Abstract
The NADPH oxidase of phagocytic leukocytes generates superoxide that plays a critical role in innate immunity and inflammatory responses. The integral membrane protein flavocytochrome b (Cyt b1, a.k.a. cytochrome b558/559) is the catalytic core of the complex and serves as a prototype for homologs important in regulating signaling networks in a wide variety of animal and plant cells. Our analysis identifies a naturally-occurring Tyr72/His72 polymorphism (p.Y72H) in the p22phox subunit of Cyt b at the protein level that has been recognized at the nucleotide level (c.214T>C, formerly C242T) and implicated in cardiovascular disease. In the present study, Cyt b was isolated from human neutrophils and reacted with chemical crosslinkers for subsequent structure analysis by MALDI mass spectrometry. Following mild chemical modification of Cyt b with two pairs of isotopically-differentiated lysine crosslinkers: BS2G-d0/d4 and BS3-d0/d4, the reaction mixtures were digested with trypsin and purified on C18ZipTips to generate samples for mass analysis. MALDI analysis of tryptic digests from each of the above reactions revealed a series of masses that could be assigned to p22phox residues 68–85, assuming an intramolecular crosslink between Lys71 and Lys78. In addition to the 30 ppm mass accuracy obtained with internal mass calibration, increased confidence in the assignment of the crosslinks was provided by the presence of the diagnostic mass patterns resulting from the isotopically-differentiated crosslinking reagent pairs and the Tyr72/His72 p22phox polymorphisms in the crosslinked peptides. This work identifies a novel, low resolution distance constraint in p22phox and suggests that the medically-relevant p.Y72H polymorphism has an invariant structural motif in this region. Because position 72 in p22phox lies outside regions identified as interactive with other oxidase components, the structural invariance also provides additional support for maturational differences as the source of the wide variation in observed reactive oxygen species production by cells expressing p.Y72H.
Keywords: p22phox, MALDI-TOF, Cytochrome b558, NADPH oxidase, intra-molecular crosslinking, c.214T>C haplotype, C242T haplotype
1.0 Introduction
In the innate immune system, phagocytic leukocytes utilize a multicomponent, NADPH oxidase complex to generate high levels of superoxide critical for the elimination of infectious agents [1;2]. The central importance of this enzyme system in host-defense is apparent from the rare genetic disorder Chronic Granulomatous disease (CGD), where the lack of a functional oxidase results in severe and recurrent infections [3]. Although critical for proper immune function, the generation of reactive oxygen species must be strictly regulated to minimize unwanted host tissue damage that has been observed in a variety of inflammatory disease states [4;5]. The broader significance of the NADPH oxidase complex is greatly enhanced by genomic and cell biological evidence of NADPH oxidase homologs in a wide variety of non-phagocytic tissues across a spectrum of animal and plant species and by the role of superoxide (and reactive oxygen species derived from superoxide) generated by the oxidase homologs required for the regulation of diverse signal transduction cascades [6–8]. Currently, a new research focus centers on the roles of non-phagocytic NADPH oxidase and ROS generation and their relationship to diseases such as those affecting gastrointestinal [9;10], limbic [11], and cardiovascular tissues [12;13].
The activated human phagocyte NADPH oxidase is composed of the heterodimeric, integral membrane protein flavocytochrome b (Cyt b; a.k.a. cytochrome b558/559, flavocytochrome b558/559) consisting of the subunits gp91phox (a.k.a. NOX2) and p22phox. Cyt b contains the catalytic site and serves as the membrane anchor for at least four bound cytosolic regulatory subunits (p40phox, p47phox, p67phox, Rac1/2) [2;14] and Rap1a, whose function in this system remains unknown [15]. In resting cells, the membrane and cytosolic components are physically separated to prevent the inappropriate generation of superoxide. Upon cell stimulation, the oxidase cytosolic factors translocate to the membrane and assemble with Cyt b to activate superoxide production. Although defects in oxidase components have been directly linked to CGD, there is now intense interest on second order effects of variants in other diseases. This interest includes the structural ramifications of single nucleotide polymorphisms [16]. Although high resolution structure determination of the cytoplasmic oxidase components has provided important insights into the nature of this enzyme complex [17], current models of the integral membrane protein Cyt b rely on a combination of computational and biochemical methods [18–20]. More detailed information on the structure of the oxidase catalytic component (Cyt b) is required to better understand the molecular mechanisms of oxidase assembly, activation, and dysfunction.
While chemical crosslinking has long been recognized as a method for obtaining low-resolution structure information [21], difficulties associated with the identification of the crosslinked residues has limited widespread use of this approach. Renewed interest in the use of crosslinking for structure analysis has been driven by recent advances in mass spectrometry, computational software, and biochemical labeling/isolation methods that facilitate the identification of modified peptides [22–26]. The combined use of conventional and isotopically-labeled crosslinking agents provides a powerful method for confident identification of even low levels of crosslinked peptides by mass spectrometry. Studies conducted on model proteins, where the atomic structure is available, have confirmed the utility of this approach for obtaining both intramolecular and intermolecular structure information [27;28]. The potential of crosslinking and mass spectrometry to obtain molecular distance constraints holds particular promise for samples that are refractory to high-resolution structure determination, including low abundance integral membrane proteins, multisubunit protein complexes, and proteins that display different conformational states [22;26;29–31].
The integral membrane protein nature of Cyt b has hampered the elucidation of its structure. However bioinformatic analyses suggest structures for gp91phox that combine elements of transmembrane electron transfer proteins, such a mitochondrial or chloroplast cytochrome bc1 or b6f, and NADPH-FAD oxido-reductases, whose X-ray crystal structures have been solved [20;32–35]. Very little information can be gained, however, from such a homology approach for the small subunit of flavocytochrome b, as there are virtually no structural homologs available to model this protein or its interaction with gp91phox. Previous studies by our group have demonstrated the utility of mass spectrometry for structure analysis of Cyt b, following affinity purification from neutrophil membranes [36]. In the present study, human neutrophil Cyt b has been modified with mixtures of conventional and isotopically-labeled crosslinking agents for subsequent structure analysis by MALDI mass spectrometry. Using this approach, an intramolecular crosslink was identified in the Cyt b subunit p22phox that was derived from a region of the protein that contains the medically-relevant, Tyr72/His72 (p.Y72H) naturally occurring polymorphism in the human population [37;38]. The present study provides a novel distance constraint in the small subunit that will facilitate modeling of the Cyt b structure, describes experimental methods that may also be fruitfully applied to other complex systems such as the assembled NADPH oxidase complex, and suggests that the local conformation of the region bearing the Tyr72/His72 polymorphism is invariant.
2.0 Experimental
2.1 Materials
Extra dry dimethylsulfoxide (supplied over molecular sieves) was obtained from Arcos Organics. Acetonitrile and trifluoroacetic acid were from J.T. Baker; and α-cyano-4-hydroxycinnamic acid (α-CHC), 2,5-dihydroxybenzoic acid (DHB) and stainless steel MALDI plates were from Bruker. The conventional and deuterated crosslinking agents BS2G-d0, BS2G-d4, BS3-d0, and BS3-d4 were from Pierce; Trypsin Gold was obtained from Promega; dodecylmaltoside was from Anatrace; PMSF and dithiothreitol were from Calbiochem; and GammaBind-sepharose and CNBr-sepharose were from GE Biosciences. Centricon concentrators and C18ZipTips were purchased from Millipore; and the 50 Sonic dismembrator probe sonicator was obtained from Fisher Scientific. The epitope-mimicking peptide for mAb CS9 (Ac-AEARKKPSEEEAA-NH2) was obtained from Global Peptides. Econo-Pac 10 DG desalting columns (30 × 10 ml) were from BioRad. All other reagents were obtained from Sigma-Aldrich. Human neutrophil membrane fractions and mAb CS9 were generated in-house by previously-described methods [18;39].
2.2 Purification of human neutrophil Cyt b
The purification of Cyt b from neutrophil membrane fractions was carried out using a recently described immunoaffinity method [40], with modifications. For purification, isolated neutrophil membranes were washed with 1 M NaCl and then extracted in 10 mM Hepes (pH-7.4), 100 mM NaCl, 10 mM KCl, 1 mM EDTA (Buffer A) containing 0.8 % dodecylmaltoside (DDM) for 50 min at 4 °C. Following centrifugation at 100,000 × g for 30 min to remove insoluble material, the resulting extracts were rotated with mAb CS9-Sepharose (that binds the p22phox subunit of Cyt b) for 1 h at 4 °C. The affinity matrix was then washed to remove unbound material, and Cyt b eluted with Buffer A, 0.8 % DDM containing the epitope-mimicking peptide Ac-AEARKKPSEEEAA-NH2 (200 µM final). To generate DDM-solubilized Cyt b for crosslinking, the above elution fraction was concentrated using a 100 kDa Centricon cutoff membrane. Peptide removal and buffer exchange was carried out by passing purified Cyt b over a 10 DG desalting column equilibrated in PBS (pH-7.4), 0.1% DDM at 4 °C. The resulting Cyt b fractions were pooled and concentrated through a 100 kDa cutoff membrane to achieve a final Cyt b concentration of 1.4 µM. The final, purified Cyt b sample was aliquoted and stored at −20 °C prior to use.
