Abstract
Mesenchymal stemcells (MSCs) have shown a great potential for clinical applications in regenerative medicine. However, it remains challenging to follow the transplanted cell grafts in vivo. Nuclear magnetic resonance spectroscopy (NMR or MRS) is capable of determining and quantifying the cellular metabolites in tissue and organs non-invasively, therefore it is an attractive method for monitoring and evaluating the differentiation and functions of transplanted stem cells in vivo. In this study, metabolic changes of MSCs undergoing adipogenic differentiation to targeted fat cells were investigated in vitro, using solid-state high-resolution magic angle spinning 1H nuclear magnetic resonance spectroscopy. Quantification of metabolite concentrations before and after differentiation of MSCs showed decreased levels of intracellular metabolites, including choline, creatine, glutamate and myo-inositol, and a substantially increased level of fatty acids, when mesenchymal stem cells were differentiated preferentially to fat cells. Intracellular creatine, myo-inositol and choline reduced from 10.4 ± 0.72, 16.2 ± 1.2 and 8.22 ± 0.51 mm to 3.27 ± 0.34, 6.1 ± 0.46 and 3.11 ± 0.32 mm, respectively, while fatty acids increased from 32.6 ± 1.5 to 91.2 ± 3.2 mm after undergoing 3 weeks of differentiation. The increase of the fatty acid concentration measured by NMR is confirmed by the observation of 80% fat cells in differentiated cells by cell counting assay, suggesting resonances from fatty acids may be used as metabolite markers for monitoring MSC differentiation to fat cells in vivo, using the magnetic resonance spectroscopic technique readily available on MRI scanners.
Keywords: stem cell, metabolite, adipogenic differentiation, magnetic resonance, imaging, spectroscopy
1. Introduction
Stem cells are considered to hold a great potential for regenerative medicine and clinical applications of the cell replacement therapy because of their unique properties of pluripotency and their ability to differentiate into a diverse range of specialized cell types (Friedmann, 2005; Englund et al., 2002). However, one of the major obstacles for developing clinical applications of stem cells is the current limitation in tracking transplanted stem cells and monitoring their activities and functions in vivo. Although cell grafts can be evaluated using postmortem histological examination in animal studies, it is difficult to address critical questions about the efficacy of the transplant, targeted differentiation, cell trafficking pattern and potential risk of developing cancer cells (Friedmann, 2005) in live animals and in human clinical trials. Therefore, it is essential to develop non-invasive approaches, especially image-based methods, to monitor the transplanted cell graft in vivo. Nuclear imaging, such as positron emission tomography (PET) using radioactive tracers (Dall et al., 2000; Pogarell et al, 2006) and magnetic resonance imaging (MRI) using magnetic nanoparticles labelling cells (Lewin et al., 2000; Frangioni et al., 2004), have been used to track transplanted stem cells in vivo. However, neither method is the solution for long-term monitoring of transplanted cells, due to their current limitations. While applications of PET may be limited by the complexity of procedures, chemophysical and radioactive properties of the tracer and poor spatial–temporal resolution, MRI tracking magnetic nanoparticle labelled cells requires the cell labeling procedure and use of exogenous reagents that may compromise the cell integrity and functions, and can not maintain the imaging contrast over certain generations of the cell division. Neither method has the capability of monitoring cell functions and metabolic activities in vivo.
Nuclear magnetic resonance spectroscopy (NMR or MRS) allows for in vivo and ex vivo measurement of metabolites in tissue or organs and permits non-invasive detection of tissue metabolic changes that may be associated with disease development or the response to a treatment (Klunk et al., 1996; Ross et al., 1997; Martinez-Granados et al., 2006). While NMR is a widely used analytical method in chemistry and biochemistry for characterizing chemicals and elucidating molecular structures, MRS has been used frequently in the clinical setting for diagnosis, using the disease-specific metabolite or a metabolite profile as the surrogate marker. This technique has recently received attention in studying transplanted cells in animal models (Shyu et al., 2007) and most recently in humans (Manganas et al., 2007). Results from these early studies showed that in vivo NMR was able to detect changes of a few metabolites associated with themetabolic activity of transplanted cells. However, those studies provided limited information on metabolite profiles of specific cell types, due to low sensitivity and low spectral resolution of in vivo NMR.
High-resolution magic angle spinning (HRMAS) solid-state NMR spectroscopy has been recently developed for ex vivo analysis of intact biological samples (Cheng et al., 1997; Swanson et al., 2004; Mao et al., 2007), including intact cells (Griffin et al., 2002) and cell grafts (Li et al., 2006) as well as differentiating cells (Chen et al., 2002). Because cellular metabolites can be identified and quantified by HRMAS NMR using intact tissue specimens or cells, this method can be used to characterize the metabolite profile of tissues and cells and to discover metabolite markers that may be specific to the diseases or functions of cells. Although this method may not be applied directly in vivo, it has been recognized as a metabolomics tool for investigating the cell metabolism and metabolite markers for their potential applications in vivo.
In the present study, 1H-NMR was used to investigate mesenchymal stem cells (MSCs) and MSC changes after undergoing adipogenic differentiation to the targeted fat cells in vitro. The results revealed that MSCs exhibited a specific metabolite profile in the NMR spectrum and the change of this metabolite profile occurred as the result of the differentiation. This study demonstrated the superb spectral resolution and sensitivity of 1H-NMR in profiling cell types, and the potential of using cell-specific metabolite markers identified by 1H-NMR for monitoring the differentiation of transplanted stem cells in vivo, using the readily available NMR method.
2. Materials and methods
2.1. Preparation of MSCs and adipogenic differentiation
A multipotent mesenchymal stem cell line, D1, cloned from Balb/c mouse bone marrow stromal cells, as described previously (Chen et al., 2005), was used in this study. D1 cells were maintained in T-medium (Life Technologies, Rockville, MD, USA) with 5% fetal bovine serum (Hyclone Laboratories, Logan, VT, USA) and 1% penicillin/streptomycin. For the adipogenic differentiation, D1 cells were treated in the basalmedium supplemented with 1 µm dexamethasone, 0.5 mm isobutyl-methylxanthine (IMBX) and 5 µg/ml insulin (Sigma, St Louis, MO, USA) for 21 days. Cells maintained in the basal medium were examined as negative controls.
