of 0.087 ± 0.008 μM and a Kiu of 2.10 ± 0.8 μM, which are defined as the equilibrium constants of dissociation from the enzyme and enzyme substrate complex, respectively. These data are consistent with mixed inhibition, which is typical of LOX inhibitors [30]. It should be noted that Kic is lower than the measured IC50 values, which is due to the fact that IC50 values assume competitive inhibition, which is not the case in this circumstance.
Figure 2.
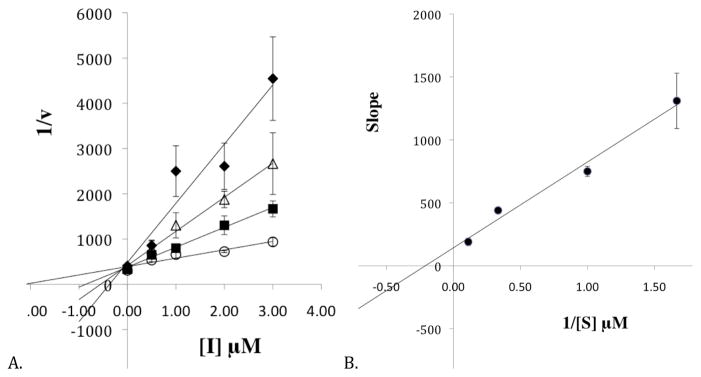
A. The Dixon plot of the primary data from the steady state inhibition kinetic experiments of h12-LOX and 15-oxo-ETE. The substrate concentrations are 0.6 μM (closed diamonds), 1 μM (open triangles), 3 μM (closed squares) and 9 μM (open circles). B. The Dixon replot of slope vs. [Inhibitor] yielded a Kic of 0.087 ± 0.008 μM and a Kiu of 2.1 ± 0.8 μM. All experiments were done in triplicate with averages and SD presented.
1.4 Conclusion
In summary, the data reveals that 15-oxo-ETE exhibited the highest potency and selectivity against h12-LOX with an IC50 = 1 ± 0.1 μM. 12-oxo-ETE had comparable potency against h15-LOX-1 (IC50 = 3.0 ± 0.1 μM) but was not selective and inhibited h12-LOX with an IC50 = 8.0 ± 1 μM. The steady state inhibition kinetics revealed a very low Kic of 0.087 ± 0.008 μM for 15-oxo-ETE against h12-LOX, which may be at a level that is biologically relevant. The time dependence incubation studies did demonstrate irreversible inhibition of LOX isozymes by oxo-lipids, however, the large concentration needed and long time required precluded any biological relevance. To our knowledge, this is the first time that an oxo-lipid has been shown to inhibit LOX and could indicate selective regulation of h12-LOX by h15-LOX activity, through 15-oxo-ETE inactivation. Future work is planned to determine if oxo-lipids regulate LOX activity in the cell.
Acknowledgments
This work was supported by the W.W. & E.M. Clark Foundation (MMA), National Science Foundation Bridges to the Doctorate (20102433 (MMA)) and the National Institutes of Health (GM56062).
The authors would like to acknowledge Steve Perry for his help with the IC50 experiments and Qiangli Zhang for her help with the mass spectrometry analysis.
Abbreviations
- LOX
lipoxygenase
- hLOX
human lipoxygenase
- h15-LOX-2
human epithelial 15-lipoxygenase-2
- h15-LOX-1
human reticulocyte 15-lipoxygenase-1
- h12-LOX
human platelet 12-lipoxygenase
- s15-LOX-1
soybean 15-lipoxygenase-1
- h5-LOX
human leukocyte 5-lipoxygenase
- r15-LOX
rabbit reticulocyte 15-LOX
- AA
arachidonic acid
- LA
linoleic acid
- 12-HETE
12-hydroxy-5,8,10,14-eicosatetraenoic acid
- 12-HpETE
12-hydroperoxyeicosatetraenoic acid
- 12-oxo-ETE
12-oxo-5,8,10,14-(Z,Z,E,Z)-eicosatetraenoic acid
- 12-oxo-ETEre
10,11-dihydro-12-oxo-ETE
- 12-HETrE
10,11-dihydro-12-HETE
- 15-HpETE
15-hydroperoxyeicosatetraenoic acid
- 15-oxo-ETE
15-oxo-5,8,11,13-(Z,Z,Z,E)-eicosatetraenoic acid
- 5-HpETE
5-hydroperoxyeicosatetraenoic acid
- 5-oxo-ETE
5-oxo-6,8,11,14-(E,Z,Z,Z)-eicosatetraenoic acid
- LA
linoleic acid
- 13-HpODE
13-(S)-hydroperoxyoctadecadienoic acid
- 13-HODE
13-hydroxy-9Z,11E-octadecadienoic acid
- 13-oxo-ODE
13-oxo-9,11-(Z,E)-octadecadienoic acid
- 5-HEDH
5-hydroxyeicosanoid dehydrogenase
- 15-PGDH
15-hydroxyprotaglandin dehydrogenase
- 12-HEDH
12-hydroxyeicosanoid dehydrogenase
- CYP2S1
human cytochrome P450
- GSH
glutathione
- fmk
fluoromethylketone-substituted ligand
- RSK1/2
p90 ribosomal protein S6 kinase
- PPARγ
Peroxisome Proliferator Activated Receptor γ
- CF
cystic fibrosis
- IEC