2.3 Chemical crosslinking of Cyt b
The chemical crosslinkers BS2G-d0, BS2G-d4, BS3-d0, and BS3-d4 were individually prepared at 7.5 mM in dry DMSO, mixed 1:1 in pairs to form the crosslinking stocks (BS2G-d0/d4 and BS3-d0/d4) and stored in single use aliquots at −80 °C. For structure analysis of Cyt b, crosslinker stocks were thawed and diluted 20-fold in distilled water immediately prior to use (350 µM crosslinker working solution in 5% DMSO). A portion of the above working solution was diluted 3-fold into 5% DMSO to generate a 117 µM crosslinker working solution. Reactions were then initiated by the addition of 2 µl of the agent under investigation (BS2G-d0/d4 or BS3-d0/d4 in 5% DMSO) to 8 µl of purified Cyt b in PBS (pH-7.4), 0.1% DDM (molar ratios of 20:1 and 60:1 crosslinker:Cyt b final). The resulting mixtures were incubated for 30–75 minutes at room temperature and then quenched with 40 mM ammonium bicarbonate for 20 minutes prior to SDS-PAGE or tryptic digestion.
2.4 Preparation of tryptic digests for mass spectrometry
Trypsin Gold was dissolved at 1 mg/ml in 50 mM acetic acid and stored at −80 °C prior to use. For solution digests, trypsin was diluted 25-fold into PBS (pH-7.4) and 1 µl of this working solution added to the Cyt b reaction mixtures described above (1:20 trypsin:Cyt b w/w). Following incubation at 37 °C for 3 h, an additional 1 µl aliquot of trypsin was added and digests were further incubated for 14–16 h at 37 °C. The resulting samples were reduced with 2 mM dithiothreitol for 30 min at 56 °C and then alkylated with 30 mM iodoacetamide for 30 min at room temperature in the dark. Reduced and alkylated digest samples were then absorbed to C18ZipTips (equilibrated according to the manufacturer’s instructions), washed with 10 µl of 5% acetonitrile, 0.1% trifluoroacetic acid, and eluted with 10 µl of 80% acetonitrile, 0.1% trifluoroacetic acid. Eluted samples were stored at −20 °C prior to mass analysis.
2.5 MALDI analysis and spectral interpretation
MALDI analysis was conducted on a Bruker BiFlexIII mass spectrometer in reflectron mode with positive ion spectra averaged over 100–600 laser shots [36]. For MALDI, digest fractions were diluted 2-fold with a saturated solution of α-CHC in 50% acetonitrile. Following a brief incubation, 1 µl of sample was spotted on a stainless steel MALDI plate and allowed to air dry. For these studies, the instrument was calibrated internally, using the singly protonated monoisotopic masses of angiotensin II and adrenocorticotropin hormone 18–36, that were added to the digests. Theoretical tryptic digests of Cyt b were generated using the program PeptideMass (http://us.expasy.org/cgi-bin/peptide-mass.pl.) with the addition of BS2G or BS3 conventional and deuterated adducts for comparison with the observed masses. The assignment of tryptic peptides was carried out manually, assuming up to 4 missed cleavage sites, given that crosslinking of two lysine residues will produce a minimum of two missed tryptic cleavages.
2.6. Molecular modeling
Prediction of the secondary structure element encompassing p22phox residues Lys71 and Lys78 was carried out by evaluating the entire p22phox primary structure (including the Tyr72/His72 polymorphism) with the program APSSP2 (http://imtech.res.in/raghava/apssp/), available at the ExPASy Proteomics Server. Visualization of the residues in a α-helix or β-strand was performed in Discover 2.0 (Accelrys, San Diego), with energy minimization at default parameter values for the energy minimization protocols.
3.0 Results
3.1 Modification of human neutrophil Cyt b with mass spectrometry compatible crosslinking agents
The current lack of a high-resolution structure for Cyt b has greatly hindered a mechanistic understanding of the control of superoxide production by the phagocyte NADPH oxidase complex. Cyt b is a relatively low abundance, large integral membrane protein with significant microheterogeneity [36;41], making it a challenging target for high resolution structure determination by x-ray crystallography or NMR spectroscopy. Crosslink localization by mass analysis of tryptic digests of crosslinked species provides an increasingly powerful approach for obtaining structural information about of integral membrane proteins or large protein complexes [22;25;26;31]. The current study describes the use of this approach for structure analysis of human neutrophil Cyt b.
To generate starting samples, Cyt b was isolated from human neutrophil membrane fractions using our recently-described mAb CS9 immunoaffinity matrix [40]. For modification, purified Cyt b was exchanged into PBS (pH-7.4), 0.1% DDM and then reacted with isotopic pairs of the crosslinkers BS2G-d0/d4 or BS3-d0/d4. As shown in Figure 1, crosslinking under these reaction conditions resulted in a relatively minor generation of higher molecular weight products, as judged by SDS-PAGE. In addition, MALDI analysis of tryptic digests indicated that the overall intensities of major peaks were largely unchanged in control (Figure 2A) and modified samples (Figures 2B and 2C). Similar results were obtained for all reaction conditions used in this study and demonstrate that undesirable excessive modification of the Cyt b surface did not occur. The relatively short reaction time and low pH used in modification reactions (pH 7.4) likely played an important role in limiting the extent of Cyt b surface modification, as primarily deprotonated Lys residues would react with the amine-reactive crosslinking agents.
Figure 1. Chemical modification of human neutrophil Cyt b with mass spectrometry compatible crosslinkers.
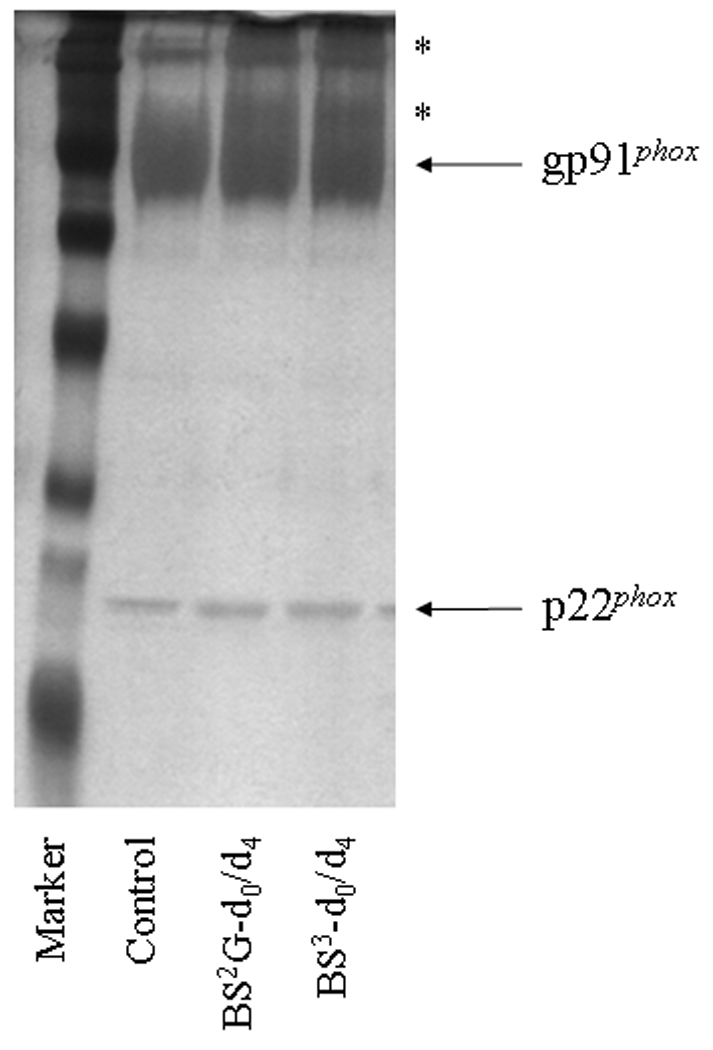
Following affinity purification of Cyt b from human neutrophil membrane fractions, chemical modification was carried out with homobifunctional, amine-reactive crosslinking agents BS2G-d0/d4 and BS3-d0/d4. In order to monitor crosslinking reactions, quenched samples were resolved by SDS-PAGE and silver stained as follows: M, molecular weight standards; Control, Cyt b following incubation with 5% DMSO for 50 min at room temperature (control sample); BS2G-d0/d4, Cyt b following incubation with a 60-fold molar excess of BS2G-d0/d4 for 50 min at room temperature; and BS3-d0/d4, Cyt b following incubation with a 60-fold molar excess of BS3-d0/d4 for 50 min at room temperature. Asterisks (*) indicate higher molecular weight species that were shown contain both gp91phox and p22phox subunits by MALDI mass spectrometry (data not shown). Qualitatively similar results were obtained for all reaction conditions used in the present study.
