Abstract
In this study, bacterial strains were isolated from soils from 30 locations of Samcheok, Gangwon province. Of the isolated strains, seven showed potential plant growth promoting and antagonistic activities. Based on cultural and morphological characterization, and 16S rRNA gene sequencing, these strains were identified as Paenibacillus species. All seven strains produced ammonia, cellulase, hydrocyanic acid, indole-3-acetic acid, protease, phosphatase, and siderophores. They also inhibited the mycelial growth of Fusarium oxysporum f. sp. radicis-lycopersici in vitro. The seven Paenibacillus strains enhanced a range of growth parameters in tomato plants under greenhouse conditions, in comparison with non-inoculated control plants. Notably, treatment of tomato plants with one identified strain, P. polymyxa SC09-21, resulted in 80.0% suppression of fusarium crown and root rot under greenhouse conditions. The plant growth promoting and antifungal activity of P. polymyxa SC09-21 identified in this study highlight its potential suitability as a bioinoculant.
Keywords: Antagonistic activity, Fusarium oxysporum f. sp. radicis-lycopersici, Paenibacillus spp., Plant growth promoting rhizobacteria
Intensive use of chemical fertilizers and pesticides in agriculture guarantees high crop yield and quality, but this approach is costly and poses risks to the environment and human health. Because of the undesirable features of these chemicals, there has recently been increasing interest in environmentally friendly, sustainable agriculture. One possible alternative to chemical fertilizers and pesticides is the use of microorganisms as biofertilizers and biological control agents.
In the rhizosphere, important and intensive interactions take place between the plant, soil, microorganisms, and soil microfauna. Bacteria are the most abundant microorganisms within soil, and those that have a beneficial effect on plant growth or health are commonly referred to as plant growth promoting rhizobacteria (PGPR) [1]. The mechanisms by which PGPR can act beneficially on plant growth have not been fully elucidated, but proposed mechanisms include production of plant growth regulators such as indole-3-acetic acid (IAA), asymbiotic nitrogen fixation, solubilization of soil phosphorus compounds and other nutrients, and antagonism against plant pathogens through the excretion of siderophores, cellulose, protease, antibiotics and cyanide [2, 3].
Among the myriads of bacteria thriving in the soil, some spore-forming PGPR, in particular gram-positive bacilli and streptomycetes, have attracted special attention due to their advantages over non-spore formers in product formulation and stable maintenance in soil [4]. Among these, Paenibacillus populations were found to be capable of suppressing several plant diseases and promoting plant growth [5, 6]. Paenibacillus populations synthesize the plant hormones auxin and cytokinin, solubilize soil phosphorus, and produce biological control substances like siderophores and antibiotics [7]. The range of beneficial properties of PGPR provides great incentive to search for new strains, which could be used as biofertilizers and biopesticides in agriculture.
The fungus Fusarium oxysporum f. sp. radicis-lycopersici (FORL) causes fusarium crown and root rot (FCRR) of tomato, which is one of the most damaging soil-borne diseases in intensive tomato production [8]. The disease results in severe loss of greenhouse and open field crops. Current practices for controlling plant diseases are based largely on disease resistant crops, cultivation management in fields and application of synthetic fungicides [4]. The relatively poor efficacy of chemical control and the lack of resistance in some commercially important tomato cultivars have focused attention on the feasibility of biological control of the pathogen. However, many biological control agents that have been evaluated against FCRR do not provide sufficient disease control [9].
Therefore, the aims of this study were to isolate and characterize novel Paenibacillus spp. from the Chinese cabbage, garlic and paddy fields, and determine their potential use for plant growth promotion and the biological control of FCRR.
MATERIALS AND METHODS
Isolation and identification of bacterial antagonists
Soil samples were collected in 2011 from Samcheok, Gangwon province, Korea. The samples were taken from 30 locations within fields where Chinese cabbage, garlic, or rice were cultivated. Each sample was air-dried at ambient temperature, passed through an 850 µm mesh sieve to remove plant debris, and then stored in a polyethylene bag at room temperature. To isolate bacteria from samples, 1 g of soil was suspended in 9 mL sterile distilled water and shaken vigorously for 15 min, and then allowed to settle for 15 min. Each suspension was serially diluted, and then 0.1 mL of 10-4, 10-5, and 10-6 dilutions was spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) and tryptic soy agar (TSA; Difco) plates in triplicate. After incubating plates at 25℃ for ten days, colonies with distinct morphological appearance were selected from the countable plates and streaked onto fresh PDA and TSA plates to obtain pure cultures. Bacteria were identified by cultural and morphological characterization, and 16S rRNA gene sequencing. To sequence 16S rRNA gene, total genomic DNA was initially extracted from colonies, as described by Nishiguchi et al. [10]. The 16S rRNA gene was amplified by PCR using universal primers 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-TACGGYTACCTTGTTACGACTT-3'). The amplified DNA was purified using a gel purification kit (Bioneer, Daejeon, Korea) and was sequenced at Bioneer. The obtained sequences were aligned using the BLAST function of the GenBank database (NCBI).
