Abstract
Twelve plant species were collected from the east coast of Korea to identify culturable endophytes present in their roots. The fungal internal transcribe spacer (ITS) region (ITS1-5.8SrRNA-ITS2) was used as a DNA barcode for identification of fungi. A total of 194 fungal strains were identified and categorized into 31 genera. The genus Penicillium accounted for the largest number of strains, followed by the genus Aspergillus. Furthermore, using 5 statistical methods, the diversity indices of the fungi were calculated at the genus level. After comprehensive evaluation, the endophytic fungal group from Phragmites australis ranked highest in diversity analyses. Several strains responsible for plant growth and survival (Penicillium citrinum, P. funiculosum, P. janthinellum, P. restrictum, and P. simplicissimum), were also identified. This study provides basic data on the sheds light on the symbiotic relationship between coastal plants and fungi.
Keywords: Coastal plants, Fungal diversity, Fungal endophytes, Korean East Coast
The seashore, located at the boundary of the land and sea, is affected by waves and tides. It has a variety of topographical and ecological characteristics in the form of coastal sand dunes, shore cliffs, and tidal flats. Repeated erosions, sedimentations, and strong winds make the landform of this region unstable. The soil in the coastal region contains high levels of salt and the sandy soil does not efficiently collect water. Frequent rain or drought also appears to be due to influences from the Taebaek Mountains and anticyclones from the East Sea. Thus, the coastal environment is harsh for plant growth.
Despite these problems, many plants inhabit the Korean East Coast. Coastal plants play important roles in the food chain and in the natural habitat of the coastal ecosystem. In addition, coastal plants help microorganisms in the soil to degrade pollutants by providing favorable components like oxygen, bioactive substances, and nutrients. The roots of coastal plants also contribute to stabilization of sandy soils, like sand dunes, via tight anchoring to the rhizosphere.
Symbiosis between plants and microorganisms is very important for the settlement of coastal plants. Soil microorganisms are also directly connected to plant diversity and productivity [1]. Fungi interact with plants and serve as, a partner in plant growth and survival. Endophytic fungi in particular play a variety of roles in their hosts such as defense from pathogenic microorganisms, growth promotion, and solubilization of essential nutrients for host plant [2, 3, 4, 5].
The aim of this study was to identify the distribution of endophytic fungi and to analyze their diversity in the roots of coastal plants. On cliffs, sand dunes, and gravelly fields of the Korean East Coast, the isolation of endophytic fungi from plant roots was performed. Every fungal colony from a root was subcultured for isolation of a single strain and recultured to obtain enough mycelia for extraction of genomic DNA. After amplification of the internal transcribed spacer (ITS) region (ITS1-5.8SrRNA-ITS2), the DNA was sequenced and BLASTed against the NCBI GenBank sequence database. Fungi were then categorized into groups based on a phylogenetic classification system and the identity of the host plants. Through statistical calculation methods, the bio-diversity of each plant was then assessed.
MATERIALS AND METHODS
Plant materials
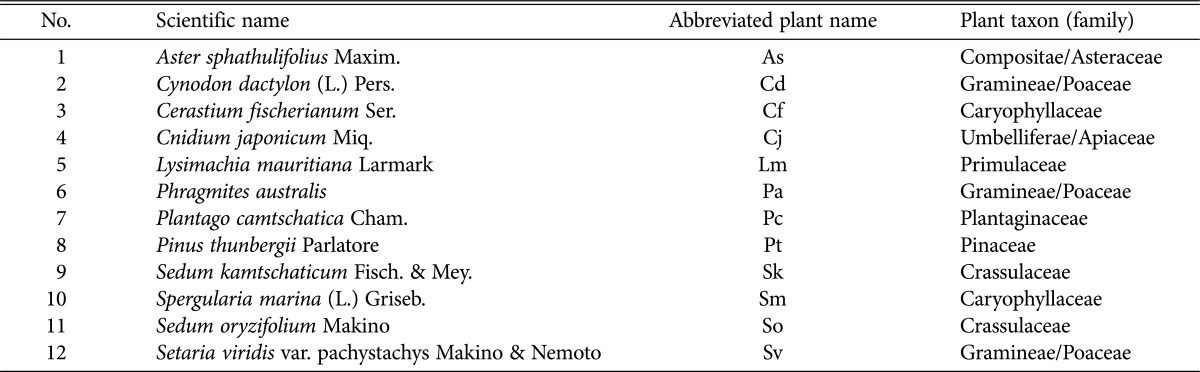
Samples from 12 plant species from the coastal cliff areas, sand dunes, and gravelly fields around Yeongdeok and Pohang were collected and used for experiments (Fig. 1). The scientific name and code of plant samples are listed in Table 1. All herbaceous plant samples were collected by more than 5 heads except for Pinus thunbergii Parlatore, which is a woody plant. Each plant sample was collected from its plant colony within 1~10 m2.
