Abstract
A methyl ester of hyaluronan in which the carboxyl groups were fully esterified was prepared using trimethylsilyl diazomethane. This derivative, while not depolymerized by hyaluronan lyases or hyaluronan hydrolases, was a substrate for both chondroitin ACI lyase (EC 4.2.2.5) from Flavobacterium heparinum and chondroitin ACII lyase (EC 4.2.2.5) from Arthrobacter aurescens. The major product isolated in these depolymerization reactions was methyl α-l-threo-hex-4-enepyranosyluronate-(1→3)-2-acetamido-2-deoxy-α,β-d-glucopyranoside as determined by 1H NMR spectroscopy and MALDITOF mass spectrometry.
Keywords: Methyl ester of hyaluronan, Chondroitin AC lyase, Chondroitinase, Flavobacterium heparinum, Arthrobacter aurescens
1. Introduction
Hyaluronan is a linear glycosaminoglycan with a repeating core of disaccharide structure comprised of a d-glucopyranosyluronic acid glycosidically linked to a 2-acetamido-2-deoxy-d-glucopyranose residue.1–3 Hyaluronan is the simplest of the glycosaminoglycans, the only one not covalently linked to a core protein, not synthesized in the Golgi, and not sulfated.1–3 Despite its lack of branching and the monotony of its saccharide composition, hyaluronan has a great number of diverse and important biological and physiological functions.4–7 Hyaluronan promotes cell motility,8 regulates cell–cell and cell–matrix adhesion,9 promotes proliferation,10 and suppresses processes such as embryological development and morphogenesis,11 promotes wound healing,12 repair and regeneration,13 and is involved in inflammation.14 Hyaluronan levels increase in response to severe stress,15 as well as in tumor progression and invasion.16 Recent studies have indicated that hyaluronan can also exist intracellularly, however, the intracellular functions of hyaluronan are unknown.17 Chondroitin has a structure similar to that of hyaluronan in which the 2-acetamido-2-deoxy-d-glucopyranose residue of hyaluronan is replaced with a 2-acetamido-2-deoxy-d-galacto-pyranose residue.
While a single enzyme is now recognized as being able to synthesize hyaluronan,18 hyaluronidases fall into three classes of enzymes based on the structure of their reaction products:19–21 (1) Bacterial hyaluronidases (EC 4.2.99.1) are endo-β-acetyl-hexosaminidases that function as eliminases affording disaccharide products.20,22 In contrast to their eukaryotic counterparts, bacterial hyaluronidases are specific for hyaluronan. (2) Hyaluronidases (EC 3.2.1.36) found in leeches, crustanceans, and some parasites, are endo-β-glucuronidases that function as hydrolases, generating tetrasaccharide and hexasaccharide products.20,23 (3) Mammalian hyaluronidases (EC 3.2.1.35) are endo-β-acetylhexosaminidases that function as hydrolases, affording tetrasaccharide as their major product.24,25 Mammalian hyaluronidases lack substrate specificity and can act at a slow rate on chondroitin sulfates. In addition, the mammalian hyaluronidases also display transglycosidase activity, and can generate complex cross-linked hybrid chains in vitro; however, this transglycosidase activity has not been documented in vivo. While polysaccharide hydrolases have been studied intensely and reaction mechanisms of these enzymes are well characterized,26,27 the reaction mechanisms for polysaccharide lyases are relatively less understood.
Chondroitin lyases are also capable of acting on hyaluronic acid. Chondroitin AC lyase (EC 4.2.2.5) from Flavobacterium heparinum (described as ‘chondroitin lyase ACI Flavo’ in this paper), degrades chondroitin, chondroitin 4-sulfate (CS-A), chondroitin 6-sulfate (CS-C), and hyaluronan.28,29 The action pattern for this enzyme has been established as random endolytic.29,30 Chondroitin AC lyase (EC 4.2.2.5) from Arthrobacter aurescens (chondroitin lyase ACII Arthro) acts in an exolytic fashion on chondroitin, CS-A, CS-C, and hyaluronan.31 Dermatan sulfate containing l-idopyranosyluronic acid has an inhibitory effect on both these AC lyases.32 While there is no absolute requirement of a metal ion for chondroitin AC lyase activity, various mono- and divalent metals have been shown to enhance enzyme activity.29
The current study demonstrates for the first time that both chondroitin lyase ACI Flavo and chondroitin lyase ACII Arthro can act on the methyl ester of hyaluronan. While the action of chondroitin lyase ACI Flavo on the methyl ester of chondroitin has been previously reported, chondroitin lyase ACII Arthro was not observed to act on this substrate.33 The discovery of this new substrate, the methyl ester of hyaluronan, and the activity of both chondroitin lyase ACI Flavo and chondroitin lyase ACII Arthro is significant in that it demonstrates that the negatively charged d-glucopyranosyluronic acid residue is not required by either enzyme, thus, further clarifying its role on the activity of both chondroitin lyases.