In vitro antifungal activity assay
The antifungal activity of isolated soil microorganisms against FORL was determined by dual culture. Briefly, a 0.65 cm diameter mycelial disc was cut from the white perimeter of a FORL colony that had been cultured on PDA plates at 25℃ for seven days. The disc was placed 1.5 cm from the edge of a fresh PDA plate, and then a sterilized toothpick was used to inoculate the test soil microorganism into the same plate, 1.5 cm from the opposite edge. Control plates without soil microorganisms were prepared simultaneously. The antagonistic effect was determined by measuring the size of the inhibition zone formed on plates after incubation at 25℃ for 14 days.
IAA production assay
IAA production by soil microorganisms was evaluated using a modification of the method described by Bric et al. [11]. Each bacterium was suspended in sterile distilled water at 108 colony-forming units (CFU) per mL (OD620 = 0.1), then 0.5mL aliquots were inoculated into 50mL of King's B broth medium supplemented with 0.1% L-tryptophan. Cultures were incubated for 72 hr at 28℃, and then centrifuged at 12,000 rpm for 10 min. Following this, 2 mL of the supernatant was mixed with 4mL of Salkowski reagent (1mL of 0.5M FeCl3 solution in 50 mL of 35% perchloric acid), which acts as an indicator for IAA production by changing the solution color to pink-red. Optical density at 530 nm was measured using a spectrophotometer, and the concentration of IAA produced by cultures was determined relative to a 10~100 µg/mL commercial IAA standard curve.
Phosphate solubilization
A pure colony from a fresh culture of each bacterial strain was streak inoculated onto Pikovskaya's phosphate (PVK) [12] and National Botanical Research Institute's Phosphate (NBRIP) [13] agar plates, using a sterile needle. Tricalcium phosphate was used as a source of insoluble inorganic phosphate. Phosphate solubilization activity was determined by the development of clear halos surrounding colonies on the PVK and NBRIP agar plates, after incubation for 14 days at 28℃.
Ammonia (NH3) production
Bacterial strains were tested for the production of NH3 in peptone water. Freshly grown cultures were inoculated into 10mL of peptone water and incubated for 72 hr at 28℃, before 0.5 of mL Nessler's reagent was added to each tube. The development of a brown-yellow color indicated NH3 production [14].
Hydrocyanic acid production
Production of hydrocyanic acid (HCN) was determined by a modified method of that described by Wei et al. [15]. Bacterial strains were grown on TSA supplemented with 4.4 g/L glycine, with filter paper strips soaked in picric acid solution (0.5% picric acid, 2% sodium carbonate) placed in the lid of each petri dish. After incubation at 28℃ for four days, the filter paper was examined for a color change, with a brown color indicative of HCN production.
Siderophore production
Siderophore production was determined using the Chrome azurol S (CAS) agar assay method [16]. A well was made in the center of the CAS agar plate using a 5 mm cork borer, and then a bacterial inoculum was dropped into the well. Plates were incubated at 28℃ for seven days, after which the development of a yellow-orange halo around the bacteria identified siderophore production.
Cellulase production
Cellulase production was determined in Casein-Yeast extract agar (CYEA; casein, 5 g; yeast extract, 2.5 g; glucose, 1 g; agar, 18 g dissolved in 1 L distilled water) medium amended with 1% carboxymethyl cellulose [17]. After 72 hr of incubation at 28℃, the agar was flooded with an aqueous solution of Congo red (1 mg/mL) for 15 min. The colonies surrounded by clear halos were considered positive for cellulase production.
Protease production
Proteolytic activity was determined by plating bacteria onto CYEA plates containing with 7% skim milk powder [12]. After 72 hr incubation at 28℃, a clear zone surrounding the colonies indicated positive proteolytic activity.
In vivo antifungal activity assay
FORL was first cultured on PDA plates at 25℃ for seven days, then in potato dextrose broth (Difco) for seven days at 25℃ in a rotary shaker at 200 rpm. After incubation, the conidial suspension was diluted in sterile distilled water to give a final concentration of 105 conidia/mL. To obtain a growth broth, each bacterial strain was cultured in tryptic soy broth (Difco) for 72 hr at 28℃ in a rotary shaker at 180 rpm. After incubation, the bacterial suspension was diluted in sterile distilled water to give a final concentration of 108 CFU/mL (OD620 = 0.1). Tomato (Solanum lycopersicum cv. Super Dotaerang) seeds were sown in horticultural soil in a seedling plate (50 holes of 4 cm diameter) and were grown in a glasshouse for one month. Each seedling was inoculated with FORL 1 hr before treatment with growth broth. Metaconazole and Tapseed (Green Biotech, Paju, Korea) were used as a positive control and water as a negative control, and each treatment was done in triplicate. The inoculated plants were placed in a moist chamber to maintain approximately 90% humidity at 25℃ for 24 hr, and were then transferred to the growth room for further incubation. After 4 wk, each root was cut with a scalpel and assessed for FCRR disease severity using a 0~3 scale [18]: 0, no internal browning; 1, slight internal browning; 2, moderate to severe internal browning of the entire tap root; 3, severe internal browning extending from the tap root into the lower stem above the soil line.