2.2. Immunohistochemistry (IHC) and histology
The cell differentiation was assessed for morphological changes using an inverted light microscope (Olympus America, Center Valley, PA, USA) and oil red O staining, as described previously (Janderova et al., 2003). Briefly, cells were fixed in acetone for 15 min, washed with the aqueous phosphate buffer and stained with 0.4% oil red O solution (Sigma) for 10 min, followed by repeated washing with distilled water, and then destained in 100% isopropanol for 15 min. The cells were then rinsed with distilled water and counterstained with haematoxylin (Zymed Laboratories, San Francisco, CA, USA) for approximately 3 min to stain the cell nuclei. Slides were then observed using an Olympus IX-70 inverted microscope configured for transmission light (bright field, DIC and polarized light) and reflected light observation. At least five randomly selected areas were assessed at × 100 or × 200 magnification. Cell nuclei were evident due to the haematoxylin counterstaining. Images were captured using a digital colour camera (Nikon D1) mounted on the microscope.
2.3. Sample preparation for 1H-NMR
To prepare each sample for NMR experiments, approximately 6 × 106 cells were harvested from the culture and washed with D2O (99%) saline and phosphate buffer five times to remove the culture medium. This process also allowed for exchanging majority of H2O in the buffer to D2O (deuterium oxide; heavy water with the isotopic form of hydrogen, deuterium or 2H, to minimize the proton signal from 1H2O). Cell pellets were made by centrifuging at 1000 r.p.m. for 10 min, then collected and weighed for NMR experiments. Each sample (usually 30–50 mg wet weight) was resuspended in 99% D2O saline before being loaded on the NMR sample holder/rotor (4 mm ZrO2). After loading a cell pellet, a 50 µl insert was placed in the sample holder to stabilize the sample and to provide the balance for the rotor. The preparation of NMR samples was done rapidly on ice to avoid sample degradation. For the external chemical shift reference, an aliquot amount of trimethylsilyl l-2,2,3,3-tetradeuteropropionic acid (TSP; sodium salt, Sigma), was added in the sample and weighed. The resonance of water proton (4.65 ppm) was also used as the internal reference.
2.4. 1H-NMR data acquisition
1H-NMR experiments were performed at 4 °C using a Bruker AVANCE 600WB solid-state NMR spectrometer (Bruker Instruments, Fremont, CA, USA) with a dedicated 4 mm 1H probe. The probe-head was pre-cooled to 4 °C before loading the sample into the instrument. The sample/probe temperature was maintained throughout the experiment (±0.1 °C) via a variable temperature control unit. The sample spinning rate was controlled in the range of 2800 kHz (±2 Hz) typically. This low sample spin rate was tested to ensure that the spin side bands do not appear in the spectrum range containing the metabolites of the interest. Spectra were acquired with and without suppression of the water signal. The presaturation of the water signal was achieved with a zqpr sequence before acquisition pulses (standard Bruker pulse sequence). The 90° pulse length was calibrated and adjusted based on each sample. The number of transients was 400 typically for collecting each onedimensional spectrum. A repetition time of 5.0 s (>5 × T1 of most of the metabolites of interest) and a spectral width of 10 kHz were typically used. The data were processed using software of XWINNMR installed on the instrument. A line broadening (1 Hz) apodization was applied to all free induction decays (FIDs) before Fourier transformation. Integrals of the selected resonances from cellular metabolites between 0.5 to 4.5 ppm in the spectra were measured for the calculation of metabolite concentrations. Each sample was measured three times over a typical 2 h experiment to monitor possible changes of the cells during the experiment.
For resonance assignment and confirmation, two-dimensional 1H J-coupled correlated spectroscopy (COSY; standard Bruker pulse sequence) was used (Martinez-Granados et al., 2006; Mao H et al., 2007) to determine J-couple correlations of resonances of the same metabolite. COSY spectra were collected with 6000 Hz spectral width (SW) in both dimensions (D1 and D2) and a 1.5 s relaxation delay; 32 transients were averaged for each of the 256 increments in D1, corresponding to a total acquisition time of ~7 h. Two-dimensional spectral data were analysed on the instrument, using zero filling to a 1 k × 1 k matrix and weighted with a shifted square sine bell function and 1 Hz exponential (D1 and D2) line broadening, followed by Fourier transformation. A one-dimensional spectrum was collected after the COSY experiment to assess possible spectral changes of the cell sample.
2.5. Metabolite assignment and quantification
After all spectra were Fourier-transformed and baseline-corrected, chemical shift values of the resonances of interest were determined using the external reference of –CH3 protons of TSP as 0 ppm at 4 °C, or using the internal reference of proton resonance of H2O at 4.65 ppm (Tong et al., 2004). Resonance assignments for the metabolites of interest were obtained based on their chemical shifts reported in the literature (Martinez-Granados et al., 2006; Griffin et al., 2002; Griffin et al., 2001; Ross et al., 1998) and further confirmed by two-dimensional COSY experiment if it could be applied.
The integral of each peak was measured using the XWINNMR program (Bruker Biospin), after performing Fourier transformation, phase and baseline corrections. The absolute concentration of each metabolite of interest in the sample was calculated from the peak integral with respect to that of the internal reference TSP, according to the equation:
| (1) |
where Cm = metabolite concentration, Im = integral of selected of 1H resonance from the metabolite of interest (e.g. from –CH3 from fatty acids), Nm = number of protons in the resonance (peak) of the metabolite (e.g. 3 for –CH3 of fatty acid), CTSP = TSP concentration, ITSP = integral of TSP peak at 0 ppm (9, since TSP has 9 protons), V = volume of TSP added in the NMR sample and W = weight of cell pellet. It is assumed that cell numbers and the concentration of TSP in the sample did not change during the NMR experiment.