Intestinal Epithelial Cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Needleman P, Turk J, Jakschik BA, Morrison AR, Lefkowith JB. Arachidonic Acid Metabolism. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- 2.Hamberg M, Samuelsson B. On the Specificity of the Oxygenation of Unsaturated Fatty Acids Catalyzed by Soybean Lipoxidase. J Biol Chem. 1967;212:5329–5335. [PubMed] [Google Scholar]
- 3.Borgeat P. Biochemistry of the Lipoxygenase Pathways in Neutrophils. Can J Physiol Pharmacol. 1989;67:936–942. doi: 10.1139/y89-147. [DOI] [PubMed] [Google Scholar]
- 4.Murphy RC, Zarini S. Glutathione Adducts of Oxyeicosanoids. Prostaglandins Other Lipid Mediat. 2002;68–69:471–482. doi: 10.1016/s0090-6980(02)00049-7. [DOI] [PubMed] [Google Scholar]
- 5.Zarini S, Murphy RC. Biosynthesis of 5-oxo-6, 8, 11, 14-eicosatetraenoic acid from 5-hydroperoxyeicosatetraenoic acid in the Murine Macrophage. J Biol Chem. 2003;278:11190–11196. doi: 10.1074/jbc.M208496200. [DOI] [PubMed] [Google Scholar]
- 6.Powell WS, Gravelle F, Gravel S. Metabolism of 5(S)-hydroxy-6,8,11,14-eicosatetraenoic acid and other 5(S)-hydroxyeicosanoids by a specific dehydrogenase in human polymorphonuclear leukocytes. J Biol Chem. 1992;267:19233–19241. [PubMed] [Google Scholar]
- 7.Bergholte JM, Soberman RJ, Hayes R, Murphy RC, Okita RT. Oxidation of 15-hydroxyeicosatetraenoic Acid and Other Hydroxy Fatty Acids by Lung Prostaglandin Dehydrogenase. Arch Biochem Biophys. 1987;257:444–450. doi: 10.1016/0003-9861(87)90589-3. [DOI] [PubMed] [Google Scholar]
- 8.Powell WS, MacLeod RJ, Gravel S, Gravelle F, Bhakar A. Metabolism and Biologic Effects of 5-oxoeicosanoids on Human Neutrophils. J Immunol. 1996;156:336–342. [PubMed] [Google Scholar]
- 9.Powell WS, Gravel S, MacLeod RJ, Mills E, Hashefi M. Stimulation of Human Neutrophils by 5-oxo-6,8,11,14-Eicosatetraenoic Acid by a Mechanism Independent of the Leukotriene B4 Receptor. J Biol Chem. 1993;268:9280–9286. [PubMed] [Google Scholar]
- 10.Hosoi T, Koguchi Y, Sugikawa E, Chikada A, Ogawa K, Tsuda N, Suto N, Tsunoda S, Taniguchi T, Ohnuki T. Identification of a Novel Human Eicosanoid Receptor Coupled to G(i/o) J Biol Chem. 2002;277:31459–31465. doi: 10.1074/jbc.M203194200. [DOI] [PubMed] [Google Scholar]
- 11.Sturm GJ, Schuligoi R, Amann R, et al. 5-oxo-6, 8, 11, 14-eicosatetraenoic acid is a Potent Chemoattractant for Human Basophils. J Allergy Clin Immunol. 2005;116:1014–1019. doi: 10.1016/j.jaci.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK. Interleukin-4-dependent production of PPAR-y Ligands in Macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 13.Hammond VJ, Morgan AH, Schopfer FJ, et al. Novel Keto-phospholipids Are Generated by Monocytes and Macrophages, Detected in Cystic Fibrosis and Activate Peroxisome Proliferator-activated Receptor-γ. J Biol Chem. 2012;287:41651–41666. doi: 10.1074/jbc.M112.405407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell WS, Hashefi M, Falck JR, Chauhan K, Rokach J, Wang SS, Mills E, MacLeod RJ. Effects of oxo and dihydro Metabolites of 12-hydroxy-5,8,10,14-eicosatetraenoic Acid on Chemotaxis and Cytosolic Calcium Levels in Human Neutrophils. J Leukocyte Biol. 1995;57:257–263. doi: 10.1002/jlb.57.2.257. [DOI] [PubMed] [Google Scholar]
- 15.Wainwright SL, Powell WS. Mechanism for the Formation of Dihydro Metabolites of 12-Hydroxyeicosanoids. J Biol Chem. 1991;266:20899–20906. [PubMed] [Google Scholar]
- 16.Naccache PH, Leblanc Y, Rokach J, Patrignani P, Fruteau de Laclos B, Borgeat P. Calcium mobilization and Right-Angle Light Scatter Responses to 12-oxo-derivatives of Arachidonic Acid in Neutrophils: Evidence for the Involvement of the Leukotriene B4 Receptor. Biochim Biophys Acta. 1991;1133:102–106. doi: 10.1016/0167-4889(91)90247-u. [DOI] [PubMed] [Google Scholar]