Fig. 1.
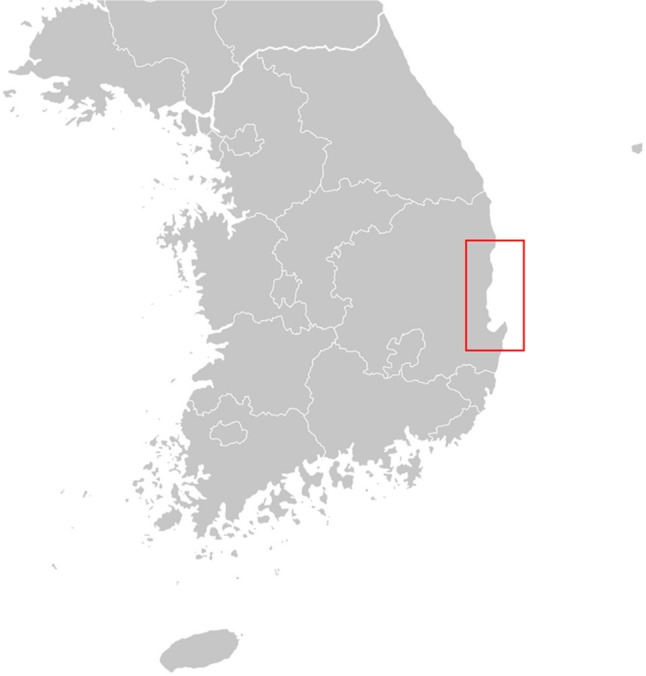
The geographical locations of sampling sites used for this study. Twelve plant samples from the Korean East Coast were collected in Uljin, Yeongdeok, and Pohang.
Table 1.
The scientific name, abbreviated plant name, and taxon (family) of 12 sampled plants
Isolation and culture of endophytic fungi
For each plant, more than 80 pieces of root were used. The soil on the plant root samples was removed using incubation with Tween 80 for 10 min, followed by washes with distilled water. Samples were then incubated twice with 1% perchloric acid for 10 min. After preprocessing, the roots were cut into 3~4 cm pieces and dehydrated. To isolate endophytic fungi, root samples were placed on Hagem minimal media (HM) containing 80 ppm streptomycin and incubated at 25℃ [6, 7]. Colonies at the tip of roots were streaked on HM and incubated again at 25℃. Fungal isolates were then transferred onto potato dextrose agar to obtain a pure culture. Pure cultures of endophytic fungi were cultivated in potato dextrose broth for 7~14 days at 25℃ and 100 rpm. Finally, the samples were lyophilized and used for identification.
DNA extraction, PCR amplification, and the identification of fungal strains
The fungal genomic DNA was extracted using the DNeasy plant mini kit (Qiagen, Valencia, CA, USA) and identified by means of sequencing of the ITS region with the universal primers ITS-1 (5'-TCC GTA GGT GAA CCT GCG G-3') and ITS-4 (5'-TCC TCC GCT TAT TGA TAT GC-3'). The reaction cycles consisted of initial denaturation (95℃, 2 min), 35 cycles of denaturation (95℃, 30 sec), annealing (55℃, 1 min), extension (72℃, 1 min), and final extension (72℃, 7 min). The PCR products were analyzed using agarose gel electrophoresis with ethidium bromide (EtBr) staining. The products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced by means of the ABI PRISM BigDye Terminator Cycle Sequencing Kit (PE Biosystems, Foster, CA, USA) on an ABI 310 DNA sequencer (Perkin Elmer, Foster, CA, USA). After preprocessing, the resulting DNA sequence was identified using the BLASTN tool of the National Center for Biotechnology Information (NCBI) nucleotide collection (nr/nt) database.