Figure 2. MALDI analysis of human neutrophil Cyt b following modification with mass spectrometry compatible crosslinkers.
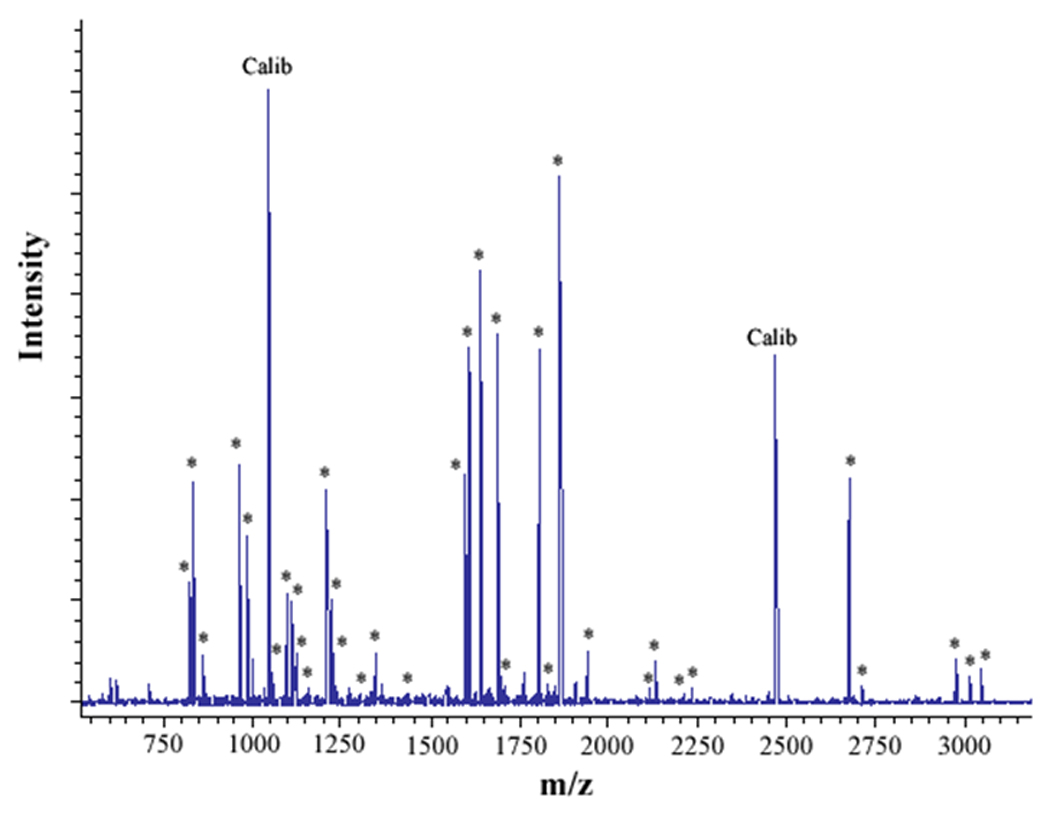
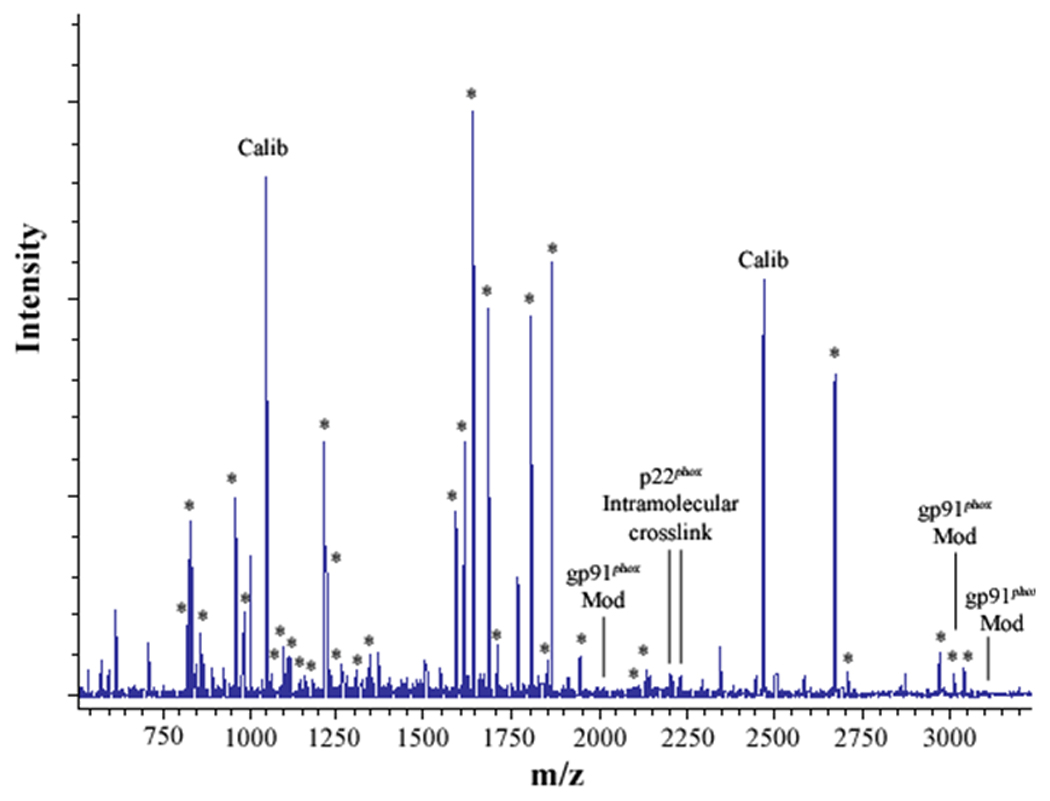
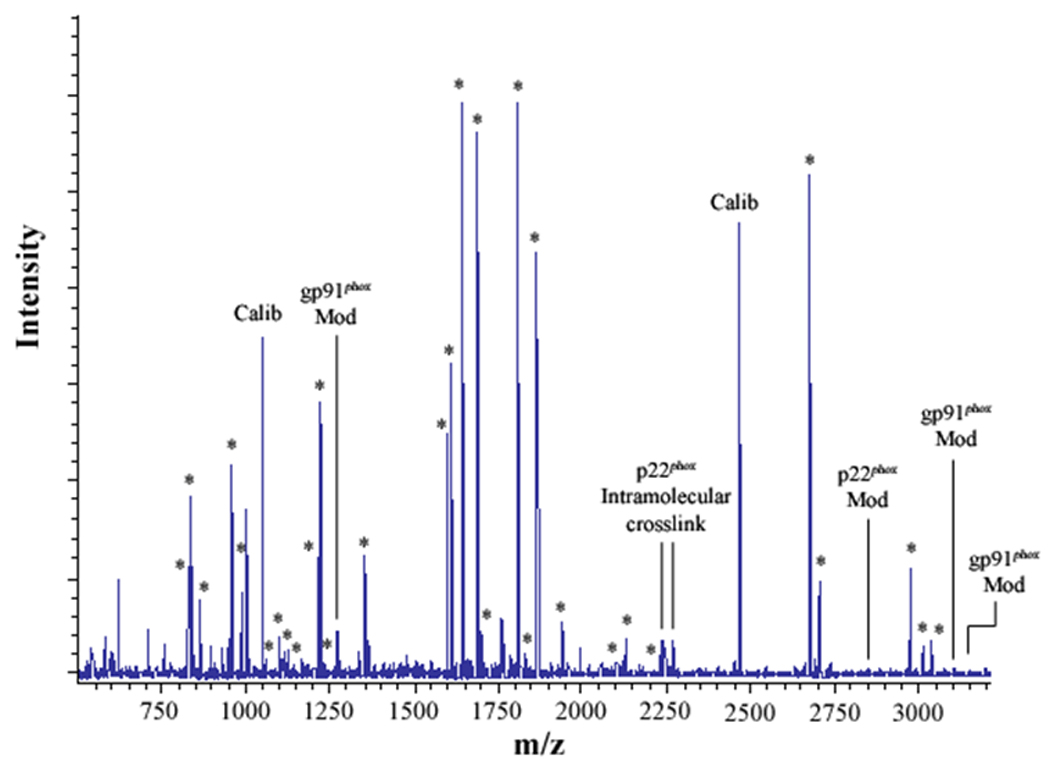
Following chemical modification as outlined in Figure 1, reactions were quenched and digested with Trypsin-gold. To prepare samples for analysis by MALDI mass spectrometry, reactions were reduced, alkylated and then purified using C18ZipTips. (2A) MALDI analysis of Cyt b tryptic digests following incubation with 5% DMSO for 50 min at room temperature (control sample). (2B) MALDI analysis of Cyt b tryptic digests following incubation with a 60-fold molar excess of BS2G-d0/d4 for 50 min at room temperature. (2C) MALDI analysis of Cyt b tryptic digests following incubation with a 60-fold molar excess of BS3-d0/d4 for 50 min at room temperature. Masses attributed to gp91phox and p22phox tryptic peptides are labeled with asterisks (Figure 2A–C), calibration peptides are labeled with text (Calib), while both modified peptides and an intramolecular p22phox crosslink are designated with text in Figures 2B and 2C. Qualitatively similar results were obtained for all reaction conditions used in the present study and indicate that the surface of Cyt b was not extensively modified.
