Abstract
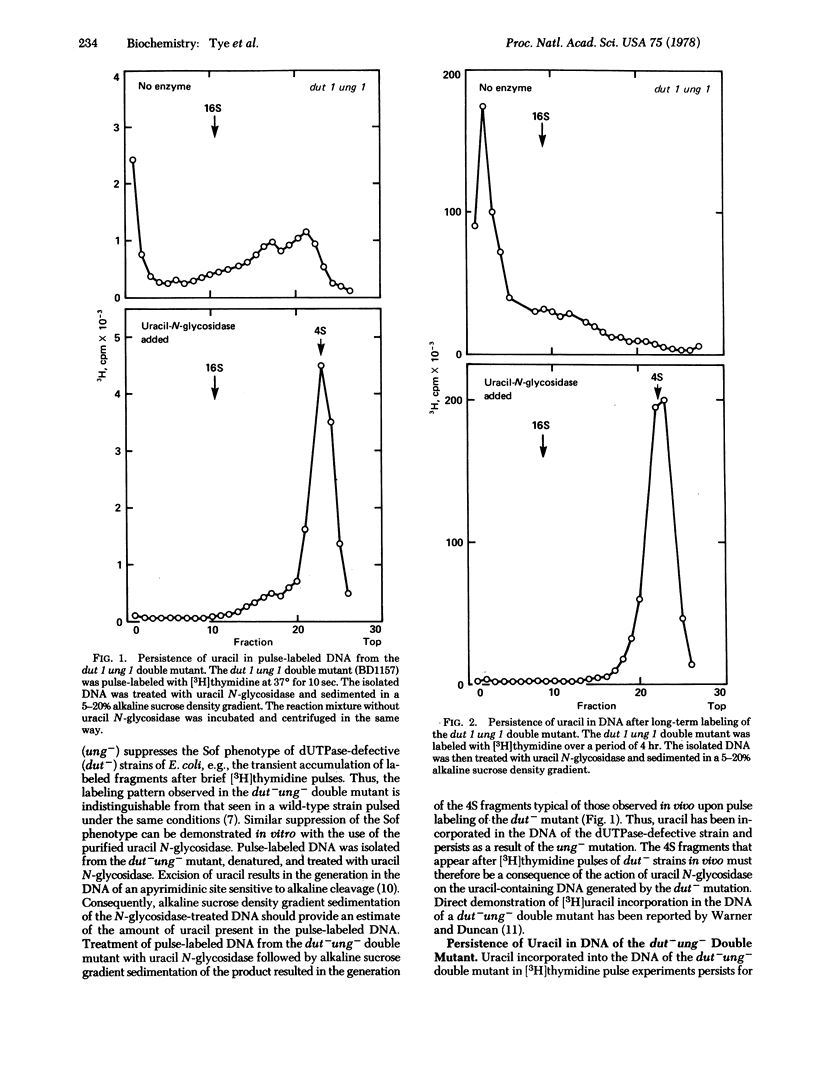
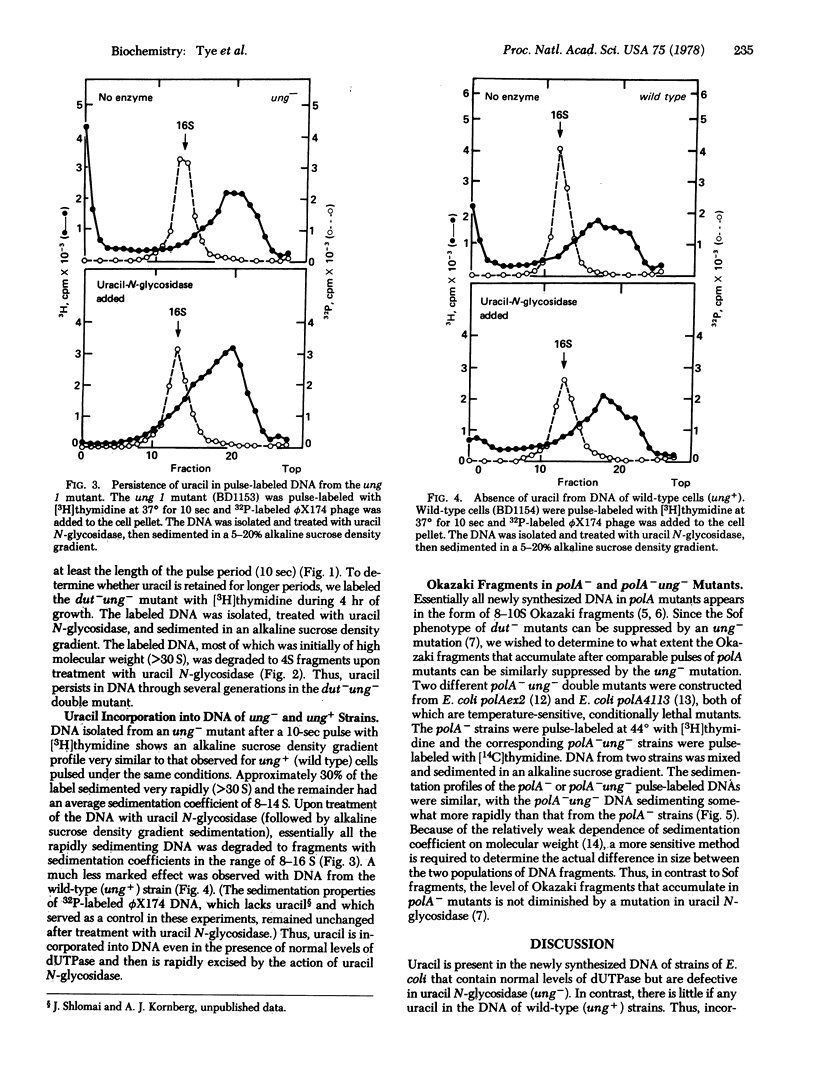
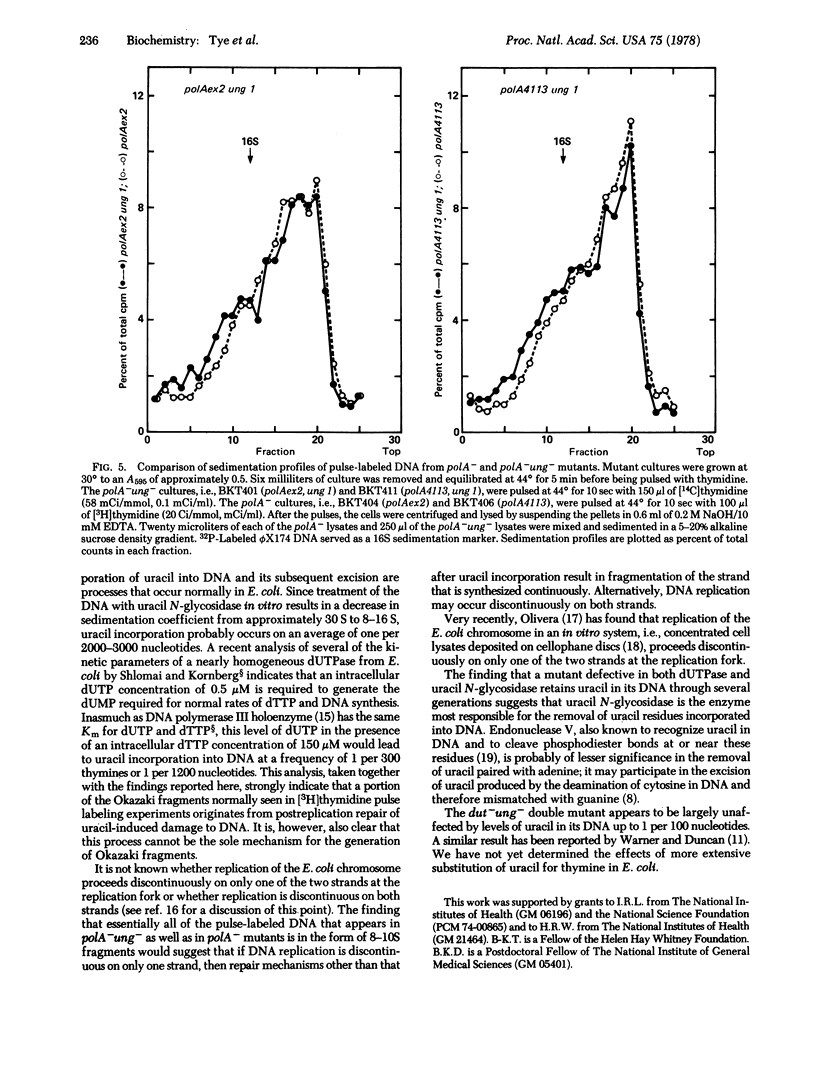
Uracil is incorporated into newly synthesized DNA by mutants of Escherichia coli with reduced levels of dUTPase (dUTP nucleotidohydrolase; EC 3.6.1.23). Excision-repair of the incorporated uracil results in the generation of labeled DNA fragments that appear after brief pulses with [3H]thymidine [Tye, B-K., Nyman, P.-D., Lehman, I. R., Hochhauser, S. & Weiss, B. (1977) Proc. Natl. Acad. Sci. USA 74, 154-157]. Uracil is also incorporated into the newly synthesized DNA of strains of E. coli that contain normal levels of dUTPase. DNA fragments generated by the postreplication excision-repair of uracil may therefore contribute to the pool of nascent DNA (Okazaki) fragments that normally appear in wild-type strains. Discontinuous DNA replication has been examined in the absence of uracil excision by comparing Okazaki fragments in strains that are defective in DNA polymerase I (polA-) and polA- strains that are also defective in uracil N-glycosidase, an enzyme required for the excision-repair of uracil in DNA (polA-ung-). Little or no difference was detected in the level of Okazaki fragments in the polA- strain as compared with the polA-ung- strain. Thus, the uracil-induced cleavage of DNA cannot be the sole mechanism for the generation of Okazaki fragments. Mutants that are defective both in dUTPase and in uracil N-glycosidase incorporate uracil into their DNA with a high frequency (up to 1 per 100 nucleotides). These uracil residues, once incorporated, persist in the DNA without an adverse affect on the growth of the cells.