Plant growth promotion assay
A loopful of each bacterial strain was transferred to a 500 mL flask containing tryptic soy broth and grown in a rotating shaker at 180 rpm for 72 hr at 28℃. The bacterial suspensions were then diluted in sterile distilled water to a final concentration of 108 CFU/mL (OD620 = 0.1). The resulting suspensions were used to treat targeted plants under greenhouse conditions. Tomato seeds were surface-sterilized for 20 min in 0.1% sodium hypochlorite, followed by four washes with sterile double distilled water, before planting in sterile soil. Seeds were sown in a seedling plate (50 holes of 4 cm diameter) filled with horticultural soil. Seedlings were grown in a greenhouse, and after four weeks, individual tomato seedlings were planted in plastic pots filled with the same commercial soil. The bacterial inoculum was applied in the form of soil drenching every seven days, four times. The plants were watered twice daily, and plants were harvested 2 wk after the last inoculation. Shoot and root length as well as fresh and dry weight measurements were compared between inoculated and uninoculated plants. Chlorophyll content was also measured, using a SPAD-502 chlorophyll meter (Konica Minolta, Tokyo, Japan).
RESULTS
Isolation and identification of antagonists
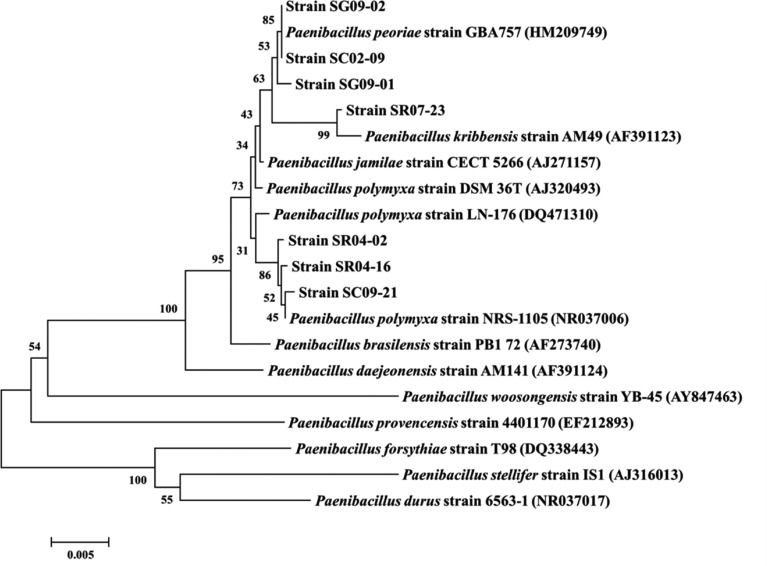
A total of 1,360 bacterial strains were selectively isolated based on macroscopic morphological traits. Seven strains, produced flat colonies and rough-surfaced spores, and were capable of inhibiting the mycelial growth of various plant pathogenic fungi, in in vitro dual culture screening. These seven strains were identified as Paenibacillus spp. according to cultural and morphological characterization (Fig. 1), and 16S rRNA gene sequence analysis. Strains SC02-09, SG09-01, and SG09-02 were identified as P. peoriae, SR07-23 was characterized as P. kribbensis, and the remaining three strains, SC09-21, SR04-02, and SR04-16, were identified as P. polymyxa (Fig. 2).
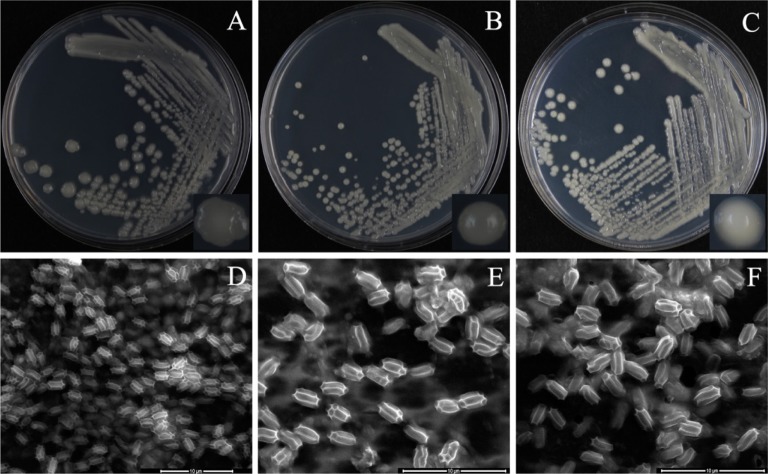
Fig. 1.
Cultural and morphological characteristics of bacterial strains. Isolated bacterial strains were streaked onto potato dextrose agar plates, to facilitate cultural and morphological characterization. Shown are SC09-21 (A, D), SG09-01 (B, E) and SR07-23 (C, F) (scale bars: D~F = 10 µm).
Fig. 2.
Phylogenetic relationship between the seven identified Paenibacillus strains and representative Paenibacillus species, based on 16S rRNA gene sequences developed with the ClustalW program in MEGA 5.20 and constructed using the neighbor-joining method with 1,000 bootstrap replicates. The values indicate the percentage of clustering matches. The scale bar indicates the number of differences in base composition among sequences.