3.2 Assignment of an intramolecular distance constraint in the Cyt b subunit p22phox
In the present study, mixtures of conventional and deuterated crosslinking agents were reacted with purified Cytb. Modified tryptic peptides could then be readily identified by: 1) the characteristic increase of 96.02 Da and 138.07 Da for the BS2G-d0 and BS3-d0 adducts, respectively, relative to unmodified Cyt b peptides; 2) a characteristic 4.02 Da mass spacing between Cyt b peptides modified with conventional (d0) and deuterated (d4) crosslinker; and 3) a characteristic 42.05 Da difference in the molecular weight of the agents BS2G and BS3. The MALDI spectra in Figures 2A–C indicated that unique masses were readily observed in the crosslinked samples, demonstrating the anticipated signatures of the pairs of crosslinking agents in tryptic digests derived from the BS2G-d0/d4 and BS3-d0/d4 reaction mixtures. Higher resolution analyses in Figure 3 show overlaid MALDI spectra from Cyt b control samples (red spectra) and the BS2G-d0/d4 or BS3-d0/d4 reaction mixtures (blue spectra). In Figure 3A, the unique 4 Da spacing of peaks in the BS2G-d0/d4 sample provides a diagnostic signature for Cyt b tryptic peptides that have been modified by crosslinking agent. Importantly, in the BS2G-d0/d4 spectra, the masses at 2199.09 Da and 2203.13 Da could be assigned to p22phox residues 68–85 assuming: 1) 2 missed trypsin cleavage sites (expected for the presence of two crosslinked lysines in the peptide); 2) a crosslink with BS2G-d0/d4; and 3) the naturally occurring His72 polymorphism [41;42]. Confidence in this spectral assignment is greatly enhanced by the masses at 2225.12 Da and 2229.13 Da, that could be assigned to p22phox residues 68–85 assuming: 1) 2 missed trypsin cleavage sites; 2) a crosslink with BS2G-d0/d4; and 3) the naturally occurring Tyr72 polymorphism [41;42]. In the above spectra, mass accuracies were greater than 30 ppm due to the internal mass calibration, which increased the confidence in the peptide assignments.
Figure 3. Identification of an intramolecular crosslink in the p22phox subunit of Cyt b.
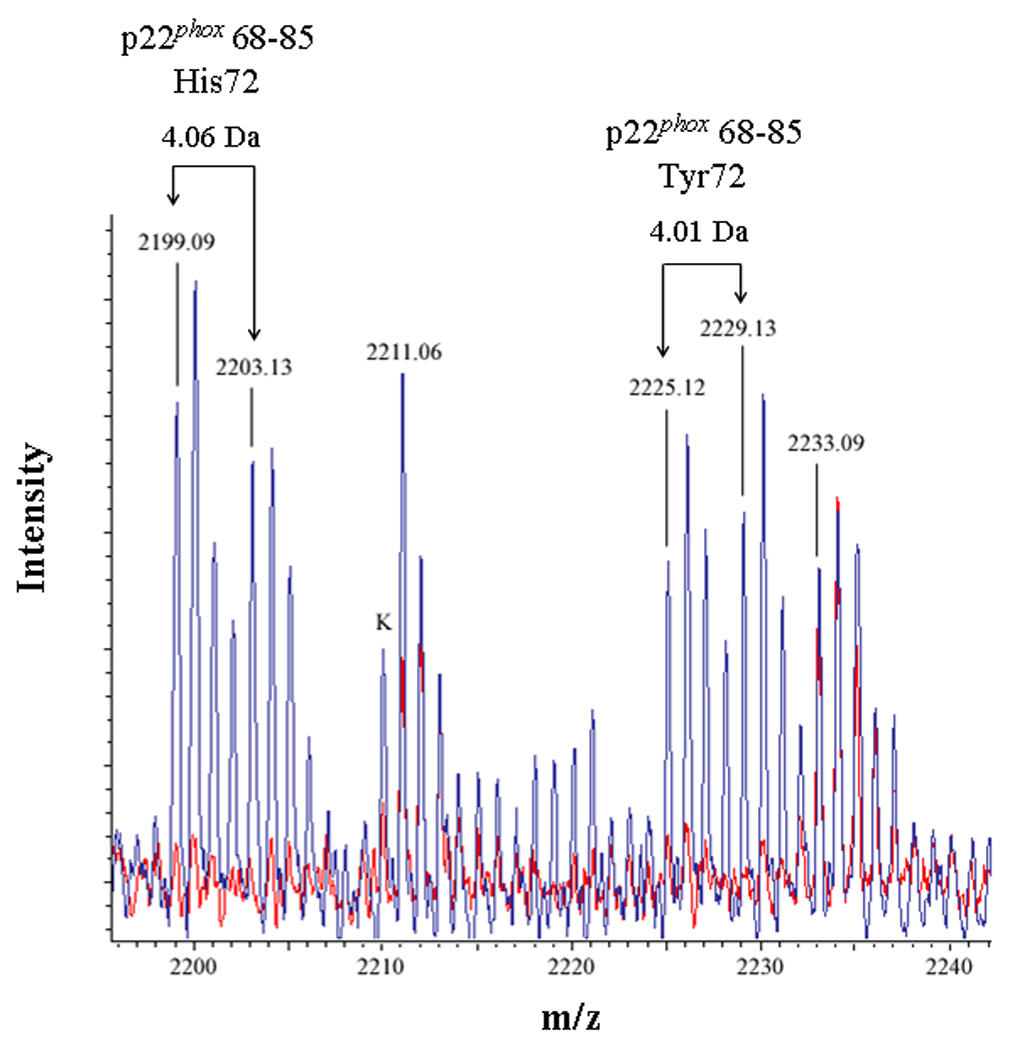
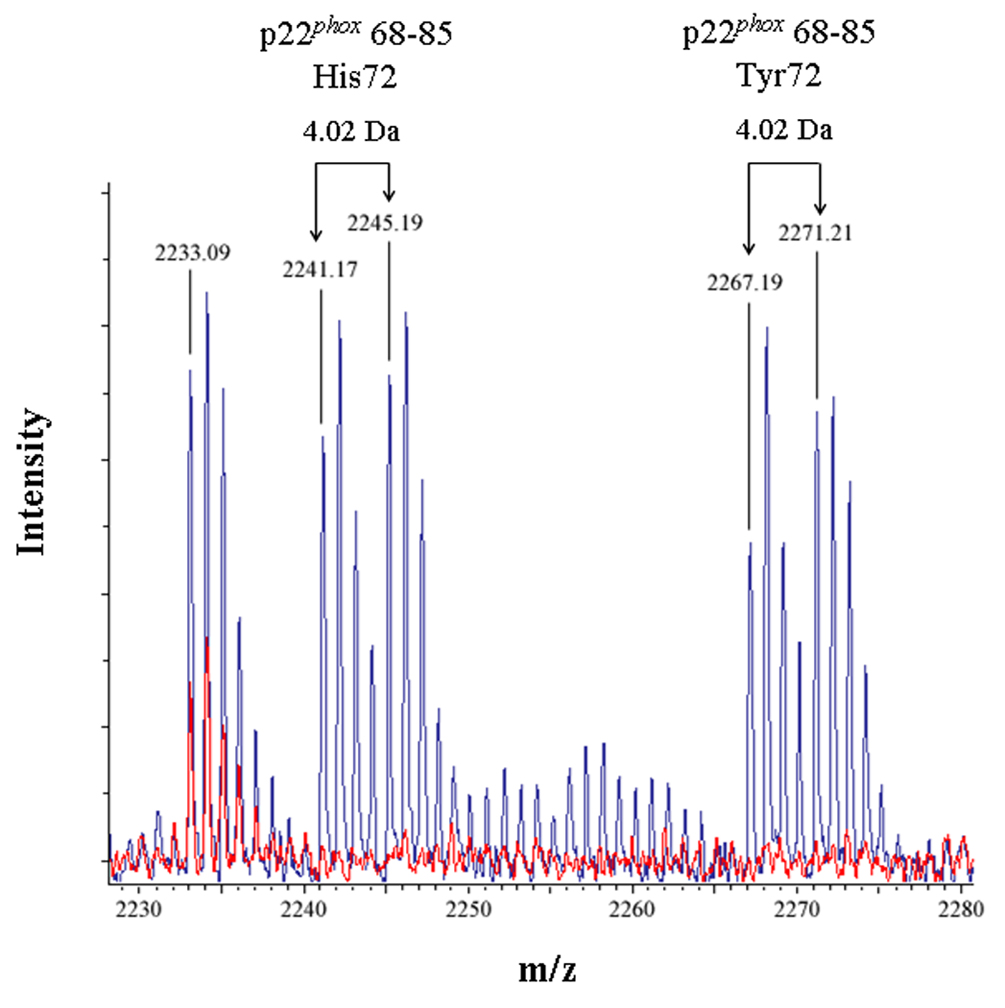
(3A) Overlaid MALDI spectra for Cyt b tryptic digests following incubation with 5% DMSO for 50 min at room temperature (red) and Cyt b tryptic digests following incubation with a 60-fold molar excess of BS2G-d0/d4 for 50 min at room temperature (blue). (3B) Overlaid MALDI spectra for Cyt b tryptic digests following incubation with 5% DMSO for 50 min at room temperature (red) and Cyt b tryptic digests following incubation with a 60-fold molar excess of BS3-d0/d4 for 50 min at room temperature (blue). Masses attributed to gp91phox and p22phox tryptic peptides are labeled numerically in 3A and 3B, and a possible keratin-derived tryptic peptide is indicated (K) in 3B. The initial mass corresponding to the do and d4 peak families in the p22phox crosslink is indicated by arrows. Theoretical masses of p22phox residues 68–85 modified with crosslinking agent are as follows: BS2G-d0 with His72-2199.15 Da; BS2G-d4 with His72-2203.17 Da; BS2G-d0 with Tyr72-2225.15 Da; BS2G-d4 with Tyr72-2229.18 Da; BS3-d0 with His72-2241.20 Da; BS3-d4 with His72-2245.22 Da; BS3-d0 with Tyr72-2267.20 Da; and BS3-d4 with Tyr72-2271.22 Da.