Statistical comparisons of the metabolite level in MSC and differentiated cells were made using the SPSS 15.0 software package, using two-way ANOVA. A value of p < 0.05 was considered to be statistically significant.
3. Results
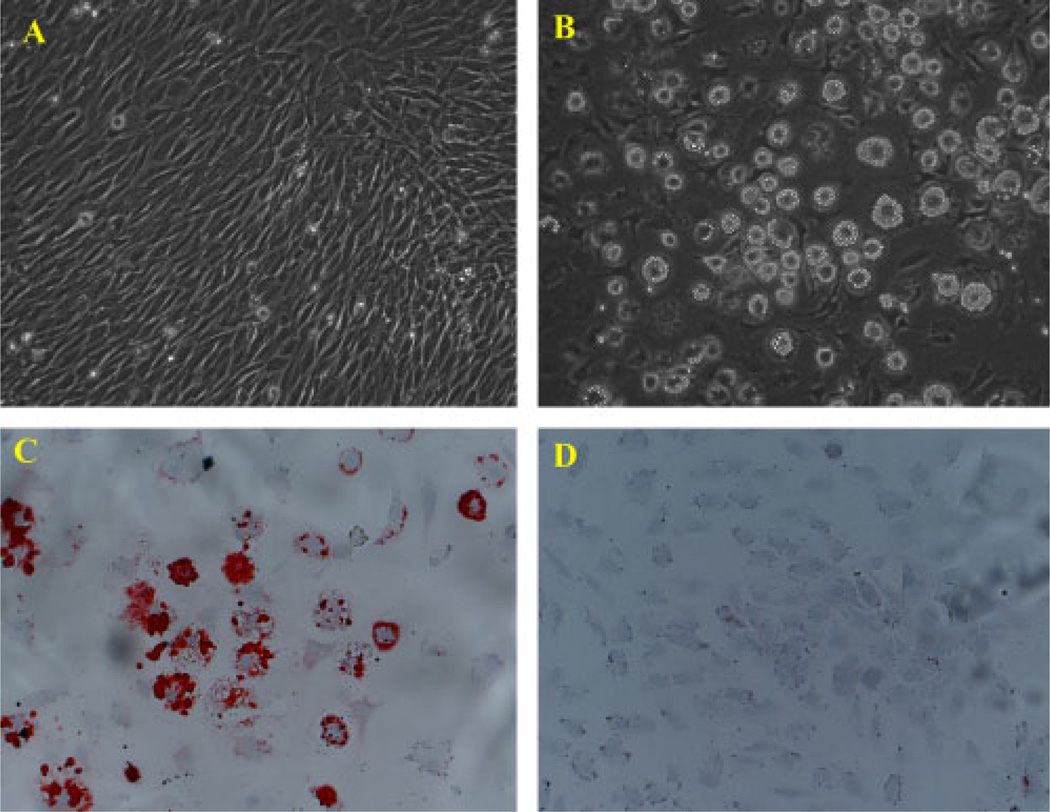
The MSC D1 cells were cloned from the mouse bone marrow with multipotency for osteogenic and adipocytic differentiation in vivo and in vitro. Figure 1A shows fibroblast-like D1 cells in the standard culture condition. After the treatment with the adipogenic medium, MSC D1 cells exhibited a significant morphological change, evidenced by the accumulation of lipid vesicles that were smaller initially and increased in size over time. Those cells containing lipid vesicles were clearly distinguishable from the surrounding cells using a phase-contrast microscope (Figure 1B). Oil red O staining further confirmed the adipogenic differentiation of D1 cells. Under inducing conditions of our experiment, about 80% of the cells were oil red O-positive at the end of the 21-day experiment (Figure 1C). In comparison, adipogenic changes were not found in D1 cells in the basal culture medium (Figure 1D).
Figure 1.
Morphological change and oil red O staining during adipogenic differentiation of MSC D1 cells. (A) D1 cells in basal culture medium. (B) Morphological change and accumulation of lipid vesicles after D1 cells were treated with adipogenic medium. (C) Cells stained with oil red O for intracellular lipid vesicles after D1 cells were treated with adipogenic medium. (D) oil red O-positive cells were not found in D1 cells in the basal culture medium without inducing agents. Cell nuclei were counterstained with haematoxylin and viewed at × 100 magnification
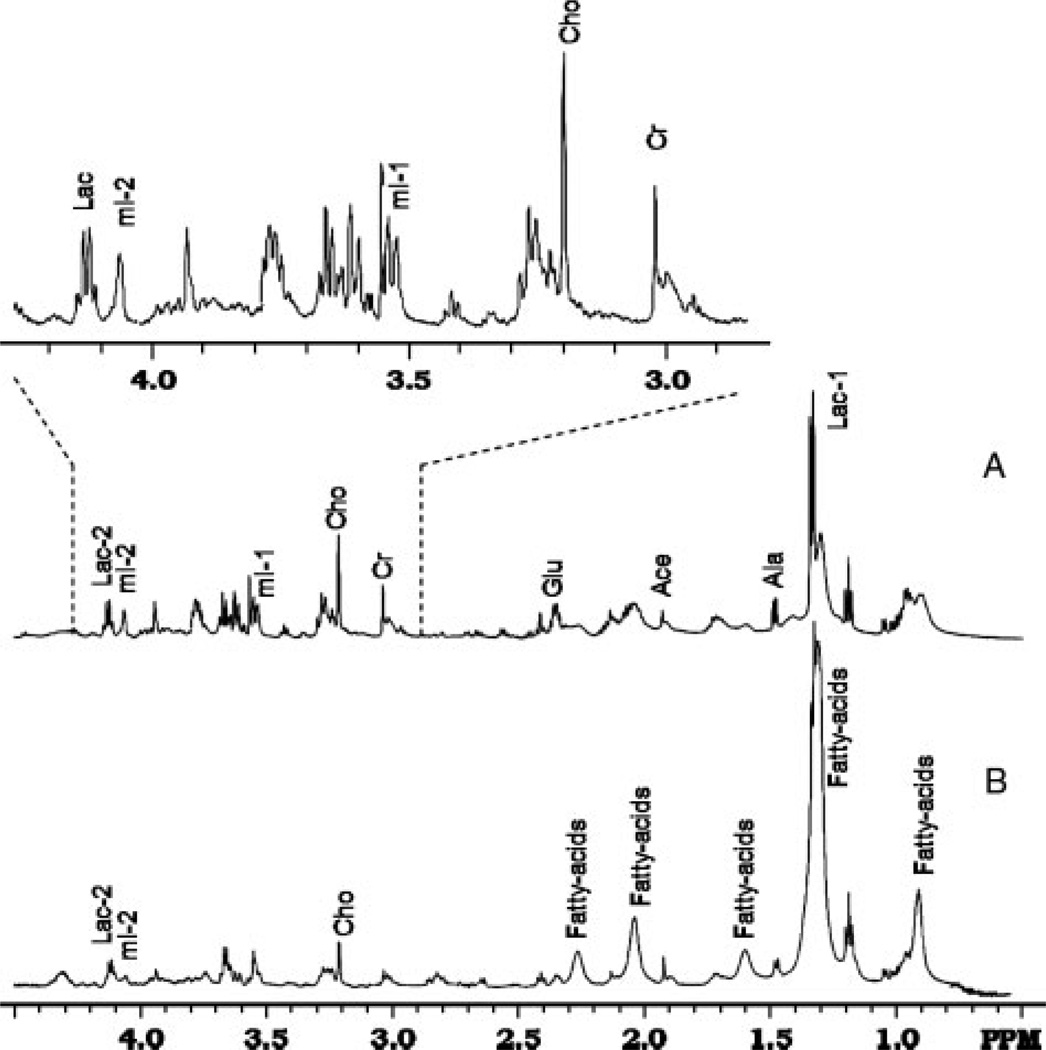
1H HRMAS NMR experiments of cell samples were mostly reproducible in our repeated experiments and during the time of collecting one-dimensional NMR spectra (usually < 2 h) with <3% changes of intensities of signals from –CH3 of Cho at 3.19 ppm and TSP at 0.0 ppm. However, there were changes in the spectra after the 7-h two-dimensional COSY experiment, likely due to the cell degradation. Therefore, two-dimensional COSY data were mostly used for confirming the assignment of signals from fatty acids which did not exhibit significant changes. Figure 2 shows one-dimensional