Antagonistic activities of strains in dual culture
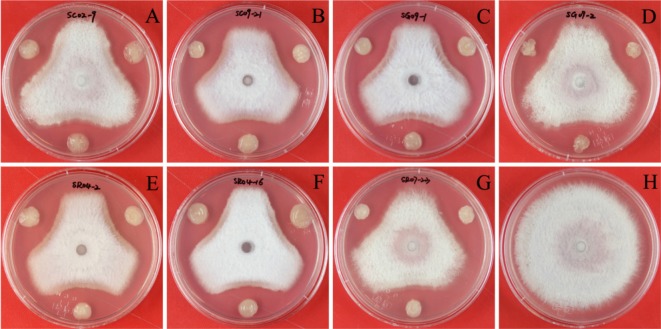
All seven bacterial strains were screened for antagonistic activity by measuring the inhibition zones present after 14 days of dual culture with FORL. The highest antagonistic activity against FORL was observed in the dual culture plate containing SC09-21, in which a 12.2 mm inhibition zone formed due to antibiosis (Table 1, Fig. 3). Strains SG09-01, SR04-02, and SR04-16 also formed an inhibition zone of > 10 mm in dual culture assays, while the remaining three strains generated inhibition zones of 8.9~9.7 mm. In contrast, no inhibition zone was seen in the control plate, where fungal mycelia covered the entire plate surface (Fig. 3).
Table 1.
Antagonistic activity of Paenibacillus strains against Fusarium oxysporum f. sp. radicis-lycopersici in dual culture assay
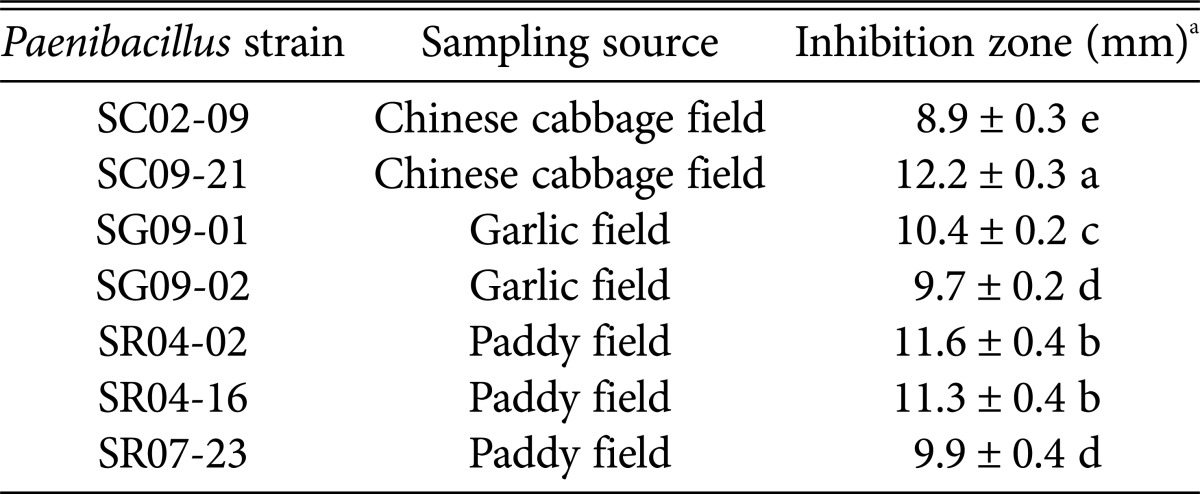
aMeans and standard errors were calculated from three independent observations. Those sharing the same letter are not significantly different, based on the Duncan's multiple range test at p = 0.05.
Fig. 3.
Dual culture assay for in vitro inhibition of mycelial growth of Fusarium oxysporum f. sp. radicis-lycopersici (FORL) by Paenibacillus strains. FORL were cultured on potato dextrose agar plates at 25℃ for seven days, then a mycelial disc was cut and placed 1.5 cm from the edge of a fresh plate. Bacterial strains were toothpick inoculated 1.5 cm from the opposite edge of the same plate, then inhibition zones were measured after incubation at 25℃ for 14 days. Results are shown for: A, SC02-09; B, SC09-21; C, SG09-01; D, SG09-02; E, SR04-02; F, SR04-16; G, SR07-23; H, Control.
IAA production and phosphate solubilization
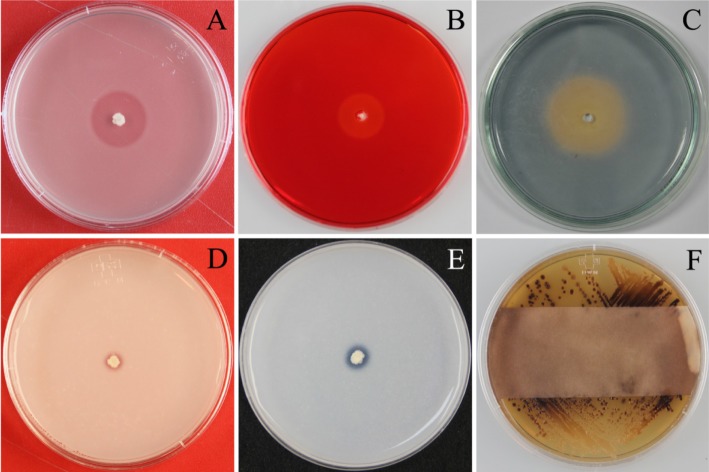
The detection of IAA production was based on a colorimetric assay. The results indicated that all seven strains of Paenibacillus showed IAA production in liquid medium supplemented with L-tryptophan after 72 hr growth. All strains produced IAA in culture broth, with concentrations of 8.7~22.1 µg/mL (Fig. 4). The highest IAA yield was recorded for strain SR04-16, which was isolated from paddy field soil. All Paenibacillus strains formed clear halos around spot inocula in PVK and NBRIP agar plates containing calcium phosphate (Fig. 5D and 5E).