2. Experimental
2.1. Chemicals and instruments
Hyaluronan sodium salt (MW 20,000) from Streptococcus zooepidemicus was purchased from Kibun Food Chemipha Co., Tokyo, Japan. Hyaluronidases from Streptomyces hyalurolyticus (lyase, EC 4.2.2.1), from bovine testis (hydrolase, EC 3.2.1.35) and from Streptococcus dysgalactiae (lyase, EC 4.2.2) for hyaluronic acid oligosaccharide preparation was purchased from Seikagaku Kogyo Co. (Tokyo, Japan), Calbiochem (Darmstadt, Germany) and Seikagaku Kogyo, respectively. Chondroitin lyase ACI Flavo (F. heparinum, EC 4.2.2.5), chondroitin lyase ACII Arthro (A. aurescens, EC 4.2.2.5) and chondroitin lyase ABC (Proteus vulgaris, EC 4.2.2.4) were from Seikagaku Kogyo. The conditions used for the degradation of hyaluronan and derivatized hyaluronan samples were according to the procedures provided by each company. Briefly, each enzyme was used for degradation of 1 mg substrate in 1.0 mL buffer under conditions as follows: 10 TRU [1 TRU (Turbidity Reducing Unit) is that amount of enzyme which causes 50% of decrease in absorbance at 660 nm in 30 min at 60 °C] S. hyalurolyticus hyaluronidase at 60 °C for 24 h in 0.04 M NaOAc buffer (pH 6.0); 0.1 U (1 U is defined as the quantity of the enzyme that liberates 1 μmol of the unsaturated disaccharide from hyaluronan per minute at 37 °C, pH 6.2) S. dysgalactiae hyaluronidase at 37 °C for 24 h in 50 mM sodium phosphate buffer (pH 6.2); 100 TRU [1 TRU (Turbidity Reducing Unit) is that amount of enzyme that causes 50% of decrease in absorbance at 600 nm in 30 min at 37 °C, pH 5.3] sheep testicular hyaluronidase at 37 °C for 24 h in 0.1 M sodium phosphate buffer (pH 5.3) containing 0.15 M NaCl; 0.1 U (1 U is defined as the quantity of the enzyme that catalyzes the formation of 1 μmol of the unsaturated disaccharide from chondroitinase C from shark cartilage per minute at 37 °C, pH 8.0) chondroitin lyase ABC at 37 °C for 24 h in 0.1 M Tris–acetate buffer (pH 6.2); 0.1 U (1 U is defined as the quantity of the enzyme that catalyzes the formation of 1 μmol of the unsaturated disaccharide from chondroitinase C from shark cartilage per minute at 37 °C, pH 7.3) chondroitin lyase ACI Flavo at 37 °C for 24 h in 0.4 M Tris–acetate buffer (pH 6.0); 0.1 U (1 U is defined as the quantity of the enzyme which catalyzes the formation of 1 μmol of the unsaturated disaccharide from chondroitinase C from shark cartilage per minute at 37 °C, pH 6.0) chondroitin lyase ACII Arthro at 37 °C for 24 h in 0.4 M acetate buffer (pH 6.0). All other chemicals were of analytical reagent grade.
The CE system was assembled with a Beckman capillary electrophoresis system (P/ACE 5010) equipped with a UV detector and an operating system using version 0.4 P/ACE station on an IBM-compatible PC, from Beckman, USA. JEOL GSX500A and ECP600 NMR instruments, equipped with a 5-mm field-gradient tunable probe with standard JEOL software, were used for 1H NMR experiments at 60 °C on 500 μL of each sample.
2.2. MALDITOF MS
MALDITOF mass spectra were collected as follows: Mass analysis was carried out in negative/positive linear and reflector mode using an Axima™ (Shimadzu Kratos Inc, Kyoto) instrument equipped with a 337-nm nitrogen laser. The acceleration voltage was set to 19 kV, and the delay time was 450 ns. A total of 100 mass spectra were acquired and summed for each sample spot. Mass calibrations were performed over several m/z ranges, using ProteoMass™Peptide Protein MALDI-MS Calibration Kit purchased from Sigma–Aldrich, Japan (Tokyo). Each sample (50 μg) was mixed with 100 μL of a solvent mixture, 30% acetonitrile containing 0.1% trifluoroacetic acid (TFA). Sample solution (1 μL) was mixed with 1 μL of a 10-μg/μL solution of 2,5- dihydroxybenzoic acid (DHB) in TFA. This preparation (1.4 μL) was placed onto a MALDI-sample plate.