NMR spectral profiles obtained from the undifferentiated MSCs (A) collected at day 1 and the mix of undifferentiated MSCs and differentiated cells after 3 weeks of differentiation (B) under the adipogenic condition, which is supplemented with 3-isobutyl-1-methylxanthine, insulin and dexamethasone. This condition is believed to promote the differentiation of MSCs into fat cells. The key resonances of major metabolites such as lactate (Lac), alanine (Ala), creatine (Cre), choline (Cho), glutamate (Glu) and myo-inositol (myo-I), were well resolved and assigned in the spectra of the samples of MSCs and differentiated cells, although the levels of those metabolites decreased to various degrees in the differentiated cells. The chemical shifts of those resonances are listed in Table 1.
Figure 2.
1D HRMAS NMR spectra of original MSCs (A) and cells differentiated from MSCs (B). MSCs had undergone differentiation for 3 weeks at the time of recording spectrum B, which represents a mix of end-point fat cells and remaining undifferentiated MSCs. An expanded spectrum from the region of 2.8–4.3 ppm of the spectrum from MSCs (A) is inserted
Table 1.
Major metabolites observed in MSCs and differentiated cells
| Metabolite | 1H or spin* | Chemical shift (ppm) |
|---|---|---|
| Fatty acids | –CH2 –CH3 | 0.89 |
| Fatty acids | –CH2 –(CH2)n –CH2 | 1.30 |
| Lactate | –CH3 | 1.33 |
| Fatty acids | –CH2 –CH3 | 1.35 |
| Alanine (Ala) | –CH3 | 1.48 |
| Fatty acids | –CH2 –CH2 –CO– | 1.58 |
| Fatty acids | –CH2 –CH═CH– | 2.02 |
| Glutamate (Glu) | –CH3 | 2.05 |
| Glutamine (Gln) | β –CH2 | 2.13 |
| Glutamine(Gln) | –CH3 | 2.15 |
| Fatty acids | CH2 –CH2 –CO– | 2.25 |
| Glutamate (Glu) | γ –CH2 | 2.36 |
| Glutamine (Gln) | γ –CH2 | 2.45 |
| Fatty acids | –CH═CH–CH2 –CH═CH– | 2.75 |
| Creatine (Cre) | –CH3 | 3.04 |
| Choline (Cho) | N–(CH3)3 | 3.19 |
| Phosphocholine (PC) | N–(CH3)3 | 3.22 |
| Glyocerphosphocholine (GPC) | –CH2 –N–(CH3)3 | 3.24 |
| Myo-inositol (myo-I) | –CH–(5) | 3.28 |
| Taurine | –CH2 –SO3 | 3.43 |
| Myo-inositol (myo-I) | –CH–(1) | 3.55 |
| Myo-inositol (myo-I) | –CH–(3) | 3.63 |
| Creatine (Cre) | –CH2 | 3.95 |
| Myo-inositol (myo-I) | –CH–(2) | 4.07 |
Resonance assignments were referenced to a previous report (Martinez-Bisbal et al., 2004).
Bold characters indicate protons assigned at Chemical Shift.