Fig. 4.
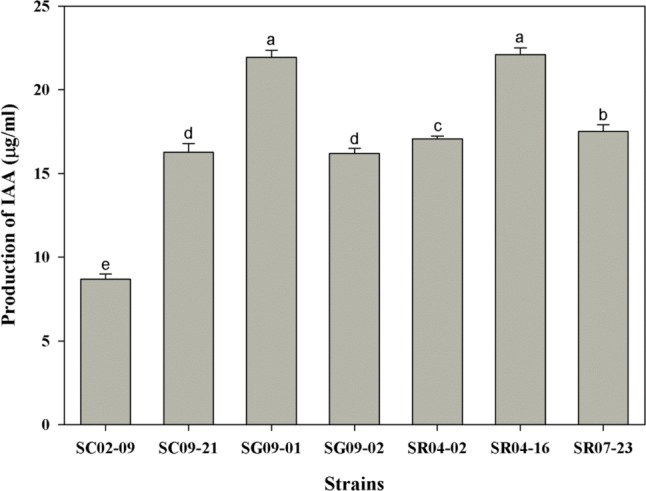
Production of indole-3-acetic acid (IAA) by Paenibacillus strains. Isolated strains were assessed for indole-3-acetic acid production after incubation in King's B broth for 72 hr. Values shown are the means ± SEs errors obtained from three replicates. The same letter above bars represents no significant difference between treatments, based on the Duncan's multiple range test at p = 0.05.
Fig. 5.
Characterization of Paenibacillus strains. A, Representative results are shown for: production of protease on Casein-Yeast extract agar (CYEA) plates with 7% skim milk after 72 hr; B, Cellulase production on CYEA plates with 1% carboxymethyl cellulose after 72 hr, followed by flooding with Congo red solution (1 mg/mL); C, Siderophore production in Chrome azurol S agar plates after seven days; tricalcium phosphate solubilization in Pikovskaya's phosphate agar medium (D) and National Botanical Research Institute's Phosphate agar medium (E) after 14 days; F, Hydrocyanic acid production on tryptic soy agar with glycine (4.4 g/L) after four days, detected using filter paper treated with picric acid solution.
Cellulase, HCN, NH3, protease, and siderophore production
All Paenibacillus strains produced cellulase and protease, as indicated by the formation of clear halos around spot inocula in CYEA plates containing carboxymethyl cellulose and skim milk (Fig. 5A and 5B). Each strain also produced HCN and NH3, as evidenced by the production of a color change in picric acid solution-treated filter paper (Fig. 5F) and Nessler's reagent (data not shown), respectively. Furthermore, all Paenibacillus strains produced orange halos in CAS agar after seven days, confirming the production of siderophores (Fig. 5C).
In vivo antifungal activity assay
The in vivo efficiency of the Paenibacillus strains for controlling FCRR in tomato was evaluated under greenhouse conditions (Table 2). Plants treated with strains SC02-09, SC09-21, and SG09-02 were healthy in appearance and had a lower incidence of FCRR, indicating significant inhibition of FORL. Specifically, treatment of tomato plants with SC09-21 resulted in > 80.0% suppression of FCRR, and treatment with SC02-09 and SG09-02 resulted in disease reduction of > 60.0%. Metaconazole and Tapseed were used as positive control treatments, and showed 97.2% and 77.2% FORL suppression, respectively.
Table 2.