- 17.Altmann R, Hausmann M, Rogler G, et al. 13-oxo-ode is an Endogenous Ligand for PPARy in human colonic epithelial cells. Biochem Pharm. 2007;74:612–622. doi: 10.1016/j.bcp.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 18.Seeley SK, Poposki JA, Maksimchuk J, Tebbe J, Gaudreau J, Mannervik B, Bull AW. Metabolism of Oxidized Linoleic Acid by Glutathione Transferases: Peroxidase Activity Toward 13-Hydroperoxyoctadecadienoic Acid. Biochim Biophys Acta. 2006;1760:1064–1070. doi: 10.1016/j.bbagen.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Bull AW, Seeley SK, Geno JL, Mannervik B. Conjugation of the Linoleic Acid Oxidation Product, 13-oxooctadeca-9,11-dienoic acid, A Bioactive Endogenous Substrate for Mammalian Glutathione Transferase. Biochim Biophys Acta. 2002;1571:77–82. doi: 10.1016/s0304-4165(02)00216-7. [DOI] [PubMed] [Google Scholar]
- 20.Podgorski IA, Bull W. Energy-dependent Export of the 13-oxooctadecadienoic acid-glutathione Conjugate from HT-29 Cells and Plasma Membrane Vesicles. Biochim Biophys Acta. 2001;1533:55–65. doi: 10.1016/s1388-1981(01)00140-8. [DOI] [PubMed] [Google Scholar]
- 21.Singh J, Petter RC, Baillie TA, Whitty A. The Resurgence of Covalent Drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 22.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural Bioinformatics-Based Design of Selective Irreversible Kinase Inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Gray NS, et al. Discovery of Potent and Selective Covalent Inhibitors of JNK. Chem Biol. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornig M, Markoutsa S, Hafner AK, George S, Wisneiwska JM, Rodl CB, Hofmann B, Maier T, Karas M, Werz O, Steinhilber D. Inhibition of 5-lipoxygenase by U73122 is Due to Covalent Binding to Cysteine 416. Mol Cell Biol Lipids. 2012;1821:279–286. doi: 10.1016/j.bbalip.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Amagata T, Whitman S, Johnson TA, Stessman CC, Loo CP, Lobkovsky E, Clardy J, Crews P, Holman TR. Exploring Sponge-Derived Terpenoids for Their Potency and Selectivity Against 12-Human, 15-Human, and 15-Soybean Lipoxygenases. J Nat Prod. 2003;66:230–235. doi: 10.1021/np020462l. [DOI] [PubMed] [Google Scholar]
- 26.Holman TR, Zhou J, Solomon EI. Spetroscopic and Functional Characterization of a Ligand Coordication Mutant of Soybean Lipoxygenase-1: First Coordination Sphere Analogue of Human 15-Lipoxygenase. J Am Chem Soc. 1998;120:12564–12572. [Google Scholar]
- 27.Martinez YV, Ohri RV, Kenyon V, Holman TR, Boza SS. Structure-activity relationship studies of Flavanoids as Potent Inhibitors of Human Platelet 12-hLO, reticulocyte 15-hLO-1, and prostate epithelial 15-hLO-2. Bioorg Med Chem. 2007;15:7408–7425. doi: 10.1016/j.bmc.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson SJ, Hoobler EK, Riener M, Loveridge ST, Tenney K, et al. Using Enzyme Assays to Evaluate the Structure and Bioactivity of Sponge-derived Meroterpenes. J Nat Prod. 2009;72:1857–1863. doi: 10.1021/np900465e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloane DL, Browner MF, Dauter Z, Wilson K, Fletterick RJ, Sigal E. Purification and Crystallization of 15-lipoxygenase from Rabbit Reticulocytes. Biochem Biophys Res Comm. 1990;173:507–513. doi: 10.1016/s0006-291x(05)80063-4. [DOI] [PubMed] [Google Scholar]
- 30.Rai G, Kenyon V, Jadhav A, Schultz L, Armstrong M, Jameson B, Hoobler E, Leister W, Simeonov A, Holman TR, Maloney DJ. Discovery of Potent and Selective Inhibitors of Human Reticulocyte 15-Lipoxygenase-1. J Med Chem. 2010;53:7392–7404. doi: 10.1021/jm1008852. [DOI] [PMC free article] [PubMed] [Google Scholar]