To confirm the assignment of MALDI spectra outlined above, purified Cyt b was also modified with the crosslinking mixture BS3-d0/d4, which has a 42.05 Da higher molecular weight than the BS2G crosslinkers. Figure 3B shows overlaid MALDI spectra from Cyt b control samples (red spectra) and the BS3-d0/d4 reaction mixture (blue spectra), where the masses at 2241.17 Da and 2245.19 Da could be assigned to p22phox residues 68–85 assuming: 1) 2 missed trypsin cleave sites; 2) a crosslink with BS3-d0/d4; and 3) the naturally occurring His72 polymorphism. Similar to the situation with shown above with BS2G-d0/d4, confidence in this spectral assignment is enhanced by the masses at 2267.19 Da and 2271.21 Da, that could be assigned to p22phox residues 68–85 assuming: 1) 2 missed trypsin cleavage sites; 2) a crosslink with BS2G-d0/d4; and 3) the naturally occurring Tyr72 polymorphism. In the above spectra, mass accuracies were greater than 20 ppm, due to internal mass calibration and also enhanced confidence in the assignments.
4.0 Discussion
4.1 Flavocytochrome b light chain: structure/function relationships
The flavocytochrome b light chain, p22phox, remains the least understood subunit of this human neutrophil integral membrane electron transferase [43], however recent studies are improving our knowledge. In our earliest studies on the protein, we showed that it was intimately linked to gp91phox in that it copurified with gp91phox and was easily crosslinked to gp91phox with either disuccinimidyl suberate or bis[2-(succinimidooxycarbonyloxy)ethyl]sulfone[44]. Subsequently we, in collaboration with Dinauer and coworkers, helped to identify it's gene sequence [45] and then showed that we could co-immunopurify gp91phox in complex with the small G-protein, rap1A, on affinity purified rabbit anti-p22phox peptide antibodies [15]. We were able to confirm and improve the immunopurification with the mAbs 44.1 and CS9 [40;46] while also proving the bis heme stoichiometry of the protein. Our interest peaked in this subunit when we were able to show that native heme colocalized with both subunits on LDS-PAGE separations at low temperature [47], a method previously used to identify heme-localization in cytochrome b6f of thylakoid membranes [48]. We proposed from that study that His 94 of p22phox may help coordinate one of the hemes of the bis heme molecule, and thus might have important consequences for its regulation. This conclusion became less likely when it was shown that gp91phox had been transiently expressed in COS cells without p22phox, while still coordinating hemes of differing electrochemical potential [49] and when it was also shown that the p22phox mutations, H94L/Y/M, did not measurably affect superoxide production[38;50]. However, these findings derived from expression in heterologous cells and did not address the potential role of the subunit's His94 as a negative regulator of flavocytochrome b activity.
Detailed structural characterization of part of p22phox was carried out by the Rittinger group [51], when they used a synthetic p22phox peptide, 149KQPPSNPPPRPPAEARKK166 and co-crystallized it with the p47 subunit of the NADPH oxidase. This achievement allowed them to obtain the first physiologically relevant X-ray crystal structure of the p47-bound stretch of p22phox 151PPSNPPPRPP160, suggesting it has an extended conformation, at least when bound in the oxidase complex. Burritt et al [52] contributed structural information about p22phox using a TrNOESY NMR analysis of the bound structure of a peptide recognized by the p22phox -specific mAb, 44.1, and concluded that it recognized two widely spaced regions of the p22phox sequence 29TAGRF33 and 183PQVNPI188. The composite peptide ATAGRFGGGPQVNPI was shown to bind 3 orders of magnitude more tightly to mAb 44.1 than either of its component sequences, with 2.5–3.5 Å proximity of atoms in 33F-185 V, 29T-32 R, 29T-33F, 30A-32R residue pairs established by the TrNOESY NMR ruler. Phage display of random peptide libraries also showed a preponderance of the consensus peptide PQVRPI which demonstrated the same inhibitory potential as the above mentioned composite peptide. These results necessarily implied that both the amino and carboxyl terminus of p22phox were intracellular and close to one another, as cells had to be permeabilized in order for mAb 44.1 to recognize these regions of p22phox [53]
p22phox is an essential component of flavocytochrome b, acting as an important regulator of structure and function [1] Maturation of flavocytochrome b is dependent on the fidelity of the p22phox sequence, as certain p22phox -specific missense and nonsense mutations in the human population result in A22 Chronic Granulomatous Disease [41;54]. A number of important polymorphisms, especially at positions Y72H and other sites [16], suggest that these residues have a wider role for the coupling of this subunit with other NADPH oxidase gp91phox homologs (NOX family of NADPH oxidases [6]) and thus may be responsible for other diseases [8].
Much has been learned by the molecular genetic manipulation of p22phox in heterologous systems. Deletion of the amino terminal residues 1MGQIEWAMWAN11, 67RWGQKH72, and 66ERWGQKHMTAVVKLFGPFTRNYYVR90 prevent Cytb maturation [55], as do lysine substitutions within 98SVPA101 [35]. Interestingly, the G102K mutation [35] supports assembly and translocation of both subunits to the plasma membrane in heterologous CHO-91-47-67 cells, but puts into question the two transmembrane domain folding model for p22phox [18]. The alternative four transmembrane domain prediction for the p22phox folding model [35] (http://www.ezim.hu/hmmtop/server/hmmtop.cgi), however, is incompatible with the binding and functional effects of mAb NS5 (see below). Another four transmembrane domain model suggested by Pick and colleagues [56] is reflected in the inhibition of NADPH oxidase activity and cytosolic factor binding observed in p22phox peptide inhibition studies [56;57], but is also incompatible with mAb 44.1 epitope placement. Other monoclonal antibodies directed against the p22phox subunit [18] confirmed the intracellular and protein surface localization of regions 78KLFGPF83 (mAb NS5), 185VNPIP189 (mAb CS6) 181GGPQVNPI188 (mAb CS8) and 162EARKKPSE169 (mAb CS9). Furthermore, the suggestion that the 29TAGRF33--183PQVNPI188 complex epitope of mAb 44.1 appears to be close enough to 162EARKKPSE169 so that their respective antibodies (e.g. CS9) will inhibit the other's binding (e.g. 44.1). Recent mutational studies [35] have suggested the 29TAGRFTQ34 to 29TAGKKKQ34 substitution and 171C truncation mutations are still recognized by mAb44.1, confirming separate recognition of each component of the complex mAb44.1epitope.
More protein surface topology information was derived from other mAb mapping studies. We interpreted the lack of recognition and functional effects of NS2 [18] that recognizes the 131WTPIEPKPR139, as evidence that this epitope was inaccessible to Ab recognition, however recently Paclet and colleagues [58] showed that mAbs 13D4 and 16G7 could recognize the same epitope region as NS2 in immunoblots, but were also able to bind permeabilized neutrophils and to immunoprecipitate the detergent solubilized protein. These latter two mAbs did not appreciably inhibit reconstituted NADPH oxidase activity, clearly suggesting that the 130QWTPIEPK137 region is cytosolic and on the protein surface but not directly involved in function. These investigators also found that mAb 12E6 was weakly positive for p22phox in immunoblots and mapped to the proline rich region 155PPPRPP160. mAb 12E6 inhibited oxidase activity and bound much more strongly to in situ flavocytochrome b in permeabilized stimulated cells compared to unstimulated cells, as well binding more strongly in detergent extracts from stimulated cells. One intriguing outcome of these experiments was the finding that part of the phage peptide library consensus sequence mapped to 557GPR559 of gp91phox, suggesting that if this 3-residue assignment was correct, this conformationally-sensitive mAb might span the interface between both subunits in the stimulated conformation.