The suppressive effects of seven Paenibacillus strains against fusarium crown and root rot in tomato
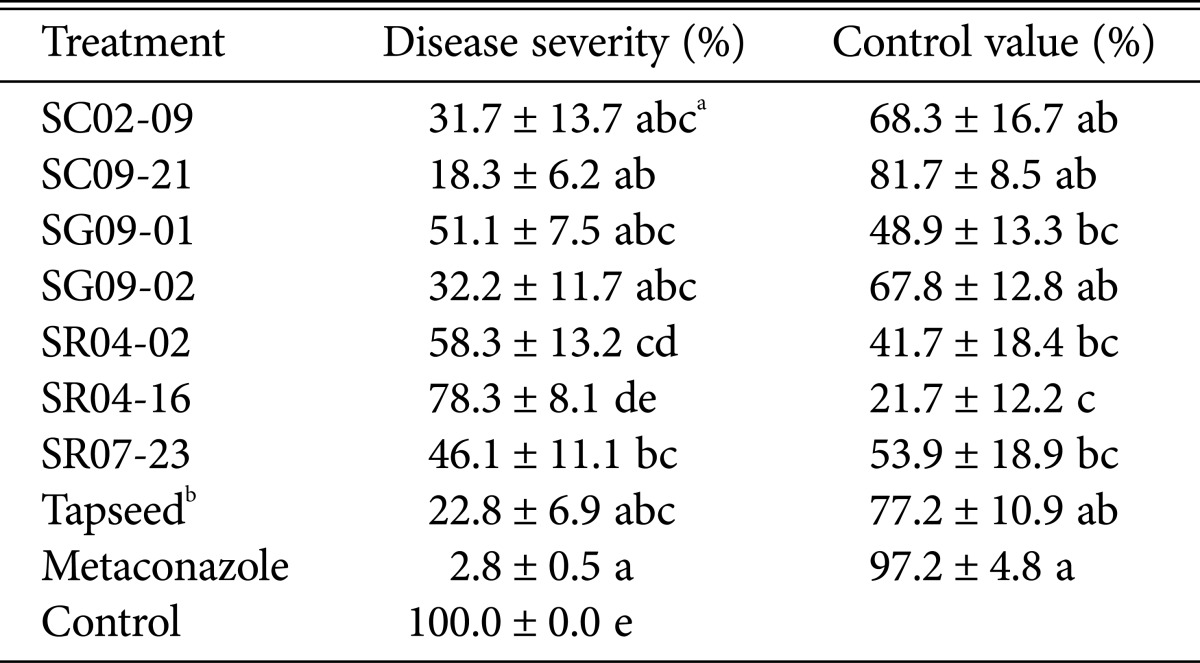
aMeans and SEs were calculated from three independent observations. Those sharing the same letter are not significantly different, based on the Duncan's multiple range test at p = 0.05.
bTapseed: Biopesticide with P. polymyxa AC-1.
Efficacy of plant growth promotion on tomato
Tomato plant growth parameters were measured to assess the growth promotion capacity of the identified Paenibacillus strains. Treatment of plants with each strain resulted in a significant increase in all observed growth parameters (Table 3, Fig. 6). Among the seven bacteria, strain SC02-09 displayed the highest growth promoting activity in terms of stem length. However, strain SG09-01 caused the greatest increase in fresh shoot weight, at 31.3%. In addition, significant root enhancement in treated plants was observed, with treated plants displaying roots that were larger and richer in biomass than those of control plants. Strain SC09-21 caused the largest increase in root length, at 14%. Further significant impact of the Paenibacillus strains was observed on fresh and dry root weight, with maximum increases of 56.6% and 13.9%, respectively. The chlorophyll content of leaves was affected by Paenibacillus strains to a small degree, with the greatest increase from control levels seen in leaves of tomato plants treated with strain SR07-23 (14%).
Table 3.
Effect of seven Paenibacillus strains on different growth parameters in tomato in greenhouse pot trials
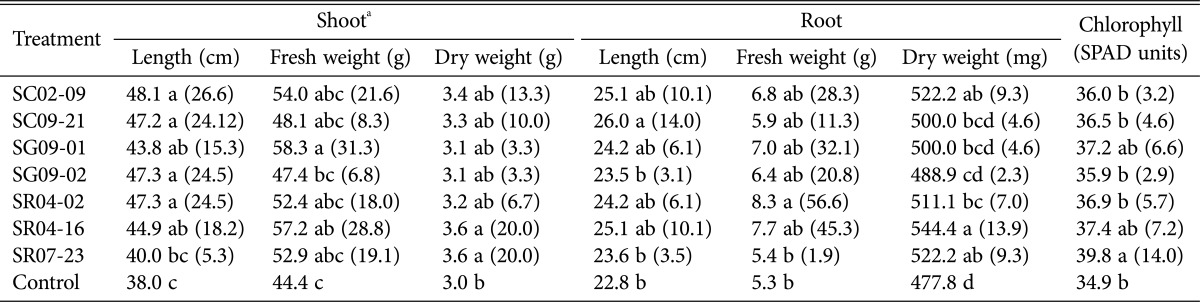
aMeans were obtained from an individual trial, with nine replicates per treatment. Each replicate consisted of a single pot, with one plant per pot. Means within a column followed by the same letter are not significantly different according to the Duncan's multiple range test at p = 0.05. Values in parentheses indicate the percentage increase over control.
Fig. 6.
Shoot growth promotion in tomato plants grown in a greenhouse, by Paenibacillus strains. Tomato seedlings were inoculated four times with bacterial strains, by soil drenching every seven days. Plants are shown two weeks after the final inoculation. A, Uninoculated control; B, SC02-09; C, SC09-21; D, SG09-01; E, SG09-02; F, SR04-02; G, SR04-16.