Keywords: Okazaki fragments, dUTPase, uracil N-glycosidase, discontinuous DNA replication
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B., Sternglanz R. Recent excitement in the DNA replication problem. Nature. 1977 Oct 20;269(5630):655–661. doi: 10.1038/269655a0. [DOI] [PubMed] [Google Scholar]
- Gates F. T., 3rd, Linn S. Endonuclease V of Escherichia coli. J Biol Chem. 1977 Mar 10;252(5):1647–1653. [PubMed] [Google Scholar]
- Gottesman M. M., Hicks M. L., Gellert M. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol. 1973 Jul 15;77(4):531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- Konrad E. B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977 Apr;130(1):167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad E. B., Modrich P., Lehman I. R. Genetic and enzymatic characterization of a conditional lethal mutant of Escherichia coli K12 with a temperature-sensitive DNA ligase. J Mol Biol. 1973 Jul 15;77(4):519–529. doi: 10.1016/0022-2836(73)90220-9. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Veomett G. E. A possible function of DNA polymerase in chromosome replication. Biochem Biophys Res Commun. 1970 Nov 25;41(4):973–980. doi: 10.1016/0006-291x(70)90180-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Okazaki R., Arisawa M., Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R., Okazaki T., Sakabe K., Sugimoto K., Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci U S A. 1968 Feb;59(2):598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera B. M., Bonhoeffer F. Discontinuous DNA replication in vitro. I. Two distinct size classes of intermediates. Nat New Biol. 1972 Dec 20;240(103):233–235. doi: 10.1038/newbio240233a0. [DOI] [PubMed] [Google Scholar]
- Olivera B. M. DNA intermediates at the Escherichia coli replication fork: effect of dUTP. Proc Natl Acad Sci U S A. 1978 Jan;75(1):238–242. doi: 10.1073/pnas.75.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera R. M., Bonhoeffer E. Replication of Escherichia coli requires DNA polymerase I. Nature. 1974 Aug 9;250(5466):513–514. doi: 10.1038/250513a0. [DOI] [PubMed] [Google Scholar]
- SHAPIRO H. S., CHARGAFF E. STUDIES ON THE NUCLEOTIDE ARRANGEMENT IN DEOXYRIBONUCLEIC ACIDS. 8. A COMPARISON OF PROCEDURES FOR THE DETERMINATION OF THE FREQUENCY OF PYRIMIDINE NUCLEOTIDE RUNS. Biochim Biophys Acta. 1964 Oct 16;91:262–270. doi: 10.1016/0926-6550(64)90250-6. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., Weiss B. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):154–157. doi: 10.1073/pnas.74.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Kornberg A. A holoenzyme form of deoxyribonucleic acid polymerase III. Isolation and properties. J Biol Chem. 1974 Oct 10;249(19):6244–6249. [PubMed] [Google Scholar]