2.3. Capillary electrophoresis (CE)
CE was performed using a Beckman P/ACE5010 system with advanced computer interface, equipped with high-voltage power supply capable of constant or gradient voltage control using a fused-silica capillary from GL Science (Tokyo). The compositional analysis of hyaluronic acid oligosaccharide mixture was confirmed by CE in the normal polarity mode using a mixture of 40 mM disodium phosphate/40 mM sodium dodecylsulfate/10 mM tetraborate adjusted to pH 9.0 with 1.0 M hydrochloric acid as described previously.34 The fused-silica capillary (75 μm I.D. × 375 μm O.D., 67 cm long) was automatically washed before use with 0.1 M sodium hydroxide, followed by nitrogen gas pressure injection (5 s) at a constant current 15 kV. The samples (1 mg/mL) were dissolved in water and loaded (7 nL) with nitrogen gas pressure injection.
2.4. NMR spectroscopy
1D and 2D 1H NMR spectroscopy was performed under conditions described previously.35 Briefly, each sample (~2.0 mg) was dissolved in 0.5 mL of 2H2O (99.9%) and freeze-dried repeatedly to remove exchangeable protons. The sample was kept in a desiccator over phosphorous pentoxide in vacuo overnight at room temperature. The thoroughly dried sample was dissolved in 500 μL of 2H2O (99.96%) and passed through 0.45-μm syringe filter and transferred to an NMR tube (5.0 mm O.D. × 25 cm). The HO2H signal was suppressed by presaturation during 3 and 1.5 s for 1D and 2D NMR experiments, respectively. To obtain 2D spectra, 1024 × 512 data matrix for a spectra width of 2000 Hz were measured, and the time domain data were multiplied after zero-filling (data matrix size 1 K × 1 K) with a shifted sine-bell window functions.
2.5. Preparation of the methyl ester of hyaluronan
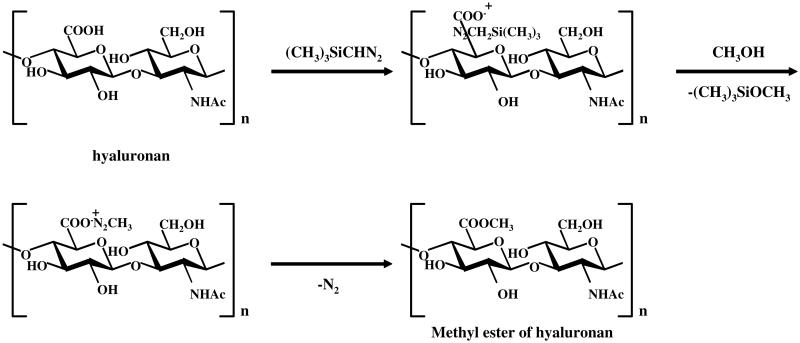
The methyl ester of hyaluronan was prepared as follows (Fig. 1). Hyaluronan (10 mg) was dissolved in 0.5 mL of water and applied to a Dowex 50X8 cation-exchange column (7.5 mm I.D. × 87 mm, H+ form), and the acidic fraction was collected and then freeze dried. The prepared hyaluronan (H+ form, 1 mg) was dissolved in a mixture of 1 mL of DMSO and 50 μL of MeOH, and then, 15 μL of 2 M trimethylsilyl diazomethane (TMSD) in hexane was added and allowed to stand for 60 min at room temperature. The hyaluronan sample used in these experiments had a relatively low molecular weight (average molecular weight 20,000 Da) allowing it to dissolve in DMSO at the concentrations used. A same aliquot of TMSD was added to the reaction mixture and let to stand for another 60 min, and 15 μL of HOAc was added to remove TMSD. To this reaction mixture, 1.8 mL of EtOH saturated with anhyd NaOAc was added and kept at 0 °C for 1 h. The reaction mixture was centrifuged and the precipitate was dissolved in 3 mL of water, and then 3 mL of EtOAc was added, mixed vigorously for 30 s, and centrifuged at 1000g. The water layer obtained after centrifugation was dialyzed against water and lyophilized. The methyl ester of hyaluronan obtained was used for further characterization and for the enzymatic degradation studies.
Figure 1.
Preparation of the methyl ester of hyaluronan.
2.6. Preparation of enzymatic degradation product from the methyl ester of hyaluronan
The methyl ester of hyaluronan (10 mg) was dissolved in 10 mL of each suitable buffer and was incubated under the optimum conditions described in Section 2.1. Hyaluronidase, or chondroitin lyase (1 or 0.1 U each) was added to the reaction medium, and the mixture was incubated for 24 h at 37 °C. Progress of the enzymatic depolymerization reaction was followed by measuring the absorbance of the sample at 232 nm. The resulting oligosaccharide products were fractionated on a Bio-Gel P4 column (2.5 cm × 80 cm; BioRad, Hercules, CA) at 0.7 mL/min. The eluent was monitored at 232 nm, and fractions corresponding to each oligosaccharide were collected.