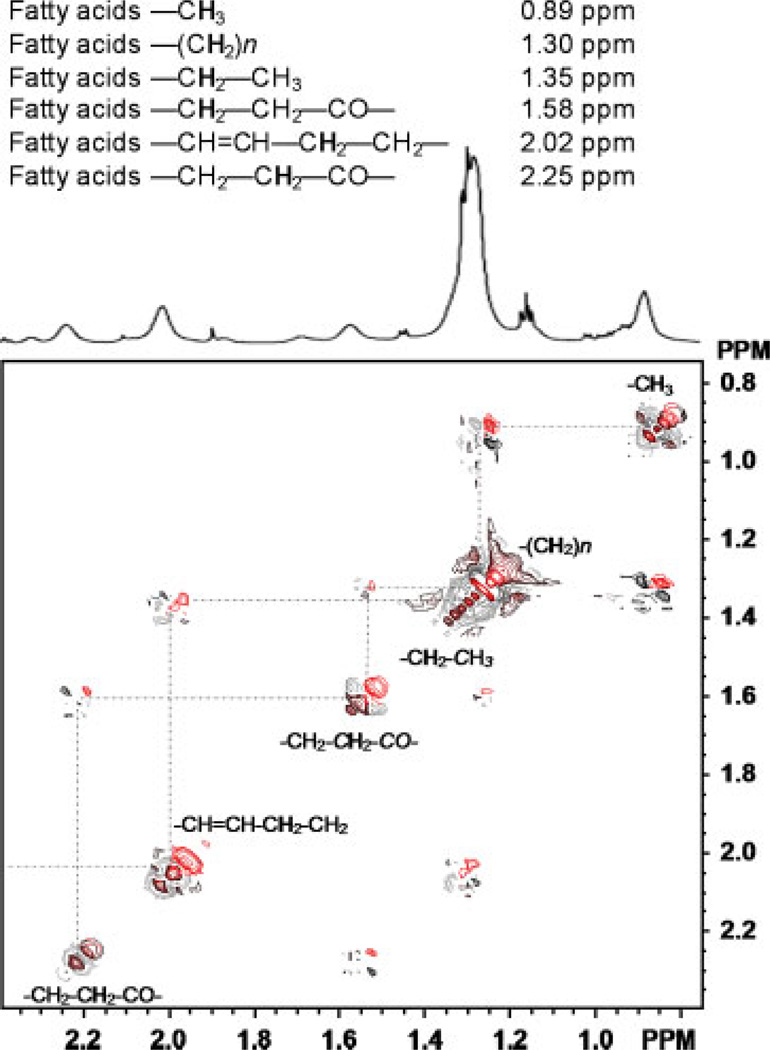
The assignments of fatty acids were confirmed by 2D COSY experiments; an expanded 2D COSY spectrum is shown in Figure 3. Resonances from fatty acids exhibit a unique J-coupling correlation and can be readily identified in chemical shifts of 0.89, 1.30, 1.35, 2.02 and 2.25 ppm, which can be attributed to the spin system of fatty acids, e.g. –CH3, –(CH2)n –, –CH═CH–CH2, –CH2–CH═CH– and –COCH2–CH2–COCH2 (proton with respective chemical shift indicated in bold). The spectra and assignments of fatty acids are generally in agreement with those reported by others (Singer et al., 1996; Griffin et al., 2001). In addition, resonances rising from glutamate and glutamine were also assigned with the assistance of 2D COSY data.
Figure 3.
The scalar J-coupling correlation of the spin system of the fatty acids of fat cells is demonstrated in the expanded plot of 2D COSY (0.8–2.3 ppm) with selected resonances. The peak from –CH3 (0.89 ppm) was used for calculating the concentration of the fatty acids
Analysis of spectra from MSC and differentiated cells revealed that there were statistically significant decreases of total Cho (p < 0.02), total Cre (p < 0.03) and Glu (p < 0.04) as well as myo-I (p < 0.02) in differentiated cells in comparison to those of original MSCs after undergoing differentiation for 3 weeks. Metabolite quantification using the NMR method showed that major metabolites: Cre, myo-I and total Cho, reduced from 10.4 ± 0.72, 16.2 ± 1.2 and 8.22 ± 0.51 mm to 3.27 ± 0.34, 6.1 ± 0.46 and 3.11 ± 0.32 mm, respectively. This was coupled with substantial increases of the signal intensity in all resonances of fatty acids, e.g. –CH3, –CH2-(CH2)n–CH2 –, –CH═CH–CH2, –CH2 –CH═CH– and –COCH2–CH2–COCH2. Fatty acids increased from 32.6 ± 1.5 mm in samples of MSCs before differentiation to 91.2 ± 3.2 mm in samples mixed with differentiated fat cells and the remaining undifferentiated MSCs. The increase of the fat content may be better evaluated with the ratio of fatty acids : creatine, which was 27.3 in differentiated cells after 3 weeks, rising from 3.14 in the MSCs. The changes of metabolite concentrations as a result of the differentiation are summarized in Table 2.
Table 2.
Concentrations of selected metabolites in MSCs and MSC differentiated cells*
| Metabolite concentrations (mm) | Metabolite ratios** | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fatty acids (0.89 ppm) |
Ala (1.48 ppm) |
Glu (2.36 ppm) |
Cre (3.04 ppm) |
Cho (3.19 ppm) |
Myo-I (4.01 ppm) |
Lip : Cre | Ala : Cre | Glu : Cre | Cho : Cre | Myo-I : Cre | ||
| MSCs | 32.6 ± 1.5 | 6.20 ± 0.42 | 5.01 ± 0.46 | 10.4 ± 0.72 | 8.22 ± 0.51 | 16.2 ± 1.2 | 3.14 | 0.59 | 0.51 | 0.79 | 1.56 | |
| Differentiated cells*** | 91.2 ± 3.2 | 2.12 ± 0.29 | 2.01 ± 0.31 | 3.27 ± 0.34 | 3.11 ± 0.32 | 6.1 ± 0.46 | 27.9 | 0.75 | 0.68 | 0.95 | 1.87 | |
Data are presented as mean ± SD from three repeated experiments.
Metabolite ratio is defined as the concentration of individual metabolite vs. concentration of creatine, i.e. [Metabolite]/[Cre].
The sample of differentiated cells may include MSC differentiated fat cells (~80%) and remaining undifferentiated MSCs.