Statistical analysis of fungal communities
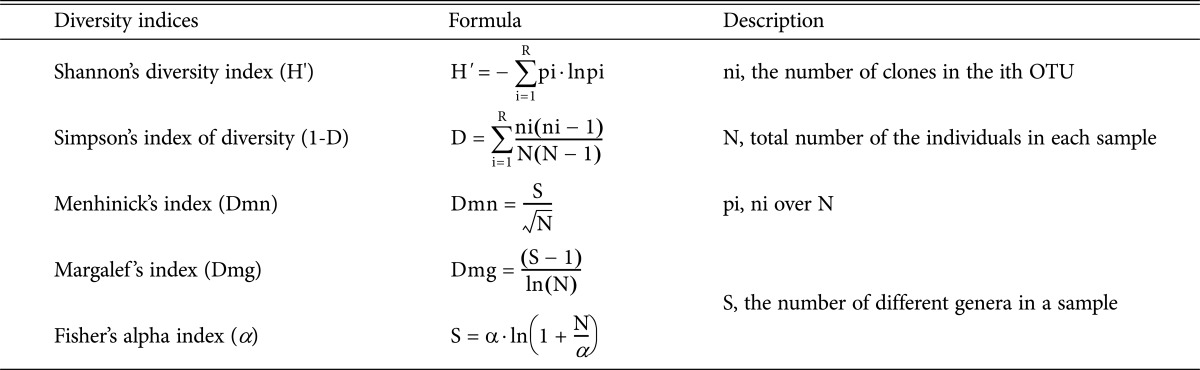
Fungal generic richness and diversity were analyzed at the genus level among the plant samples. Margalef 's index (Dmg) and Menhinick's index (Dmn) were used to assess genus richness [8]. Fisher's alpha index (α), Shannon diversity index (H'), and Simpson's index of diversity were used to evaluate genus diversity [9, 10, 11]. The formulas for all calculations method are listed in Table 2.
Table 2.
The diversity index formulas used in this study
RESULTS AND DISCUSSION
Identification of endophytic fungi
The nucleotide sequences of endophytic fungi were registered in the GenBank database of the National Center for Biotechnology Information (accession Nos. JX238717~JX238739, JX238758~JX238905, JX238907~JX238967, JX238969~JX239035, and KJ511463~KJ511464) (Table 3). A total of 194 fungal isolates were confirmed from plants in the Korean East Coast and classified into 31 genera and 69 species through comparisons with sequences in GenBank.
Table 3.
Partial identification of the 194 fungal isolates using the internal transcribed spacer sequence analysis
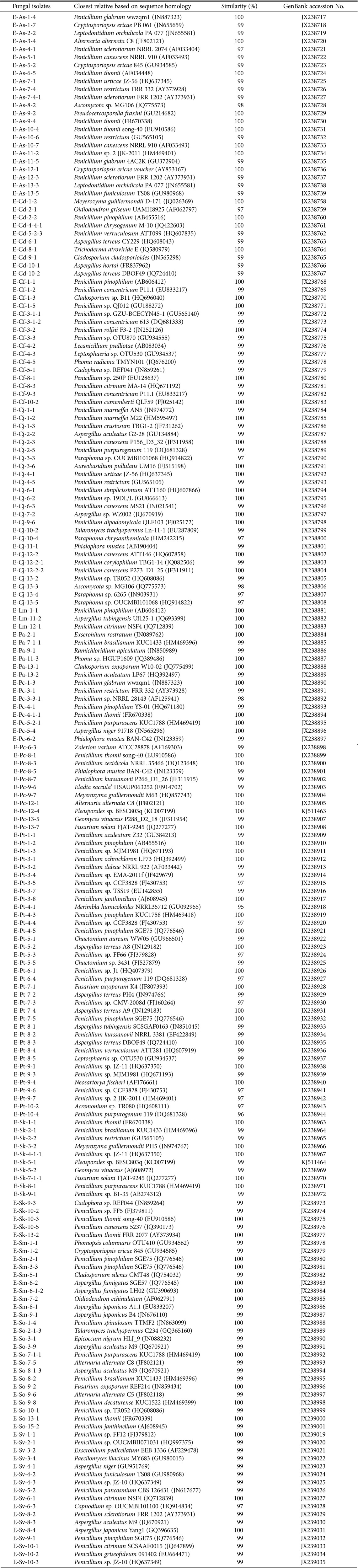
aEladia saccula = Penicillium sacculum.