2.7. Determination of the kinetic constants for the enzymes acting on the methyl ester of hyaluronan
The Michaelis–Menten constants (KM) and the maximum velocities (Vmax) of chondroitin AC lyases were measured for two substrates, hyaluronan and methyl-esterified hyaluronan. Each substrate was used at seven concentrations (5, 10, 20, 40, 60, 80, and 100 mM) in 50 mM Tris–HCl–NaOAc buffer (pH 8.0). Chondroitin AC lyase (10 mU) was added, and the reaction was incubated at 37 °C. Beginning at initiation of the enzymatic reaction, the change in absorbance at 232 nm was measured over 3 min, and the linear region corresponding to <10% of total depolymerization was used to calculate initial velocity of the reaction. The initial velocity determinations at different concentrations were used with the Michaelis–Menten equation to determine Vmax and KM of the enzyme for each substrate.
3. Results and discussion
3.1. Preparation and characterization of the methyl ester of hyaluronan
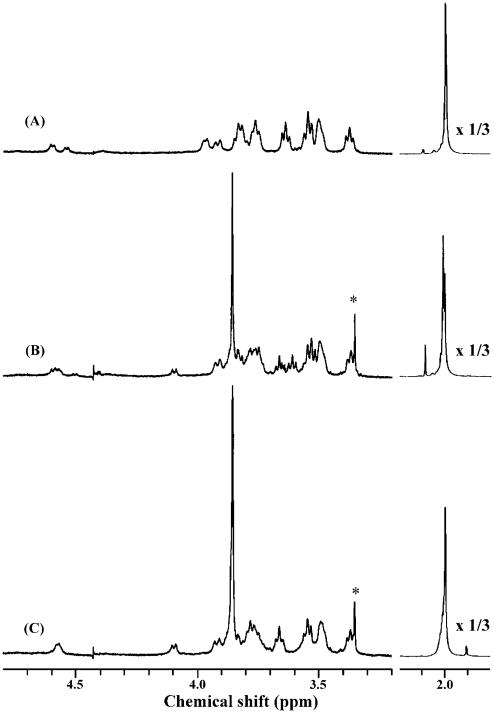
The production of the methyl ester of chondroitin was previously observed by Kantor and Schubert36 using acidic methanol. However, this method requires a prolonged reaction time, results in ~80% methyl-esterification, and causes significant depolymerization of the polysaccharide (data not shown). In contrast, trimethylsilyl diazomethane (TMSD) is a mild, high-yielding esterification procedure that has been successfully used for the methylation of the carboxyl groups in a variety of compounds.37 The methyl-esterification of hyaluronan was optimized using trimethylsilyl diazomethane. The optimal conditions for esterification of carboxyl groups of glucuronate residues were established using 1D NMR characterization of the derivatives. Figure 2 shows the 1H NMR spectra of hyaluronan and the partially esterified (~80%) and fully esterified hyaluronan. The singlet at 3.85 ppm corresponding to three protons clearly shows that carboxyl groups of glucuronate residues have been methyl-esterified. The intensity of this signal was compared to the N-acetyl (CH3–) signal observed at 2 ppm to calculate percent esterification. Furthermore, the doublet at 4.1 ppm corresponding to H-5 proton of glucuronate was downfield shifted in the presence of the methyl-esterified carboxyl group. These data clearly show that the hyaluronan is fully methyl-esterified in the optimized procedure.
Figure 2.
1H NMR spectra of hyaluronan and methyl ester of hyaluronan derivatives. (A) Hyaluronan, (B) partially methyl-esterified hyaluronan (~80%), (C) fully methyl-esterified hyaluronan. (*) Impurities from a vacuum pump.