4. Discussion
4.1. Metabolites associated with cell activities and functions
Cellular metabolism is closely associated with cell functions. While some metabolites are essential to all cell types, some may only be required by the specific function of certain cell types. The metabolic products of those functions may also vary in different cells. Different metabolite profiles of MSCs and fat cells differentiated from MSCs, as reported in this study, provide another example that it is possible to use NMR methods to identify cell type-specific metabolite profiles.
While detailed profiling and assignments of all resonances and their metabolites in different cells are not possible at this time, due to the sensitivity and spectral resolution of the current NMR method and the complexity of the cellular metabolism of each cell type, most in vivo 1H-NMR spectra of cells typically consist of several detectable resonances that are key to cellular functions. These include: choline and its derivatives, creatine, lactate, glutamate/glutamine and myo-inositol (Ross et al., 1997; Griffin et al., 2002; Li et al., 2006). The Cho resonances, often a sum of choline and its derivatives, including phosphocholine (PC) and glycerophosphocholine (GPC), due to the difficulty of resolving individual derivatives in the spectrum, are generally considered to be related to the membrane metabolism (Ross et al., 1997). An increase in Cho, or total choline, is considered to represent the metabolic processes associated with the high turnover of the cell membrane. The resonance from Cre is composed of creatine and its derivative phosphocreatine, and is related to the cellular energy storage. Lactate is the product of glycolysis, and is often observed under hypoxic conditions. Glutamate (Glu) and glutamine (Gln) are considered to be major nitrogen resources for cell functions (Ross et al., 1997). Myo-I is considered to play the role of osmolyte (Ross et al., 1991, 1997). Resonances from the fatty acids, appearing with scalar J-coupling pattern, represent mostly intracellular phospholipids and fatty acids, such as triacylglycerols in the fat cells (Szczepaniak et al., 1999).
Interestingly, observed reductions of Cre, Cho and myo-I as well as Glu from MSCs to differentiated fat cells may reflect the change of the cellular metabolism required by different cell types. Possible interpretations of such observations are as follows. First, it is expected that fat cells have much lower cellular metabolism compared to MSCs. Furthermore, because it is unlikely that MSCs were completely differentiated to targeted fat cells, it is possible that the levels of cell metabolites observed in the differentiated cells represent those of non-fat cells or cells that were not differentiated.
Results from this study showed that fatty acids, as a part of fat cells and a measurement of the fat content, are increased when MSCs differentiate to fat cells, while most of the essential cell metabolites required by cellular metabolism, such as Cre, Cho and myo-I, are reduced. Therefore, it is possible to follow MSCs or other transplanted cells differentiate to fat cells non-invasively by the NMR method in vivo, using signals from fatty acids. Given the potential of identifying NMR-detectable cell-specific metabolite profiles, the implication of this study is that the magnetic resonance spectroscopy method may provide an excellent tool for in vivo monitoring of functions and activities of transplanted stem cells or other therapeutic cells in live animals or patients.
However, we recognize that our study is limited in using a large number of cultured cells for the NMR analysis in vitro. It is possible that the results may differ when using NMR analysing transplanted cells in vivo, given that stem cell differentiation in vivo occurs in the complex tissue environment and possible interfering NMR signals from the tissue background. Therefore, NMR-sensitive cell specific metabolite markers and the NMR detection method will need to be eventually tested in animal models before they can be applied in humans.
4.2. Adipogenic differentiation of MSCs and its clinical relevance
The adipose lineage is a well-known mesenchymal lineage originated from multipotential human MSCs. Adipose tissue has been recognized as an active organ that can store and release energy, maintain glucose homeostasis and secrete hormones and cytokines (Trayhurn et al., 2001). Adipogenic trans-differentiation may occur in in vivo conditions and can contribute to age-related diseases such as osteoporosis and osteopenia, which are accompanied by increased adipose tissue and a decreased number of osteoblasts in the bone marrow (Nuttall et al., 2000). Establishing specific metabolite markers of adipogenic differentiation may allow for non-invasive monitoring of such events in vivo using the image-guided NMR method. Further development and testing of this methodology in vivo may lead to a very important step toward the application of cell transplantation methods in regenerative medicine, since current strategies of assessing the efficacy of cell implants are only able to follow cell differentiation by analysis of cell type-specific gene expression and histochemical staining (Schilling et al., 2007).
Adipogenic differentiation can be characterized by the strong expression of lipoprotein lipase (LPL) and peroxisome proliferator-activated receptor γ 2 (PPARγ 2) at the early stage and the formation of lipid vesicles in the cytoplasm at the late stage (Barak et al., 1999). It has been extensively confirmed that medium supplemented with 3-isobutyl-1-methylxanthine, insulin and dexamethasone is able to effectively induce adipogenic differentiation (Pittenger et al., 1999). Here, we used the in vitro medium containing 3-isobutyl-1-methylxanthine, insulin and dexamethasone to induce the adipogenic differentiation in MSCs and demonstrated that 1H-NMR offers a powerful tool for investigating the adipogenic differentiation of MSCs by monitoring changes in cellular metabolites and metabolite profiles, specifically signals from fatty acids.