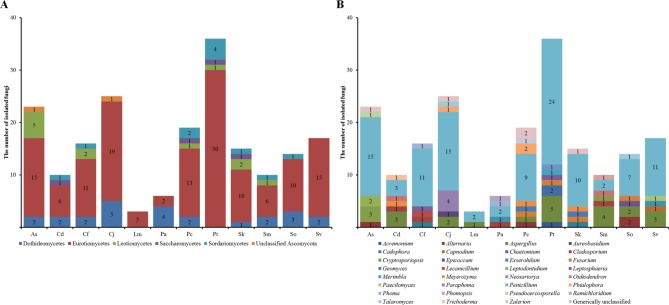
A total of 194 strains were categorized into the phylum Ascomycota. The class Eurotiomycetes (140 strains) accounted for the highest number of strains followed by the class Dothideomycetes (25 strains), Leotiomycetes (12 strains), Sordariomycetes (11 strains), Saccharomycetes (4 strains), and unclassified Ascomycota (2 strains). At the genus level, Penicillium accounted for the highest proportion (112 strains) followed by Aspergillus (21 strains).
The genus of each strain was noted and the proportion of each group at the class and genus levels was analyzed (Fig. 2). Eurotiomycetes accounted the highest percentage at the class level; except for the plant Pa, Eurotiomycetes accounted for more than half of the fungi in every plant sample. At the genus level, Penicillium was the most prevalent (57.7%), followed by Aspergillus (10.8%), of the total fungal isolates in plant samples (except for Sm). The rest of the genera constituted 0.5~2.6%. The distribution of endophytic fungi from roots of coastal plants in the East Coast was similar to that of Ulleung Island [12]. Several plants, including Aster sphathulifolius, Plantago camtschatica, Sedum oryzifolium, and Setaria viridis Makino & Nemoto, inhabit the Ulleung island in the East Sea. Ulleung Island in the East Sea, which 130 km from the Korean Peninsula. The climate and vegetation of Ulleung Island are thus very similar to the East Coast and members of the genus Penicillium accounted for the highest percentage there as well.
Fig. 2.
Distribution of fungal isolates in different plant samples at the class (A) and genus (B) levels. As, Aster sphathulifolius Maxim.; Cd, Cynodon dactylon (L.) Pers.; Cf, Cerastium fischerianum Ser.; Cj, Cnidium japonicum Miq.; Lm, Lysimachia mauritiana Larmark; Pa, Phragmites australis; Pc, Plantago camtschatica Cham.; Pt, Pinus thunbergii Parlatore; Sk, Sedum kamtschaticum Fisch. & Mey.; Sm, Spergularia marina (L.) Griseb.; So, Sedum oryzifolium Makino; Sv, Setaria viridis var. pachystachys Makino & Nemoto.
Fungal diversity at the genus level in the sampled plants.
Depending on the plants, fungal isolates were categorized into 5 genera and 11 species from As, 6 genera and 9 species from Cd, 6 genera and 7 species from Cf, 6 genera and 14 species from Cj, 2 genera and 3 species from Lm, 5 genera and 5 species from Pa, 8 genera and 15 species from Pc, 8 genera and 14 species from Pt, 5 genera and 8 species from Sk, 6 genera and 7 species from Sm, 6 genera and 11 species from So, and 5 genera and 11 species from Sv (Table 4).
Table 4.