4.2 Implications of mass analysis on p22phox structure
In the present study we add another distance constraint, obtained by mass spectrometry and crosslinking analysis, to our understanding of p22phox structure. We used a combination of chemical modification and reflectron MALDI mass spectrometry to confidently identify a tryptic peptide derived from the Cyt b subunit p22phox (residues 68–85) that contained an intramolecular crosslink with two different isotopically-paired crosslinking agents BS2G-d0/d4 and BS3-d0/d4. Confident assignments of the derived MALDI spectra were facilitated by a number of factors including: 1) the extremely high purity of human neutrophil Cyt b [40]; 2) chemical modification reactions using mixtures of conventional and isotopically-labeled crosslinking agents; 3) the use of two distinct crosslinking agents of different molecular weight (BS2G-d0 adds 96.02 Da and BS3-d0 adds 138.07 Da) but similar crosslinking distances (see below); 4) 30 ppm mass accuracy due to internal calibration of the reflectron MALDI; and 5) a naturally occurring polymorphism in the p22phox segment that was identified. In addition, mass analysis was carried out on samples that lacked extensive surface modification by the crosslinking agents, due to the mild crosslinking conditions employed (pH-7.4, relatively short reaction times and relatively low crosslinker:Cyt b molar ratios). A lack of extensive surface modification represented an important consideration for both preserving native protein structure and limiting the complexity of the tryptic peptide digests to be analyzed.
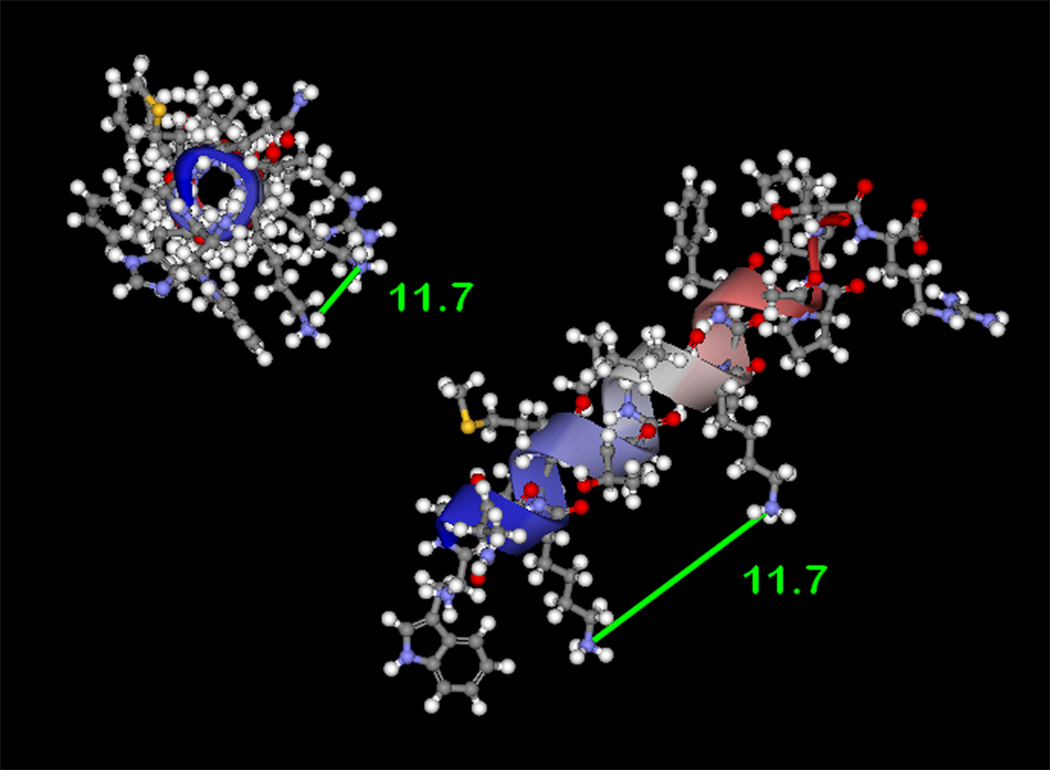
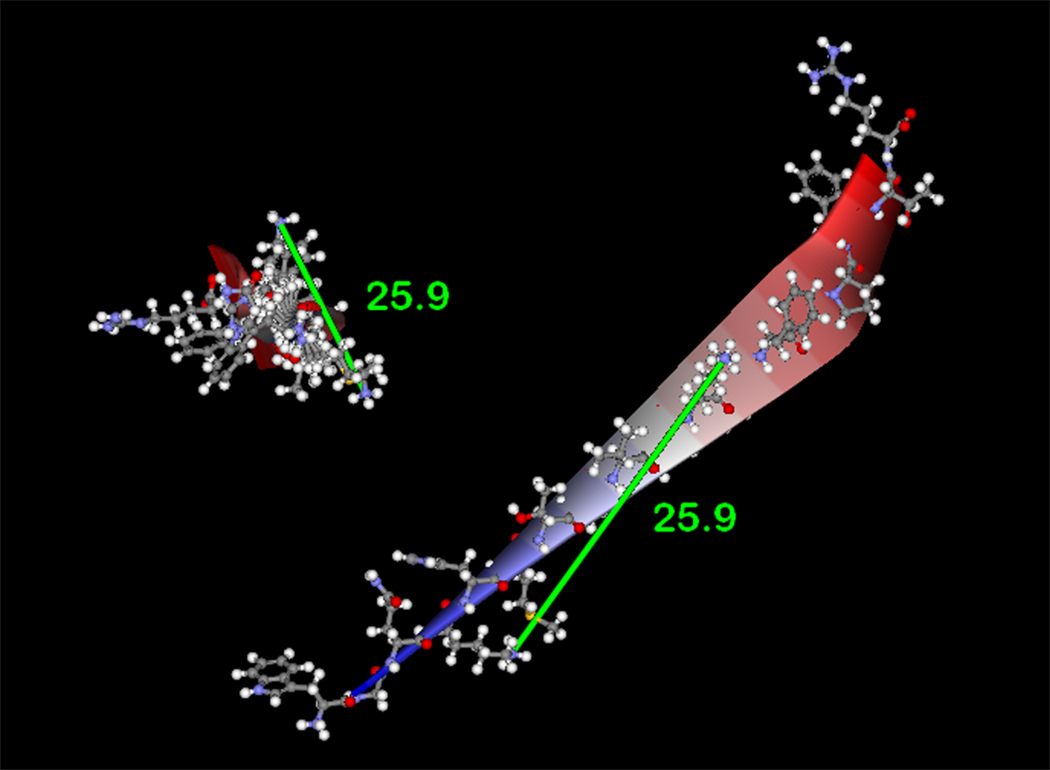
Since homobifunctional, amine-reactive crosslinking agents were used in this study, MALDI assignments identifying the formation of intramolecular crosslinks in p22phox residues 68–85 provides a novel, molecular distance constraint between Lys71 and Lys78 of p22phox. To facilitate interpretation of the above results, the program Discover 2.0 was used to generate models of p22phox residues 68–85 as canonical α-helix and β-sheet secondary structure elements. As shown in Figure 4A, the epsilon amino groups of Lys 71 and Lys78 are on the same side of the model α-helix and occur 11.7 Å apart, if the side chains are extended perpendicular to the helix axis. This proposed structure motif is consistent with the molecular crosslinking distance expected for both BS2G (7.7 Å) and BS3 (11.4 Å), allowing for some flexibility in the conformation of the Lys side chains [59]. Support for the above model was obtained using the secondary structure prediction program APSSP2 (http://www.imtech.res.in/raghava/apssp2/), which predicts an α-helical secondary structure for the entire segment of p22phox encompassing Lys71 and Lys78. While the above results cannot rule out a flexible loop structure for p22phox residues 68–85, the structure model in Figure 4B indicates that a canonical β-sheet structure for this region is unlikely, since that would produce a 25.9 Å spacing between Lys71 and Lys78. Even allowing for the maximum flexibility of the lysine side chains, the BS3 crosslinker could not accomplish a crosslink between Lys71 and Lys78 with the β-sheet structure for this region, as the distance from α-carbon to ε-amino nitrogen, for an extended lysine residue, is 6.3Å.
Figure 4. Computational modeling of p22phox residues identified by crosslinking and mass spectrometry.
(4A) Model of p22phox residues 68–85 as an α-helix showing a stacked configuration of Lys71 and Lys78, with a minimized distance between the reactive amines of 11.7 Å. (4B) Model of p22phox residues 68–85 as a β-strand, showing an elongated distance of 25.9 Å between the reactive amine groups of Lys71 and Lys78. In both 4A and 4B, the left structures represent side views of the structure model, while the right structures represent the amino to carboxy-terminal end views. Colorized solid ribbons showing secondary structure are blue and red at the amino and carboxy-terminal ends, respectively, with graded color proceeding in that direction.