3.2. Enzymatic degradation of the methyl ester of hyaluronan and characterization of the products
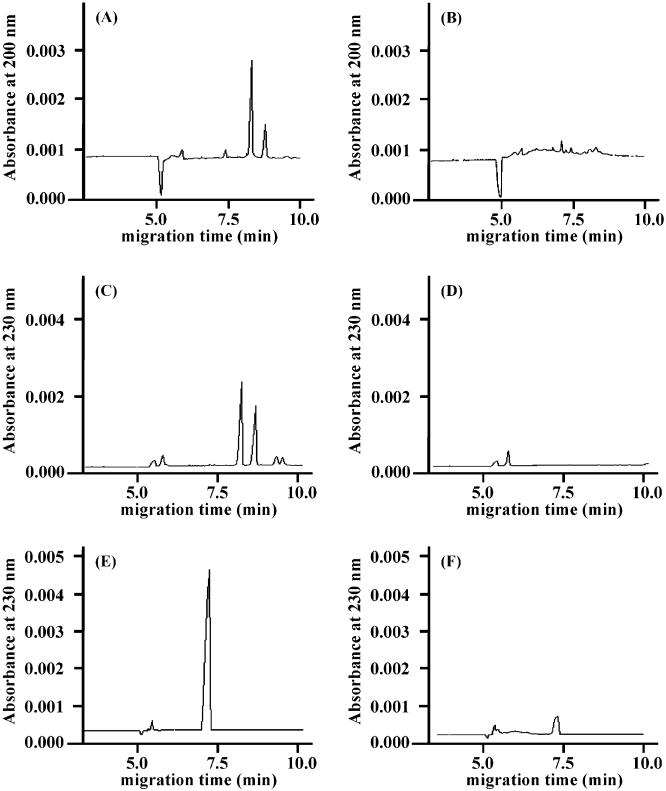
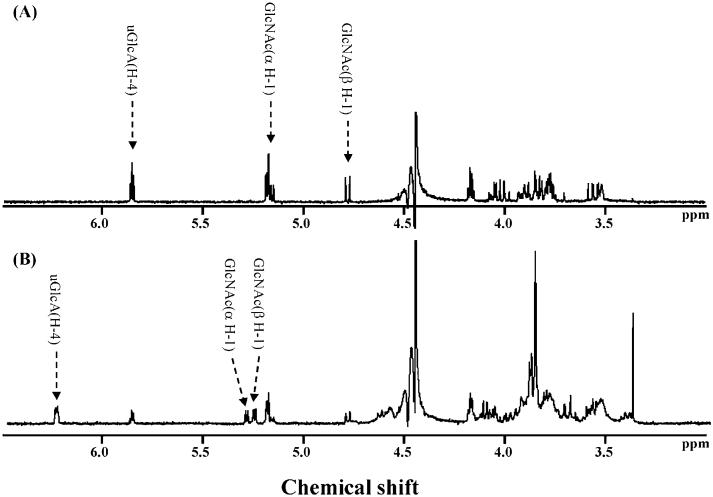
The methyl ester of hyaluronan prepared using the optimized conditions was treated with hyaluronidases and chondroitin lyases, and the products that were formed were analyzed by CE (Figs. 3 and 4). The UV absorbance was determined at 230 nm for the lyases and at 200 nm for the hydrolase enzyme. While three hyaluronidases and chondroitin lyase ABC showed products from hyaluronan, no products were observed using the methyl ester of hyaluronan as a substrate (Fig. 3). Chondroitin lyase ACI Flavo and chondroitin lyase ACII Arthro both afforded a single major product from hyaluronan migrating at 7.5 min and a different major product migrating at 5.2 min when using the methyl ester of hyaluronan as a substrate. Since methyl-esterification of a carboxy group at the nonreducing end decreases the absorbtivity coefficient of the product compared to the product from intact hyaluronan at 230 nm as indicated previously,33 the response on CE profile of each product from methyl-esterified hyaluronan was smaller than those of the products from intact hyaluronan. The migration velocity of molecules on normal polarity is ordered as those of cationic, neutral, and anionic molecules. As expected, the product obtained from the fully methyl ester of hyaluronan migrated faster than that obtained from intact hyaluronan (Fig. 4). F Based on these results, each product obtained from chondroitin lyase-treated methyl ester of hyaluronan was purified by gel-filtration HPLC. Only one product, corresponding to a disaccharide on gel-filtration chromatography, was obtained from each enzymatic degradation mixture, consistent with CE analyses. The structures of these products were further characterized by both 500-MHz 1H NMR spectroscopy and MALDITOF-MS spectrometry. The 1D NMR spectra of the products obtained using chondroitin ACII Arthro lyase acting on hyaluronan and on the methyl ester of hyaluronan are shown in Figure 5. Spectrum A is consistent with the structure of ΔUA-GlcpNAc (where ΔUA is 4-deoxy-α-l-threo-hex-4-enopyranosyluronic acid and GlcpNAc is 2-acetamido-2-deoxy-d-glucopyranose), while spectrum B is consistent with that of the methyl ester of ΔUA-GlcpNAc. Identical results were obtained on the analysis of the chondroitin lyase ACI Flavo products. A small amount of ΔUA-GlcpNAc was observed in both MALDITOF-MS (not shown) and 1H NMR spectra in the product obtained from the methyl ester of hyaluronan, suggesting some hydrolysis had taken place during the purification procedures. This is consistent with the observation of a minor peak in CE analysis migrating at 7.5 min corresponding to ΔUA-GlcpNAc (Fig. 4B and D) that appeared only after prolonged incubation at 37 °C. The identity of the methyl ester of ΔUA-GlcpNAc was confirmed as a peak at 416.0 amu in MALDI MS.
Figure 3.