Diversity of fungal isolates according to host plant
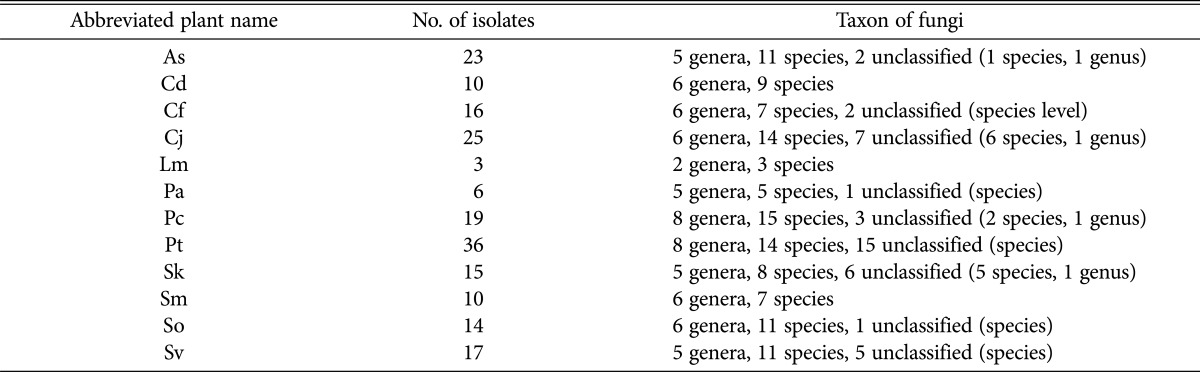
As, Aster sphathulifolius Maxim.; Cd, Cynodon dactylon (L.) Pers.; Cf, Cerastium fischerianum Ser.; Cj, Cnidium japonicum Miq.; Lm, Lysimachia mauritiana Larmark; Pa, Phragmites australis; Pc, Plantago camtschatica Cham.; Pt, Pinus thunbergii Parlatore; Sk, Sedum kamtschaticum Fisch. & Mey.; Sm, Spergularia marina (L.) Griseb.; So, Sedum oryzifolium Makino; Sv, Setaria viridis var. pachystachys Makino & Nemoto.
Based on counting of genera by plant samples, generic richness and diversity were calculated (Table 5). In terms of generic richness, Pc had the highest score in Margalef 's index (2.38), and Pa had the highest score in Menhinick's index (2.04). In generic diversity, Pa exhibited the highest score according to Fisher's α (14.12) and Simpson's index of diversity (0.93), and Pc had the highest score according to Shannon's index (1.66). This result is likely due to the higher sensitivity of Fisher's α and Simpson's to evenness than Shannon's index [8]. Because the genus Penicillium was the dominant genus in all plant samples (except for Sm), comparison of evenness does not mean much. When looking only at results of calculations of diversity index formulas, Pa is regarded as the environment that the most diverse endophytic fungi can inhabit.
Table 5.