4.3. Potentials and limitations of NMR in monitoring transplanted stem cells
The method of NMR has been widely used in research and clinical diagnosis of various diseases, particularly brain diseases, using specific metabolites as ‘surrogate markers’ associated cell types. When applied in vivo, this method can provide insights into metabolic processes non-invasively. Comparing other imaging modalities available for imaging of animals and humans, NMR has particular advantages, including: its ability to detect and analyse multiple endogenous chemicals or metabolites; capability of studying tissue or cells transplanted deeply inside the body; and providing a good spatial resolution (~5 mm in human and ~100 µm in animal experiments) with accurate sampling when combined with MRI. However, one significant limitation of magnetic resonance detection is its relatively low sensitivity, particularly to those cellular metabolites at low concentrations (below mm). Although this work provides a proof-of-principle example that one can identify a NMR/MRS detectable cell-specific metabolite profile for monitoring stem cell differentiation, using NMR methods to follow the differentiation of transplanted stem cells in vivo will encounter the challenges of localizing, detecting and eventually analysing a small number of transplanted stem cells which may also migrate after the transplantation. Furthermore, additional questions may need to be addressed, such as: (a) is the NMR sensitive enough to detect a small number of cells?; (b) how can NMR distinguish the differentiated stem cells from mature cells that are already present within the body with similar profiles? Despite these challenges, there are an increasing number of studies that demonstrate the potentials of using NMR for in vivo tracking of transplanted stem cells. One previous in vivo NMR study of transplanted stem cells in treating stroke in a rat model showed that a change of N-acetyl-aspartate (NAA) level, a metabolite marker for neuronal cells, could be used as an indicator for improved neuronal activity, and that NMR could be applied in a living system repeatedly (Shyu et al., 2007). A more recent study during the preparation of this manuscript has shown that this approach can be applied in patients to monitor the therapeutic progenitor cells in the brain (Manganas et al., 2007). It is worth noting that the MR spectroscopic approach will be complementary to anatomical MRI and cell labelling-based MRI cell tracking methods, which can provide great spatial resolution for localizing the region or tissue of interest but lack the ability to provide functional and metabolic information on targeted cells.
4.4. Advantages of the HRMAS NMR method in identifying cell-specific markers
To develop the in vivo application of NMR in monitoring transplanted cells, one critical step is to test and validate the method and results using histological or ex vivo analytical analysis. The effort has been made to investigate possible metabolite profiles of different stem cells using cell extractions and the method of solution NMR (Jansen et al., 2006), although the use of samples from cell extractions require destructive sample preparation. Solid-state HRMAS NMR analysis therefore has the significant advantage of using intact biological samples from either tissue specimens or cells to profile metabolites that may associate to specific pathology and diseases (Cheng et al., 2002). With remarkably high spectral resolution and sensitivity, HRMAS NMR can be used to characterize unknown or newly discovered resonances or chemicals, as well as to assist the interpretation and quantification of spectra obtained in vivo, therefore providing biochemical information complementary to histological and pathological analysis. In addition, one advantage of using the ex vivo NMR method as an investigative tool to study the cell-specific metabolite profiles in cultured cells or collected tissue samples is that it allows for cell metabolite profiles to be examined and analysed quantitatively, since stem cells transplanted in vivo will likely migrate and may be difficult to locate. Given the fact that the NMR method is already available in clinical diagnosis and treatment monitoring in patients, the results from HRMAS NMR analysis may become important to further development of the in vivo NMR method for studying the animal models and eventually for clinical applications of stem cells in the future.
5. Conclusion
1H-NMR analysis of MSCs before and after differentiation showed distinguishable metabolite profiles of different cell types. Observed increased levels of fatty acids and decreased levels of other intracellular metabolites are the results of the preferential differentiation of MSCs to fat cells. Therefore, it is possible to use the signals of fatty acids to follow the differentiation of MSCs to fat cells by the method of NMR spectroscopy. This study also demonstrated that HRMAS NMR offers a tool for identifying cell type-specific metabolite ‘surrogate’ markers that can be potentially used for in vivo monitoring of the metabolic activities and differentiation of transplanted stem cells with the clinically available NMR method.
Acknowledgements
This work was partially supported (to C.S.) by the NSFC programme (Grant No. 30400188 and 30870966), the ‘973’ programme (Grant No. 2005CB522605), the FANEDD programme (Grant No. 200777) and the PCSIRT program (Grant No. IRT0712) from China, and (to H.M.) by a pilot grant from Emory Alzheimer’s Disease Research Center under the programme project Grant No. P50AG025688) from NIH, USA.
References
- Barak NMC, Ong ES, Jones YZ, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Chen CH, Ho ML, Chang JK, et al. Green tea catechin enhances osteogenesis in a bone marrow mesenchymal stem cell line. Osteoporos Int. 2005;16:2039–2045. doi: 10.1007/s00198-005-1995-0. [DOI] [PubMed] [Google Scholar]
- Chen JH, Enloe BM, Weybright P, et al. Biochemical correlates of thiazolidinedione-induced adipocyte differentiation by high-resolution magic angle spinning NMR spectroscopy. Magn Reson Med. 2002;48:602–610. doi: 10.1002/mrm.10256. [DOI] [PubMed] [Google Scholar]