Capillary electropherograms of the products after the enzymatic digestion by hyaluronidases. (A) Hyaluronan treated with hyaluronidase from bovine testis, (B) methyl-esterified hyaluronan treated by hyaluronidase from bovine testis, (C) hyaluronan treated with hyaluronidase from Streptomyces hyalurolyticus, (D) methyl-esterified hyaluronan treated with hyaluronidase from Streptomyces hyalurolyticus, (E) hyaluronan treated with hyaluronidase from Streptococcus dysgalactiae, (F) methyl-esterified hyaluronan treated with hyaluronidase from Streptococcus dysgalactiae.
Figure 4.
Capillary electropherograms of the products after the enzymatic digestion by chondroitin lyases. (A) Hyaluronan treated with chondroitin lyase ACI Flavo, (B) methyl-esterified hyaluronan treated with chondroitin lyase ACI Flavo, (C) hyaluronan treated with chondroitin lyase ACII Arthro, (D) methyl-esterified hyaluronan treated with chondroitin lyase ACII Arthro, (E) hyaluronan treated with chondroitin lyase ABC, (F) methyl-esterified hyaluronan treated with chondroitin lyase ABC.
Figure 5.
1H NMR spectra of the end products derived from the methyl ester of hyaluronan by chondroitin lyase ACII Arthro. (A) Hyaluronan treated by chondroitin lyase ACII Arthro, (B) methyl-esterified hyaluronan treated with chondroitin lyase ACII Arthro.
It is interesting to note that the products obtained from the methyl ester of chondroitin by chondroitin lyase ACI Flavo were reportedly a mixture of oligosaccharides following complete enzymatic digestion.33 This is in contrast to the formation of only a disaccharide from treatment of methyl ester of hyaluronan with chondroitin lyase ACI Flavo. Furthermore, chondroitin lyase ACII Arthro showed no observable activities on the methyl ester of chondroitin, whereas it showed substantial activity on methyl-esterified hyaluronan. These results might be rationalized by the difference in the C-4 configuration (gluco vs galacto) in these two substrates, and a thorough study of the X-ray crystal study for these enzyme–substrate complexes might provide some additional insight.
In our previous study, we showed that the methyl ester of chondroitin that was depolymerized by chondroitin lyase ACI Flavo resulted in formation of 60% disaccharide and 20% tetrasaccharide products,33 and chondroitin lyase ACII Arthro acted very poorly on the same substrate leading to <10% completion of the depolymerization reaction (data not shown). Thus, it was unclear whether chondroitin lyase ACII Arthro was acting on methyl-esterified chains or just a fraction of chains that still contained free carboxyl groups (20–30% residual carboxyl groups were present in our substrate).33
3.3. Determination of the kinetic constants for chondroitin ACI and ACII lyases acting on methyl-esterified of hyaluronan
The catalytic mechanism of chondroitin lyase ACI Flavo on chondroitin sulfate has been proposed to involve general acid–base type catalysis, and it has been suggested that this catalytic mechanism is stepwise38 requiring the stabilization of the doubly charged enolic intermediate. This must be achieved with a positively charged amino acid residue at the active site binding to the carboxyl group or through binding of a metal cation.39–42 The esterification of the carboxyl group in both methyl-esterified chondroitin and hyaluronan might alter the interaction of anion-stabilizing elements in an enzymatic reaction, thus adversely impacting catalysis. Kinetic studies show that the KM on the methyl ester of hyaluronan is comparable to that for intact hyaluronan and lower than that observed on chondroitin (Table 1). In contrast, the Vmax on the methyl ester of hyaluronan is significantly lower than that on hyaluronan, suggesting that methylation of the carboxyl group less adversely impacts the binding step than the catalytic step. The low KM observed for the methyl ester of hyaluronan might be ascribed to the contribution of hydrophobic interactions between the methyl ester of the methyl ester of hyaluronan and the enzyme.
Table 1.