DISCUSSION
The increasing importance of beneficial bacteria in agriculture has resulted in many efforts to isolate and identify bacteria associated with the soil and rhizosphere of plants, in order to elucidate their roles in plant growth promotion and protection against pathogens. The application of PGPR is a potentially attractive approach to disease management and improved crop productivity in sustainable agriculture [19]. In the present investigation, a total of 1,360 bacterial strains were selectively isolated based on macroscopic morphological traits. Seven strains produced flat colonies and rough-surfaced spores, and were capable of inhibiting the mycelial growth of FORL in in vitro dual culture screening. All seven strains were identified as Paenibacillus spp., according to cultural and morphological characterization, and 16S rRNA gene sequence analysis.
This study was carried out to evaluate the efficacy of seven Paenibacillus strains in the reduction of FCRR disease and the promotion of growth in tomato under greenhouse conditions. Results showed that all tested strains significantly reduced the severity of disease caused by artificial inoculation of the plant pathogen FORL into host plants, compared with the control. Strain SC09-21 reduced the disease severity of FCRR by > 80.0%, whereas strains SC02-09 and SG09-02 resulted in disease reduction of > 60.0%. This protection of tomato plants could be due to the action of antagonistic strains on the growth of plant pathogens. However, many factors are likely to affect the biological control of plant diseases in vivo [20].
In the present study, all seven strains were found to be capable of enhancing different growth parameters in comparison with non-inoculated control plants during pot experiments. An important effect on fresh and dry root weight was observed, with maximum increases of 56.6% and 13.9%, respectively. Among the strains, treatment with SC09-21 resulted in the most significant increase in root length, at 14%. Many reports have similarly shown that bacteria can increase root length in different plants [21], and such an increase may confer advantages to the host plant with respect to health and overall growth.
Paenibacillus spp. are resistant to environmental stresses due to their formation of endospores, making them ideal for using as commercial microbial agents with a long shelf-life [4, 22]. Recently, numerous studies have described advantages of using endospore-forming Paenibacillus spp. in agriculture, including plant growth promotion and the suppression of plant diseases [5, 6]. However, Paenibacillus spp. have been considered less than Pseudomonas spp. as potential PGPR [4]. In the present study, all seven Paenibacillus strains showed antagonism against FORL and promoted the growth of tomato in the greenhouse environment. These results show that Paenibacillus spp. are suited to use in the cultivation of vegetables in greenhouses.
The production of phytohormones by bacteria is one of the most important elements of plant growth promotion. In the present investigation, all strains were able to produce IAA in the presence of L-tryptophan, to varying degrees. Previous studies have similarly found that strains belonging to the same genus can produce different amounts of IAA [2]. IAA production is an important feature because it enables plants to develop highly organized root systems, by which the uptake of nutrients is more efficient [23].
Phosphorus is an essential nutrient in plants. Although there is a large reserve of phosphorus in soil, it is mostly in the form of insoluble compounds that cannot be absorbed by plants, which limits potential plant growth. In contrast, many soil microorganisms are able to solubilize phosphate from relatively insoluble inorganic and organic phosphate compounds, and there are considerable populations of these phosphate-solubilizing bacteria in the soil and rhizosphere [24]. Bacteria capable of phosphate solubilization are believed to promote plant growth by increasing phosphorous uptake [25]. In the present investigation, all Paenibacillus strains were able to solubilize calcium phosphate, as observed with PVK and NBRIP agar plates. Phosphate-solubilizing bacteria are considered to play an important ecophysiological role in mobilizing insoluble inorganic phosphates from their mineral matrix to the bulk soil, where they can be absorbed by plant roots [26]. Thus, it is imperative to identify microorganisms producing phosphatase in the soil.
Iron, an essential nutrient for all forms of life, is unavailable for direct assimilation by plants and microorganisms in the soil because ferric iron (Fe3+) is only sparingly soluble. Siderophores are low-molecular-weight iron chelators that bind Fe3+ and are produced by various soil microorganisms. These microorganisms make siderophores available for their own and other species in the soil microbial community, and act indirectly as the biocontrol agents of plant pathogens [27]. Raza et al. [28] showed that the addition of Fe3+, up to a certain concentration, increases the growth of P. ploymyxa SQR-21 and the production of fusaricidin-type compounds. In the current study, all strains produced siderophores, which is an important feature for the suppression of plant pathogens and the promotion of plant growth.
Other influential traits of PGPR that can indirectly enhance plant growth are the production of NH3 cellulose, HCN, and protease, which protect plants against pathogens [3]. In the present investigation, these important traits were assessed in vitro, and it was found that all seven identified Paenibacillus strains produced these compounds. HCN is a secondary metabolite that plays a role in disease suppression [29]. Microorganisms with cellulose and protease activity not only play a role in organic matter decomposition and plant growth promotion, but also aid in the suppression of disease by the inhibition of fungal growth [2]. NH3 production is another attribute of PGPR that promotes plant growth indirectly, by suppressing plant pathogens [30].
Taken together, the results presented here show that, in laboratory and greenhouse conditions, seven Paenibacillus strains behave as PGPR. It is well known that the capacity of any bacterium to act as a biological control agent could vary between laboratory and field conditions. Therefore, it is critical to assess fungicidal ability and PGP activity of potential PGPR strains in the field, as we have done in this study.