- Cheng LL, Ma MJ, Becerra L, et al. Quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 1997;94:6408–6413. doi: 10.1073/pnas.94.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LL, Newella K, Mallorya AE, et al. Quantification of neurons in Alzheimer and control brains with ex vivo high resolution magic angle spinning proton magnetic resonance spectroscopy and stereology. Magn Reson Imaging. 2002;20:527–533. doi: 10.1016/s0730-725x(02)00512-x. [DOI] [PubMed] [Google Scholar]
- Dall AM, Danielsen EH, Sorensen JC, et al. Danish Neuronal Xenografting Group. Quantitative [18F] fluorodopa/PET and histology of fetal mesencephalic dopaminergic grafts to the striatum of MPTP-poisoned minipigs. Cell Transpl. 2002;11:733–746. [PubMed] [Google Scholar]
- Englund U, Bjorklund A, Wictorin K, et al. Grafted neural stem cells develop into functional pyramidal neurons and integrate into host cortical circuitry. Proc Natl Acad Sci USA. 2002;99:17089–17094. doi: 10.1073/pnas.252589099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni JV, Hajjar RJ. In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation. 2004;110:3378–3383. doi: 10.1161/01.CIR.0000149840.46523.FC. [DOI] [PubMed] [Google Scholar]
- Friedmann T. Lessons for the stem cell discourse from the gene therapy experience. Perspect Biol Med. 2005;48:585–591. doi: 10.1353/pbm.2005.0089. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Bollard M, Nicholson JK, et al. Spectral profiles of cultured neuronal and glial cells derived from HRMAS 1H-NMR spectroscopy. NMR Biomed. 2002;15:375–384. doi: 10.1002/nbm.792. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Williams HJ, Sang E, et al. Abnormal lipid profile of dystrophic cardiac tissue as demonstrated by one- and two-dimensional magic-angle spinning 1H-NMR spectroscopy. Magn Reson Med. 2001;46:249–255. doi: 10.1002/mrm.1185. [DOI] [PubMed] [Google Scholar]
- Janderova L, McNeil M, Murrell AN, et al. Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obesity Res. 2003;11:65–74. doi: 10.1038/oby.2003.11. [DOI] [PubMed] [Google Scholar]
- Jansen JF, Shamblott MJ, van Zijl PC, et al. Stem cell profiling by nuclear magnetic resonance spectroscopy. Magn Reson Med. 2006;56:666–670. doi: 10.1002/mrm.20968. [DOI] [PubMed] [Google Scholar]
- Klunk WE, Xu C, Panchalingam K, et al. Quantitative 1H and 31P NMR of PCA extracts of postmortem Alzheimer’s disease brain. Neurobiol Aging. 1996;17:349–357. doi: 10.1016/0197-4580(96)00035-8. [DOI] [PubMed] [Google Scholar]
- Lewin M, Cariesso N, Tung CH, et al. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- Li W. Multidimensional HRMAS NMR: a platform for in vivo studies using intact bacterial cells. Analyst. 2006;131:777–781. doi: 10.1039/b605110c. [DOI] [PubMed] [Google Scholar]
- Mao H, Wang X, Lacreuse A, et al. HRMAS NMR measured metabolic changes in kainic acid-induced hippocampal injury in rat. Exp Brain Res. 2007;183:477–485. doi: 10.1007/s00221-007-1061-6. [DOI] [PubMed] [Google Scholar]
- Martínez-Bisbal MC, Martí-Bonmatí L, Piquer J, et al. 1H and 13C HR-MAS spectroscopy of intact biopsy samples ex vivo and in vivo 1H-NMR study of human high grade gliomas. NMR Biomed. 2004;17:191–205. doi: 10.1002/nbm.888. [DOI] [PubMed] [Google Scholar]
- Martinez-Granados B, Monleon D, Martinez-Bisbal MC, et al. Metabolite identification in human liver needle biopsies by high-resolution magic angle spinning 1H-NMR spectroscopy. NMR Biomed. 2006;19:90–100. doi: 10.1002/nbm.1005. [DOI] [PubMed] [Google Scholar]
- Manganas LN, Zhang X, Li Y, et al. Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain. Science. 2007;318:980–985. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama K, Kumar K, Wuthrich K, et al. Experimental techniques of two-dimensional correlated spectroscopy. J Magn Reson. 1980;40:321–334. [Google Scholar]
- Nuttall ME, Gimble JM. Is there a therapeutic opportunity to either prevent or treat osteopenic disorders by inhibiting marrow adipogenesis? Bone. 2000;27:177–184. doi: 10.1016/s8756-3282(00)00317-3. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Pogarell O, Koch W, Gildehaus FJ, et al. Long-term assessment of striatal dopamine transporters in Parkinsonian patients with intrastriatal embryonic mesencephalic grafts. Eur J Nucl Med Mol Imaging. 2006;33:407–411. doi: 10.1007/s00259-005-0032-z. [DOI] [PubMed] [Google Scholar]
- Ross BD. Biochemical considerations in 1H spectroscopy. Glutamate and glutamine; myo-inositol and related metabolites. NMR Biomed. 1991;4:59–63. doi: 10.1002/nbm.1940040205. [DOI] [PubMed] [Google Scholar]
- Ross BD, Bluml S, Cowan R. In vivo MRspectroscopy of human dementia. Neuroimag Clin N Am. 1998;8:809–822. [PubMed] [Google Scholar]
- Ross BD, Bluml S, Cowan R, et al. In vivo magnetic resonance spectroscopy of human brain: the biophysical basis of dementia. Biophys Chem. 1997;68:161–172. doi: 10.1016/s0301-4622(97)00032-x. [DOI] [PubMed] [Google Scholar]
- Schilling T, Noth U, Klein-Hitpass L, et al. Plasticity in adipogenesis and osteogenesis of human mesenchymal stem cells. Mol Cell Endocrinol. 2007;271:1–17. doi: 10.1016/j.mce.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Shyu WC, Chen CP, Lin SZ, et al. Efficient tracking of non-iron-labelled mesenchymal stem cells with serial MRI in chronic stroke rats. Stroke. 2007;38:367–374. doi: 10.1161/01.STR.0000254463.24655.14. [DOI] [PubMed] [Google Scholar]
- Singer S, Sivaranja M, Souza K, et al. 1H-NMR detectable fatty acyl chain unsaturation in excised leiomyosarcoma correlate with grade and mitotic activity. J Clin Invest. 1996;98:244–250. doi: 10.1172/JCI118785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MG, Vigneron DB, Tabatabai ZL, et al. Proton HR-MAS spectroscopy and quantitative pathologic analysis of MRI/3D-NMRI-targeted postsurgical prostate tissues. Magn Reson Med. 2003;50:944–954. doi: 10.1002/mrm.10614. [DOI] [PubMed] [Google Scholar]
- Szczepaniak LS, Babcock EE, Schick F, et al. Measurement of intracellular triglyceride stores by 1H spectroscopy: validation in vivo. Am J Physiol. 1999;276:977–989. doi: 10.1152/ajpendo.1999.276.5.E977. [DOI] [PubMed] [Google Scholar]
- Tong Z, Yamaki Y, Harada K, et al. In vivo quantification of the metabolites in normal brain and brain tumors by proton MR spectroscopy using water as an internal standard. Magn Reson Imaging. 2004;22:1017–1024. doi: 10.1016/j.mri.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Trayhurn P, Beattie JH. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. Proc Nutr Soc. 2001;60:329–339. doi: 10.1079/pns200194. [DOI] [PubMed] [Google Scholar]