Kinetic parameters of chondroitin lyase ACI Flavo and chondroitin lyase ACII Arthro
| Substrate | KM (μM) | Vmax (μmol/min) | Vmax/KM | |
|---|---|---|---|---|
| Chondroitin lyase ACI Flavo | Hyaluronan | 11.4 | 0.91 | 0.080 |
| O-Methyl-esterified hyaluronan | 0.82 | 0.19 | 0.23 | |
| Chondroitin | 26.7 | 8.40 | 0.31 | |
| O-Methyl-esterified chondroitin | 6.17 | 2.90 | 0.47 | |
| Chondroitin lyase ACII Arthro | Hyaluronan | 175.6 | 11.8 | 0.067 |
| O-Methyl-esterified hyaluronan | 1.19 | 0.14 | 0.12 |
The catalytic activity of chondroitin lyase ACI Flavo acting on four different substrates can be conveniently compared by its Vmax/KM values (Table 1). It is interesting to note that the best substrate for this substrate is the unnatural methyl ester of chondroitin. Furthermore, both of the unnatural substrate methyl esters of chondroitin and hyaluronan are better substrates than their natural counterparts, chondroitin and hyaluronan. It must be noted, however, that the standard substrates for this enzyme chondroitin 4-sulfate and chondroitin 6-sulfate are 2- to 5-fold better substrates than unsulfated chondroitin and hyaluronan.33,43
Crystal structures of both chondroitin lyase ACI Flavo42,43 and chondroitin lyase ACII Arthro44 complexed with GAG oligosaccharides have been published. Soaking with various oligosaccharides indicated that chondroitin lyase ACII Arthro displays higher affinity toward hyaluronan than to chondroitin sulfate.44 We have determined the kinetic parameters of chondroitin lyase ACII Arthro with chondroitin sulfate and hyaluronan oligosaccharides and found that Vmax for hyaluronan is twice as high as that for chondroitin sulfate, while the KM calculated on a per monomer base is about three times lower (data not shown). These values suggest a small contribution made by the sulfate groups to the total binding energy of the substrate.44
Acknowledgements
This work was supported in part by Grants-in-Aid from the Ministry of Culture and Education of Japan (11672136 and 14572029, T.T.). We also thank Seikagaku Kogyo Co. Ltd (Tokyo) for their help in providing the information of the enzymes.
References
- 1.Meyer K, Palmer JW. J. Biol. Chem. 1934;107:629–634. [Google Scholar]
- 2.Toole BP. Hyaluronan. In: Iozzo RV, editor. Proteoglycans. Marcel Dekker; New York: 2000. pp. 61–92. [Google Scholar]
- 3.Laurent TC, editor. The Chemistry, Biology and Medical Applications of Hyaluronan and its Derivatives. Vol. 72. Portland Press; London: 1998. Wenner–Gren International Series. [Google Scholar]
- 4.Stern R. Eur. J. Cell Biol. 2004;83:317–325. doi: 10.1078/0171-9335-00392. [DOI] [PubMed] [Google Scholar]
- 5.Toole BP. Glycobiology. 2002;12:37R–42R. doi: 10.1093/glycob/12.3.37r. [DOI] [PubMed] [Google Scholar]
- 6.Adamia S, Maxwell CA, Pilarski LM. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2005;5:3–14. doi: 10.2174/1568006053005056. [DOI] [PubMed] [Google Scholar]
- 7.Knudson CB, Knudson W. Clin. Orthop. Relat. Res. Suppl. 2004;427:S152–S162. [PubMed] [Google Scholar]
- 8.Liu N. Lymphat. Res. Biol. 2003;1:67–70. doi: 10.1089/15396850360495718. [DOI] [PubMed] [Google Scholar]
- 9.Skubitz AP. Cancer Treat. Res. 2002;107:305–329. doi: 10.1007/978-1-4757-3587-1_15. [DOI] [PubMed] [Google Scholar]
- 10.Hascall VC, Majors AK, De La Motte CA, Evanko SP, Wang A, Drazba JA, Strong SA, Wight TN. Biochim. Biophys. Acta. 2004;1673:3–12. doi: 10.1016/j.bbagen.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Spicer AP, Tien JY. Birth Defects Res. C. Embryo Today. 2004;72:89–108. doi: 10.1002/bdrc.20006. [DOI] [PubMed] [Google Scholar]
- 12.Brown JA. J. Wound Care. 2004;13:48–51. doi: 10.12968/jowc.2004.13.2.26573. [DOI] [PubMed] [Google Scholar]
- 13.Turino GM, Cantor JO. Am. J. Resp. Crit. Care Med. 2003;167:1169–1175. doi: 10.1164/rccm.200205-449PP. [DOI] [PubMed] [Google Scholar]