Fungal diversity analysis using 5 diversity indices at the genus level
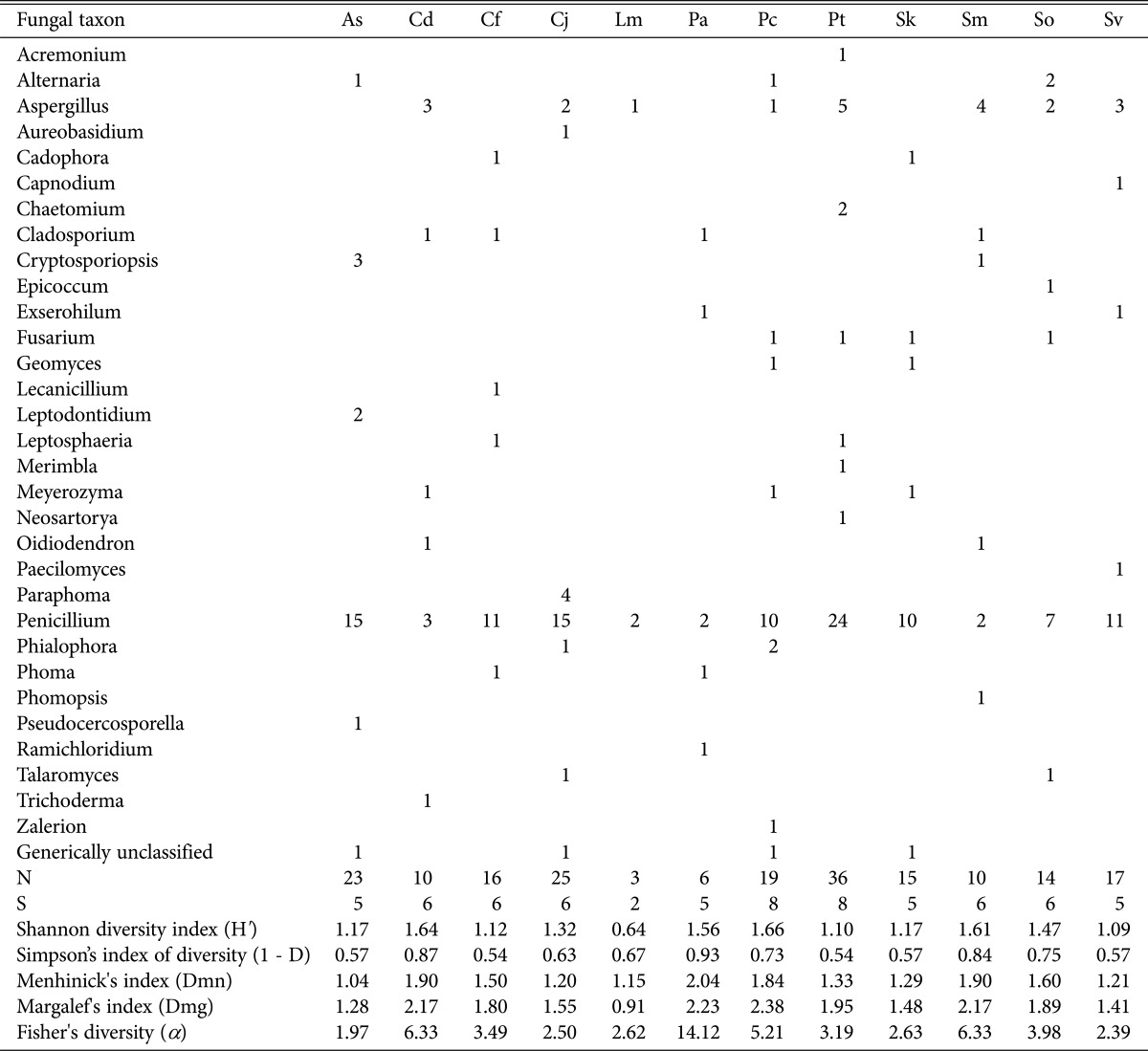
As, Aster sphathulifolius Maxim.; Cd, Cynodon dactylon (L.) Pers.; Cf, Cerastium fischerianum Ser.; Cj, Cnidium japonicum Miq.; Lm, Lysimachia mauritiana Larmark; Pa, Phragmites australis; Pc, Plantago camtschatica Cham.; Pt, Pinus thunbergii Parlatore; Sk, Sedum kamtschaticum Fisch. & Mey.; Sm, Spergularia marina (L.) Griseb.; So, Sedum oryzifolium Makino; Sv, Setaria viridis var. pachystachys Makino & Nemoto.
Symbiotic fungi can help plants overcome abiotic stressors like drought, heat, and salinity [13]. In particular, coastal plants are frequently exposed to salt stress from scattered seawater or via permeation of saline ground water. P. funiculosum and P. janthinellum, fungal strains identified there, reportedly promote resistance to salt stress in the host [14, 15].
Penicillium citrinum was isolated from Cf, Lm, and Sv. P. citrinum reportedly produces gibberellins for the host plant [16]. Gibberellins are essential for developmental stages, including leaf expansion, pollen maturation, seed germination, stem elongation [17], and affect the growth and settlement during the early stage of plant growth. Thus, these 2 species likely help their plant host to absorb nutrients, and they also promote host's growth.