4.3 Significance
The results reported here demonstrate the potential of crosslinking and mass spectrometry as a tool for augmenting the structural analysis of human phagocyte Cyt b. In addition to this structural information, the mass analyses described provides the first demonstration of the naturally-occurring Tyr72/His72 polymorphism, expressed at the protein level in the human population (information that was previously hypothesized from nucleotide sequence analysis) [41;42]. The Tyr72/His72 p22phox polymorphism appears to play a relevant role in human physiology, as individuals homozygous for Tyr72 appear to have diminished superoxide production by the NADPH oxidase and may be less susceptible to adverse cardiovascular events [37;38]. Our results also suggest that the local conformation in the region bearing this polymorphism must be relatively invariant, having minimal effect on the crosslinking and thus implying that functional differences between the polymorphic forms of Cytb may result from the amino acid residues directly in contact with this position. Such interactions could result from intramolecular or inter-subunit interactions. Position 72 is close to but not within regions shown to mediate interaction with p47phox or p67phox [56;57] and thus might be more important for maturation of Cytb [55]. Thus although the polymorphism might have second order effects in the function of the protein, it may have larger maturational effects in the context of haplotypes such as within the haplogroup C1 that also contains p. V 174A (nucleotide designation c.521T>C) and c.*24G>A (in the 3’untranslated region) as outlined by Bedard [16].
In light of such findings, it will be of interest to better define structural consequences of these amino acid substitutions on the structure and function of Cyt b[35]. Additional low-resolution distance constraints provided by crosslinking and mass spectrometry could greatly facilitate modeling the structure of the NADPH oxidase complex and provide important independent cross checks for larger structural analysis emerging from molecular modeling of X-ray crystal solutions to the structure of flavocytochrome b. Therefore current efforts are focused on the identification of additional intramolecular crosslinks, intermolecular crosslinks between the Cyt b subunits, and intermolecular crosslinks between Cyt b and the cytosolic oxidase subunits.
5.0 Conclusions
-
5.1
Mass analysis of intramolecular crosslinking, using two different crosslinking reagents, suggest that the epsilon amino groups of Lys71 and Lys 78 of p22phox are approximately 7 to 12 Å apart.
-
5.2
The crosslinkers BS2G-d0/d4 and BS3-d0/d4, are good candidates for intra- and inter-molecular crosslinking mass analyses for other integral membrane proteins as well as flavocytochrome b.
-
5.3
The local conformation of p22phox encompassing amino acid residue 72 is invariant for the p.Y72H polymorphisms
-
5.4
Our results support functional differences in ROS generation for certain haplogroups arising from differences in p22phox expression.
Acknowledgments
Funding
This work was supported by American Heart Association Scientist Development Grants 0630253N (to R. M. T.) and National Institutes of Health (NIH) Grants [5R01AI026711-21] and [3R01AI026711-21S1] (to A. J. J, and [R01 AI 64107], [R01 GM 62547] (to E. A. D). The Montana State University Mass Spectrometry Facility is supported by National Science Foundation Grant [MRI 0321267], the Murdock Charitable Trust, and the NIH CoBRE Grant [1P20RR024237] (to E.A.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations 1: flavocytochrome b, Cytb; α-cyano-4-hydroxycinnamic acid (α-CHC), 2,5-dihydroxybenzoic acid, DHB; dodecylmaltoside, DDM, chronic granulomatous disease, CGD
References
- 1.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 2.Nauseef WM. Assembly of the phagocyte NADPH oxidase. Histochem. Cell Biol. 2004;122:277–291. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 3.Dinauer MC. Chronic granulomatous disease and other disorders of phagocyte function. Hematology. Am. Soc. Hematol. Educ. Program. 2005:89–95. doi: 10.1182/asheducation-2005.1.89. [DOI] [PubMed] [Google Scholar]
- 4.Weiss SJ. Tissue destruction by neutrophils. N. Engl. J. Med. 1989;320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 5.Babior BM. Phagocytes and oxidative stress. Am. J. Med. 2000;109:33–44. doi: 10.1016/s0002-9343(00)00481-2. [DOI] [PubMed] [Google Scholar]
- 6.Aguirre J, Lambeth JD. Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic. Biol. Med. 2010;49:1342–1353. doi: 10.1016/j.freeradbiomed.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nauseef WM. Biological roles for the NOX family NADPH oxidases. J. Biol. Chem. 2008;%20(283):16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 9.Matute JD, Arias AA, Wright NA, Wrobel I, Waterhouse CC, Li XJ, Marchal CC, Stull ND, Lewis DB, Steele M, Kellner JD, Yu W, Meroueh SO, Nauseef WM, Dinauer MC. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schappi MG, Smith VV, Goldblatt D, Lindley KJ, Milla PJ. Colitis in chronic granulomatous disease. Arch. Dis. Child. 2001;84:147–151. doi: 10.1136/adc.84.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano Y, Longo-Guess CM, Bergstrom DE, Nauseef WM, Jones SM, Banfi B. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J. Clin. Invest. 2008;118:1176–1185. doi: 10.1172/JCI33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griendling KK, Sorescu D, Ushio-Fukai M. NAD(P)H oxidase: role in cardiovascular biology and disease. Circ. Res. 2000;86:494–501. doi: 10.1161/01.res.86.5.494. [DOI] [PubMed] [Google Scholar]
- 13.Lassegue B, Griendling KK. NADPH oxidases: functions and pathologies in the vasculature. Arterioscler. Thromb. Vasc. Biol. 2010;30:653–661. doi: 10.1161/ATVBAHA.108.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLeo FR, Quinn MT. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J. Leukoc. Biol. 1996;60:677–691. doi: 10.1002/jlb.60.6.677. [DOI] [PubMed] [Google Scholar]
- 15.Quinn MT, Parkos CA, Walker L, Orkin SH, Dinauer MC, Jesaitis AJ. Association of a Ras-related protein with cytochrome b of human neutrophils. Nature. 1989;342:198–200. doi: 10.1038/342198a0. [DOI] [PubMed] [Google Scholar]
- 16.Bedard K, Attar H, Bonnefont J, Jaquet V, Borel C, Plastre O, Stasia MJ, Antonarakis SE, Krause KH. Three common polymorphisms in the CYBA gene form a haplotype associated with decreased ROS generation. Hum. Mutat. 2009;30:1123–1133. doi: 10.1002/humu.21029. [DOI] [PubMed] [Google Scholar]
- 17.Groemping Y, Rittinger K. Activation and assembly of the NADPH oxidase: a structural perspective. Biochem. J. 2005;386:401–416. doi: 10.1042/BJ20041835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor RM, Burritt JB, Baniulis D, Foubert TR, Lord CI, Dinauer MC, Parkos CA, Jesaitis AJ. Site-specific inhibitors of NADPH oxidase activity and structural probes of flavocytochrome b: characterization of six monoclonal antibodies to the p22phox subunit. J. Immunol. 2004;173:7349–7357. doi: 10.4049/jimmunol.173.12.7349. [DOI] [PubMed] [Google Scholar]
- 19.Taylor RM, Lord CI, Riesselman MH, Gripentrog JM, Leto TL, McPhail LC, Berdichevsky Y, Pick E, Jesaitis AJ. Characterization of surface structure and p47phox SH3 domain-mediated conformational changes for human neutrophil flavocytochrome b. Biochemistry. 2007;46:14291–14304. doi: 10.1021/bi701626p. [DOI] [PubMed] [Google Scholar]
- 20.Taylor WR, Jones DT, Segal AW. A structural model for the nucleotide binding domains of the flavocytochrome b-245 beta-chain. Protein Sci. 1993;2:1675–1685. doi: 10.1002/pro.5560021013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters K, Richards FM. Chemical cross-linking: reagents and problems in studies of membrane structure. Annu. Rev. Biochem. 1977;46:523–551. doi: 10.1146/annurev.bi.46.070177.002515. 523–551. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen RB, Sale KL, Ayson MJ, Novak P, Hong J, Lane P, Wood NL, Kruppa GH, Young MM, Schoeniger JS. Structure and dynamics of dark-state bovine rhodopsin revealed by chemical cross-linking and high-resolution mass spectrometry. Protein Sci. 2006;15:1303–1317. doi: 10.1110/ps.052040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young JL. Mass Spectrometry-based Cross-linking Sites Mapping for Structural Elucidation of Proteins and Protein Complexes”. Molecular BioSystems. 2008;4:816–823. doi: 10.1039/b801810c. [DOI] [PubMed] [Google Scholar]