References
- 1.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 2.Kavamura VN, Santos SN, da Silva JL, Parma MM, Ávila LA, Visconti A, Zucchi TD, Taketani RG, Andreote FD, de Melo IS. Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiol Res. 2013;168:183–191. doi: 10.1016/j.micres.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Kumar P, Dubey RC, Maheshwari DK. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res. 2012;167:493–499. doi: 10.1016/j.micres.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Emmert EA, Handelsman J. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol Lett. 1999;171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 5.Von der Weid I, Alviano DS, Santos AL, Soares RM, Alviano CS, Seldin L. Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. J Appl Microbiol. 2003;95:1143–1151. doi: 10.1046/j.1365-2672.2003.02097.x. [DOI] [PubMed] [Google Scholar]
- 6.Deng Y, Lu Z, Lu F, Wang Y, Bie X. Study on an antimicrobial protein produced by Paenibacillus polymyxa JSa-9 isolated from soil. World J Microbiol Biotechnol. 2011;27:1803–1807. [Google Scholar]
- 7.Raza W, Yang W, Shen QR. Paenibacillus polymyxa: antibiotics, hydrolytic enzymes and hazard assessment. J Plant Pathol. 2008;90:419–430. [Google Scholar]
- 8.Ozbay N, Newman SE. Fusarium crown and root rot of tomato and control methods. Plant Pathol J. 2004;3:9–18. [Google Scholar]
- 9.Omar I, O'Neill TM, Rossall S. Biological control of fusarium crown and root rot of tomato with antagonistic bacteria and integrated control when combined with the fungicide carbendazim. Plant Pathol. 2006;55:92–99. [Google Scholar]
- 10.Nishiguchi MK, Doukakis P, Egan M, Kizirian D, Phillips A, Prendini L, Rosenbaum HC, Torres E, Wyner Y, DeSalle R, et al. DNA isolation procedures. In: DeSalle R, Giribet G, Wheeler W, editors. Methods and tools in biosciences and medicine: techniques in molecular systematics and evolution. Basel: Birkhäuser Verlag; 2002. pp. 249–287. [Google Scholar]
- 11.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;57:535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar RS, Ayyadurai N, Pandiaraja P, Reddy AV, Venkateswarlu Y, Prakash O, Sakthivel N. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J Appl Microbiol. 2005;98:145–154. doi: 10.1111/j.1365-2672.2004.02435.x. [DOI] [PubMed] [Google Scholar]
- 13.Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 14.Dey R, Pal KK, Bhatt DM, Chauhan SM. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol Res. 2004;159:371–394. doi: 10.1016/j.micres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Wei G, Kloepper JW, Tuzun S. Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology. 1991;81:1508–1512. [Google Scholar]
- 16.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 17.Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe RC. Comparative pathogenicity and host ranges of Fusarium oxysporum isolates causing crown and root rot of greenhouse and field-grown tomatoes in North America and Japan. Phytopathology. 1980;70:1143–1148. [Google Scholar]
- 19.Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot. 2001;20:1–11. [Google Scholar]
- 20.Dhanasekaran D, Sivamani P, Panneerselvam A, Thajuddin N, Rajakumar G, Selvamani S. Biological control of tomato seedling damping off with Streptomyces sp. Plant Pathol J. 2005;4:91–95. [Google Scholar]
- 21.Lamsal K, Kim SW, Kim YS, Lee YS. Biocontrol of late blight and plant growth promotion in tomato using rhizobacterial isolates. J Microbiol Biotechnol. 2013;23:897–904. doi: 10.4014/jmb.1209.09069. [DOI] [PubMed] [Google Scholar]
- 22.McSpadden Gardener BB. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology. 2004;94:1252–1258. doi: 10.1094/PHYTO.2004.94.11.1252. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro CM, Cardoso EJ. Isolation, selection and characterization of root-associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia) Microbiol Res. 2012;167:69–78. doi: 10.1016/j.micres.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17:319–339. doi: 10.1016/s0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 25.Hameeda B, Harini G, Rupela OP, Wani SP, Reddy G. Growth promotion of maize by phosphate-solubilizing bacteria isolated from composts and macrofauna. Microbiol Res. 2008;163:234–242. doi: 10.1016/j.micres.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Pérez E, Sulbarán M, Ball MM, Yarzábal LA. Isolation and characterization of mineral phosphate-solubilizing bacteria naturally colonizing a limonitic crust in the south-eastern Venezuelan region. Soil Biol Biochem. 2007;39:2905–2914. [Google Scholar]
- 27.Siddiqui ZA. PGPR: prospective biocontrol agents of plant pathogens. In: Siddiqui ZA, editor. PGPR: biocontrol and biofertilization. Dordrecht: Springer; 2006. pp. 111–142. [Google Scholar]
- 28.Raza W, Hongsheng W, Qirong S. Response of Paenibacillus polymyxa to iron: alternations in cellular chemical composition and the production of fusaricidin type antimicrobial compounds. Braz Arch Biol Technol. 2010;53:1145–1154. [Google Scholar]
- 29.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 30.Minaxi LN, Yadav RC, Saxena J. Characterization of multifaceted Bacillus sp. RM-2 for its use as plant growth promoting bioinoculant for crops grown in semi arid deserts. Appl Soil Ecol. 2012;59:124–135. [Google Scholar]