Two of the identified species are known to improve the resilience of plant-host defense systems against pathogens. P. simplicissimum has been reported to activate multiple host defense signals [18] and P. restrictum exerts antagonistic action on pathogenic fungi [19]. Because plants are exposed to a large number of pathogenic microorganisms in the soil or air, it is important for plants to possess such defense systems.
In summary, a total of 194 fungal strains were isolated from 12 plants inhabiting the East Coast and were classified into 1 phylum, 5 classes, 11 orders, 16 families, and 31 genera. Penicillium (class Eurotiomycetes) was the most dominant genus followed by Aspergillus. The group of fungi isolated from Phragmites australis was the most diverse according to diversity analysis. Species helping plant growth and survival such as P. citrinum, P. funiculosum, P. janthinellum, P. restrictum, and P. simplicissimum were also identified. This study provides basic data on the symbiosis of coastal plants and fungi.
ACKNOWLEDGEMENTS
This research work was supported by Eco-Innovation Project, Korean Government's R&D program on Environmental Technology & Development.
References
- 1.van der Heijden MG, Bardgett RD, van Straalen NM. The unseen majority:soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 2.Varma A, Verma S, Sudha, Sahay N, Bütehorn B, Franken P. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol. 1999;65:2741–2744. doi: 10.1128/aem.65.6.2741-2744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mejía LC, Rojas EI, Maynard Z, Bael SV, Arnold AE, Hebbar P, Samuels GJ, Robbins N, Herre EA. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biol Control. 2008;46:4–14. [Google Scholar]
- 4.Mack KM, Rudgers JA. Balancing multiple mutualists: asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos. 2008;117:310–320. [Google Scholar]
- 5.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–26544. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Vázquez MM, César S, Azcón R, Barea JM. Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl Soil Ecol. 2000;15:261–272. [Google Scholar]
- 7.Yamada A, Ogura T, Degawa Y, Ohmasa M. Isolation of Tricholoma matsutake and T. bakamatsutake cultures from field-collected ectomycorrhizas. Mycoscience. 2001;42:43–50. [Google Scholar]
- 8.Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 9.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. [Google Scholar]
- 10.Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- 11.Fisher RA, Steven Corbet A, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol. 1943;12:42–58. [Google Scholar]
- 12.Kim M, You YH, Yoon H, Kim H, Seo Y, Khalmuratova I, Shin JH, Lee IJ, Choo YS, Kim JG. Genetic diversitiy of endophytic fungal strains isolated from the roots of coastal plants in Ulleung island for restoration of coastal ecosystem. J Life Sci. 2012;22:1384–1391. [Google Scholar]
- 13.Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim YO, Redman RS. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 14.Khan AL, Hamayun M, Kim YH, Kang SM, Lee IJ. Ameliorative symbiosis of endophyte (Penicillium funiculosum LHL06) under salt stress elevated plant growth of Glycine max L. Plant Physiol Biochem. 2011;49:852–861. doi: 10.1016/j.plaphy.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Khan AL, Waqas M, Khan AR, Hussain J, Kang SM, Gilani SA, Hamayun M, Shin JH, Kamran M, Al-Harrasi A, et al. Fungal endophyte Penicillium janthinellum LK5 improves growth of ABA-deficient tomato under salinity. World J Microbiol Biotechnol. 2013;29:2133–2144. doi: 10.1007/s11274-013-1378-1. [DOI] [PubMed] [Google Scholar]
- 16.Khan SA, Hamayun M, Yoon H, Kim HY, Suh SJ, Hwang SK, Kim JM, Lee IJ, Choo YS, Yoon UH, et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008;8:231. doi: 10.1186/1471-2180-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 18.Hossain MM, Sultana F, Kubota M, Koyama H, Hyakumachi M. The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol. 2007;48:1724–1736. doi: 10.1093/pcp/pcm144. [DOI] [PubMed] [Google Scholar]
- 19.Nicoletti R, De Stefano M. Penicillium restrictum as an antagonist of plant pathogenic fungi. Dyn Biochem Process Biotechnol Mol Biol. 2012;6:61–69. [Google Scholar]