- 24.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 25.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 26.Singh P, Panchaud A, Goodlett DR. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal. Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 27.Pearson KM, Pannell LK, Fales HM. Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 2002;16:149–159. doi: 10.1002/rcm.554. [DOI] [PubMed] [Google Scholar]
- 28.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome. Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 29.Back JW, Sanz MA, De JL, De Koning LJ, Nijtmans LG, De Koster CG, Grivell LA, Van Der SH, Muijsers AO. A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 2002;11:2471–2478. doi: 10.1110/ps.0212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 31.Weerasekera R, She YM, Markham KA, Bai Y, Opalka N, Orlicky S, Sicheri F, Kislinger T, Schmitt-Ulms G. Interactome and interface protocol (2IP): a novel strategy for high sensitivity topology mapping of protein complexes. Proteomics. 2007;7:3835–3852. doi: 10.1002/pmic.200700688. [DOI] [PubMed] [Google Scholar]
- 32.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 33.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 34.Burritt JB, Foubert TR, Baniulis D, Lord CI, Taylor RM, Mills JS, Baughan TD, Roos D, Parkos CA, Jesaitis AJ. Functional epitope on human neutrophil flavocytochrome b558. J. Immunol. 2003;170:6082–6089. doi: 10.4049/jimmunol.170.12.6082. [DOI] [PubMed] [Google Scholar]
- 35.von Lohneysen K, Noack D, Jesaitis AJ, Dinauer MC, Knaus UG. Mutational analysis reveals distinct features of the Nox4-p22 phox complex. J. Biol. Chem. 2008;283:35273–35282. doi: 10.1074/jbc.M804200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor RM, Baniulis D, Burritt JB, Gripentrog JM, Lord CI, Riesselman MH, Maaty WS, Bothner BP, Angel TE, Dratz EA, Linton GF, Malech HL, Jesaitis AJ. Analysis of human phagocyte flavocytochrome b(558) by mass spectrometry. J. Biol. Chem. 2006;281:37045–37056. doi: 10.1074/jbc.M607354200. [DOI] [PubMed] [Google Scholar]
- 37.Arca M, Conti B, Montali A, Pignatelli P, Campagna F, Barilla F, Tanzilli G, Verna R, Vestri A, Gaudio C, Violi F. C242T polymorphism of NADPH oxidase p22phox and recurrence of cardiovascular events in coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2008;28:752–757. doi: 10.1161/ATVBAHA.107.154823. [DOI] [PubMed] [Google Scholar]
- 38.Wyche KE, Wang SS, Griendling KK, Dikalov SI, Austin H, Rao S, Fink B, Harrison DG, Zafari AM. C242T CYBA polymorphism of the NADPH oxidase is associated with reduced respiratory burst in human neutrophils. Hypertension. 2004;43:1246–1251. doi: 10.1161/01.HYP.0000126579.50711.62. [DOI] [PubMed] [Google Scholar]
- 39.Parkos CA, Allen RA, Cochrane CG, Jesaitis AJ. Purified cytochrome b from human granulocyte plasma membrane is comprised of two polypeptides with relative molecular weights of 91,000 and 22,000. J. Clin. Invest. 1987;80:732–742. doi: 10.1172/JCI113128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lord CI, Riesselman MH, Gripentrog JM, Burritt JB, Jesaitis AJ, Taylor RM. Single-step immunoaffinity purification and functional reconstitution of human phagocyte flavocytochrome b. J. Immunol. Methods. 2008;329:201–207. doi: 10.1016/j.jim.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Rae J, Noack D, Heyworth PG, Ellis BA, Curnutte JT, Cross AR. Molecular analysis of 9 new families with chronic granulomatous disease caused by mutations in CYBA, the gene encoding p22(phox) Blood. 2000;96:1106–1112. [PubMed] [Google Scholar]
- 42.Dinauer MC, Pierce EA, Bruns GA, Curnutte JT, Orkin SH. Human neutrophil cytochrome b light chain (p22-phox). Gene structure, chromosomal location, and mutations in cytochrome-negative autosomal recessive chronic granulomatous disease. J. Clin. Invest. 1990;86:1729–1737. doi: 10.1172/JCI114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol. Life Sci. 2002;59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkos CA, Allen RA, Cochrane CG, Jesaitis AJ. The quaternary structure of the plasma membrane b-type cytochrome of human granulocytes. Biochim. Biophys. Acta. 1988;932:71–83. doi: 10.1016/0005-2728(88)90140-5. [DOI] [PubMed] [Google Scholar]
- 45.Parkos CA, Dinauer MC, Walker LE, Allen RA, Jesaitis AJ, Orkin SH. Primary structure and unique expression of the 22-kilodalton light chain of human neutrophil cytochrome b. Proc. Natl. Acad. Sci. U. S. A. 1988;85:3319–3323. doi: 10.1073/pnas.85.10.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor RM, Burritt JB, Foubert TR, Snodgrass MA, Stone KC, Baniulis D, Gripentrog JM, Lord C, Jesaitis AJ. Single-step immunoaffinity purification and characterization of dodecylmaltoside-solubilized human neutrophil flavocytochrome b. Biochim. Biophys. Acta. 2003;1612:65–75. doi: 10.1016/s0005-2736(03)00086-5. [DOI] [PubMed] [Google Scholar]
- 47.Quinn MT, Mullen ML, Jesaitis AJ. Human neutrophil cytochrome b contains multiple hemes. Evidence for heme associated with both subunits. J. Biol. Chem. 1992;267:7303–7309. [PubMed] [Google Scholar]
- 48.Widger WR, Cramer WA, Hermodson M, Meyer D, Gullifor M. Purification and partial amino acid sequence of the chloroplast cytochrome b-559. J. Biol. Chem. 1984;259:3870–3876. [PubMed] [Google Scholar]
- 49.Yu L, Quinn MT, Cross AR, Dinauer MC. Gp91(phox) is the heme binding subunit of the superoxide-generating NADPH oxidase. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7993–7998. doi: 10.1073/pnas.95.14.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biberstine-Kinkade KJ, Yu L, Stull N, LeRoy B, Bennett S, Cross A, Dinauer MC. Mutagenesis of p22(phox) histidine 94. A histidine in this position is not required for flavocytochrome b558 function. J. Biol. Chem. 2002;277:30368–30374. doi: 10.1074/jbc.M203993200. [DOI] [PubMed] [Google Scholar]
- 51.Groemping Y, Lapouge K, Smerdon SJ, Rittinger K. Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell. 2003;113:343–355. doi: 10.1016/s0092-8674(03)00314-3. [DOI] [PubMed] [Google Scholar]
- 52.Burritt JB, Busse SC, Gizachew D, Siemsen DW, Quinn MT, Bond CW, Dratz EA, Jesaitis AJ. Antibody imprint of a membrane protein surface. Phagocyte flavocytochrome b. J. Biol. Chem. 1998;273:24847–24852. doi: 10.1074/jbc.273.38.24847. [DOI] [PubMed] [Google Scholar]
- 53.Burritt JB, Quinn MT, Jutila MA, Bond CW, Jesaitis AJ. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J. Biol. Chem. 1995;270:16974–16980. doi: 10.1074/jbc.270.28.16974. [DOI] [PubMed] [Google Scholar]
- 54.Roos D, de BM, Kuribayashi F, Meischl C, Weening RS, Segal AW, Ahlin A, Nemet K, Hossle JP, Bernatowska-Matuszkiewicz E, Middleton-Price H. Mutations in the X-linked and autosomal recessive forms of chronic granulomatous disease. Blood. 1996;87:1663–1681. [PubMed] [Google Scholar]
- 55.Zhu Y, Marchal CC, Casbon AJ, Stull N, von LK, Knaus UG, Jesaitis AJ, McCormick S, Nauseef WM, Dinauer MC. Deletion mutagenesis of p22phox subunit of flavocytochrome b558: identification of regions critical for gp91phox maturation and NADPH oxidase activity. J. Biol. Chem. 2006;281:30336–30346. doi: 10.1074/jbc.M607191200. [DOI] [PubMed] [Google Scholar]
- 56.Dahan I, Issaeva I, Gorzalczany Y, Sigal N, Hirshberg M, Pick E. Mapping of functional domains in the p22(phox) subunit of flavocytochrome b(559) participating in the assembly of the NADPH oxidase complex by "peptide walking". J. Biol. Chem. 2002;277:8421–8432. doi: 10.1074/jbc.M109778200. [DOI] [PubMed] [Google Scholar]
- 57.El-Benna J, Dang PM, Perianin A. Peptide-based inhibitors of the phagocyte NADPH oxidase. Biochem. Pharmacol. 2010;80:778–785. doi: 10.1016/j.bcp.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 58.Campion Y, Jesaitis AJ, Nguyen MV, Grichine A, Herenger Y, Baillet A, Berthier S, Morel F, Paclet MH. New p22-phox monoclonal antibodies: identification of a conformational probe for cytochrome b 558. J. Innate. Immun. 2009;1:556–569. doi: 10.1159/000231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DM, Sinz A. Isotope-labeled cross-linkers and Fourier transform ion cyclotron resonance mass spectrometry for structural analysis of a protein/peptide complex. J. Am. Soc. Mass Spectrom. 2006;17:1100–1113. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]