- 14.Zhuo L, Hascall VC, Kimata K. J. Biol. Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- 15.Eboumbou C, Steghens JP, Abdallahi OM, Mirghani A, Gallian P, van Kappel A, Qurashi A, Gharib B, De Reggi M. Acta Trop. 2005;94:99–106. doi: 10.1016/j.actatropica.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Hotta T, Taniguchi K, Kobayashi Y, Johata K, Sahara M, Naka T, Tanimura H, Tsubota YT. Hepatogastroenterology. 2004;51:1073–1083. [PubMed] [Google Scholar]
- 17.Afify AM, Stern R, Michael CW. Diagn. Cytopathol. 2005;32:145–150. doi: 10.1002/dc.20201. [DOI] [PubMed] [Google Scholar]
- 18.Spicer AP, Nguyen TK. Biochem. Soc. Trans. 1999;27:109–115. doi: 10.1042/bst0270109. [DOI] [PubMed] [Google Scholar]
- 19.Jedrzejas MJ. Crit. Rev. Biochem. Mol. Biol. 2000;35:221–251. doi: 10.1080/10409230091169195. [DOI] [PubMed] [Google Scholar]
- 20.Sutherland IW. FEMS Microbiol. Rev. 1995;16:323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 21.Menzel EJ, Farr C. Cancer Lett. 1998;131:3–11. doi: 10.1016/s0304-3835(98)00195-5. [DOI] [PubMed] [Google Scholar]
- 22.Jedrzejas MJ, Mello LV, de Groot BL, Li S. J. Biol. Chem. 2002;277:28287–28297. doi: 10.1074/jbc.M112009200. [DOI] [PubMed] [Google Scholar]
- 23.Hovingh P, Linker A. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999;124:319–326. doi: 10.1016/s0305-0491(99)00128-5. [DOI] [PubMed] [Google Scholar]
- 24.Meyer K, Boyer PD. The Enzymes. Academic Press; New York: 1971. Hyaluronidases; pp. 307–320. [Google Scholar]
- 25.Frost GI, Csoka TB, Wong T, Stern R. Biochem. Biophys. Res. Commun. 1997;236:10–15. doi: 10.1006/bbrc.1997.6773. [DOI] [PubMed] [Google Scholar]
- 26.Hrmova M, Fincher GB. Plant Mol. Biol. 2001;47:73–91. [PubMed] [Google Scholar]
- 27.Papanikolau Y, Prag G, Tavlas G, Vorgias CE, Oppenheim AB, Petratos K. Biochemistry. 2001;40:11338–11343. doi: 10.1021/bi010505h. [DOI] [PubMed] [Google Scholar]
- 28.Yamagata T, Saito H, Habuchi O, Suzuki S. J. Biol. Chem. 1968;243:1523–1535. [PubMed] [Google Scholar]
- 29.Gu K, Linhardt RJ, Laliberte M, Gu K, Zimmermann J. Biochem. J. 1995;312:569–577. doi: 10.1042/bj3120569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jandik KA, Gu K, Linhardt RJ. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- 31.Hiyama K, Okada S. J. Biochem. 1976;80:1201–1207. doi: 10.1093/oxfordjournals.jbchem.a131390. [DOI] [PubMed] [Google Scholar]
- 32.Gu K, Liu J, Pervin A, Linhardt RJ. Carbohydr. Res. 1993;244:369–377. doi: 10.1016/0008-6215(83)85014-9. [DOI] [PubMed] [Google Scholar]
- 33.Avci FY, Toida T, Linhardt RJ. Carbohydr. Res. 2003;338:2101–2104. doi: 10.1016/s0008-6215(03)00348-3. [DOI] [PubMed] [Google Scholar]
- 34.Al-Hakim A, Linhardt RJ. Anal Biochem. 1991;195:68–73. doi: 10.1016/0003-2697(91)90296-6. [DOI] [PubMed] [Google Scholar]
- 35.Toida T, Yoshida H, Toyoda H, Koshiishi I, Imanari T, Hileman RE, Fromm JR, Linhardt RJ. Biochem. J. 1997;322:499–506. doi: 10.1042/bj3220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kantor TG, Schubert M. J. Am. Chem. Soc. 1957;79:152–153. [Google Scholar]
- 37.Moy TW, Brumley WC. J. Chromatogr. Sci. 2003;41:343–349. doi: 10.1093/chromsci/41.7.343. [DOI] [PubMed] [Google Scholar]
- 38.Rye CS, Withers SG. J. Am. Chem. Soc. 2002;124:9756–9767. doi: 10.1021/ja020627c. [DOI] [PubMed] [Google Scholar]
- 39.Gacesa P. FEBS Lett. 1987;212:199–202. [Google Scholar]
- 40.Gerlt JA, Gassman PJ. J. Am. Chem. Soc. 1992;114:5928–5934. [Google Scholar]
- 41.Guthrie JP, Kluger R. J. Am. Chem. Soc. 1993;115:11569–11572. [Google Scholar]
- 42.Huang W, Boju L, Tkalec L, Su H, Yang H, Gunay NS, Linhardt RJ, Kim YS, Matte A, Cygler M. Biochemistry. 2001;40:2359–2372. doi: 10.1021/bi0024254. [DOI] [PubMed] [Google Scholar]
- 43.Capila I, Wu Y, Rethwisch DW, Matte A, Cygler M, Linhardt RJ. Biochim. Biophys. Acta. 2002;1597:260–270. doi: 10.1016/s0167-4838(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 44.Lunin VV, Li Y, Linhardt RJ, Miyazono H, Kyogashima M, Kaneko T, Bell AW, Cygler M. J. Mol. Biol. 2004;337:367–386. doi: 10.1016/j.jmb.2003.12.071. [DOI] [PubMed] [Google Scholar]