SUMMARY
Lung squamous cell carcinoma (SCC) is a deadly disease for which current treatments are inadequate. We demonstrate that biallelic inactivation of Lkb1 and Pten in the mouse lung leads to SCC that recapitulates the histology, gene expression, and microenvironment found in human disease. Lkb1;Pten null (LP) tumors expressed the squamous markers KRT5, p63 and SOX2, and transcriptionally resembled the basal subtype of human SCC. In contrast to mouse adenocarcinomas, the LP tumors contained immune populations enriched for tumor-associated neutrophils. SCA1+NGFR+ fractions were enriched for tumor-propagating cells (TPCs) that could serially transplant the disease in orthotopic assays. TPCs in the LP model and NGFR+ cells in human SCCs highly expressed Pd-ligand-1 (PD-L1), suggesting a mechanism of immune escape for TPCs.
INTRODUCTION
Lung squamous cell carcinoma (SCC) is a common type of non-small-cell lung cancer and the second leading cause of death related to lung cancer, causing approximately 400,000 deaths per year worldwide (Cancer Genome Atlas Research Network, 2012; Siegel et al., 2013). Unlike lung adenocarcinoma (ADC), for which many relevant oncogenic mutations have been defined and used to develop strategies for targeted therapies, the genomic landscape of lung SCC is only now emerging. There are not yet any approved targeted therapies for lung SCC. Unfortunately, therapeutic targets in lung ADC, such as EGFR and EML4-ALK, do not appear to play major roles in lung SCC (Rekhtman et al., 2012). This fact underscores the need to develop a preclinical model of lung SCC in which to define and test novel therapeutic approaches.
Currently, the field lacks a mouse model in which the introduction of genetic alterations found in human squamous lung cancers leads to tumors of purely squamous phenotype. We previously reported that simultaneous activation of KrasG12D(Kras) and inactivation of Lkb1 (also known as serine-threonine kinase 11 [Stk11]) in the lung gave rise to multiple lung cancer histologies, including SCCs (Ji et al., 2007); however, KRAS mutations are very rarely found in human squamous lung tumors. Recently, it was reported that kinase-dead Ikkα knockin mice developed spontaneous lung SCCs characterized by Ikkα downregulation and marked pulmonary inflammation (Xiao et al., 2013). Significant downregulation of Lkb1 was found in IkkαKA/KA lung SCCs and adjacent lung tissues compared with wild-type lungs. Together these results indicate that Lkb1 reduction is likely an important determinant of lung squamous tumorigenesis.
Despite indications that Lkb1 loss may be central to the generation of squamous cell cancers, deletion of Lkb1 alone is unable to drive tumor formation (Ji et al., 2007). PTEN (phosphatase and tensin homolog) is another commonly mutated, deleted, or epigenetically silenced tumor suppressor in human lung cancers (Salmena et al., 2008). Importantly, PTEN is altered in 15% of human SCCs (Cancer Genome Atlas Research Network, 2012). PTEN negatively regulates the phosphatidylinositol 3-kinase (PI3K)/AKT pathway, and PI3K pathway gene alterations are found in more than half of human lung SCCs (Cancer Genome Atlas Research Network, 2012). In the mouse model, Pten deletion alone in airway basal cells can initiate lung tumor formation, but with low tumor incidence, long latency, and mixed ADC and SCC phenotype (Malkoski et al., 2013).
One key feature of tumor development that autochthonous genetically engineered mouse models provide is a physiologically relevant tumor microenvironment. All of the models of lung SCC to date, including the Ikkα knockin mice and a model driven by chronic tuberculosis infection, showed marked pulmonary inflammation (Nalbandian et al., 2009; Xiao et al., 2013), suggesting that an inflammatory microenvironment is central to the development of lung SCCs. This is not surprising given that nearly all humans with lung SCCs have histories of tobacco use that drives squamous metaplasia, and the development of SCCs is associated with inflammatory diseases and chronic immunosuppression. Both tumor-associated macrophages (TAMs) and tumor-associated neutrophils (TANs) comprise significant proportions of the inflammatory infiltrates in a wide variety of mouse tumor models and human cancers (Murdoch et al., 2008). Neutrophils were shown to predominate in human head and neck squamous carcinomas (Trellakis et al., 2011). Neutrophils found in mouse tumors are phenotypically characterized as polymorphonuclear CD11b+Ly6G+ cells and may be related to a subtype of myeloid-derived suppressive cells (MDSCs). MDSCs encompass a heterogeneous population of myeloid cells, which share the ability to suppress T cells through the production of arginase, the expression of inducible nitric oxide synthase, and other mechanisms (Dumitru et al., 2012). In the tumor microenvironment, accumulated MDSCs are thought to promote tumor progression through enhancing matrix degradation, tumor cell proliferation, metastasis, and angiogenesis (Welch et al., 1989). MDSCs have also been shown to antagonize effector T cell function, support the generation of immunosuppressive T cell populations, and inhibit the lysis of tumor cells by cytotoxic T cells or natural killer (NK) cells (Dumitru et al., 2012). Some MDSCs have neutrophilic features, but the precise relationship between these cells and normal polymorphonuclear leukocytes remains under active investigation. In this paper, we refer to polymorphonuclear cells infiltrating lung cancers as TANs.
Tumors can also evade immune surveillance by expressing molecules that maintain immune tolerance in peripheral tissues, such as Pd-ligand-1 (PD-L1), which interacts with the immune receptor programmed cell death-1 (PDCD1 or PD-1) (Barber et al., 2006). The PD-1/PD-L1 interaction inhibits CD8+ cytotoxic T lymphocyte (CTL) proliferation, survival, and effector function and can induce apoptosis of tumor-infiltrating T cells (Barber et al., 2006); PD-1/PD-L1 interactions can also promote the differentiation of CD4+ T cells into FOXP3+ Tregs (Francisco et al., 2009), which are known to further suppress the immune system and cause peripheral immune tolerance in lung cancer patients (Adeegbe and Nishikawa, 2013). Ectopic PD-L1 expression in tumor cells in a syngeneic transplant model facilitated the escape of the tumor cells from CTL control (Iwai et al., 2002). Consistent with these findings in preclinical systems, infusing lung cancer patients with blocking anti-PD-1/PD-L1 monoclonal antibodies has shown efficacy in early stage trials, despite limited activity of prior immunotherapies for lung malignancies (Brahmer et al., 2012; Topalian et al., 2012).
Tumor-propagating cells (TPCs) have the ability to self-renew and differentiate into the bulk population of the tumor and are thought to drive both disease recurrence and metastatic spread (Visvader and Lindeman, 2012). Stem cell antigen-1 (Sca1 or Ly6a) was reported as a bronchioalveolar stem cell (BASC) marker in the distal lung and is also enriched in bronchiolar progenitor cells (Kim et al., 2005; Lee et al., 2014). SCA1+ cells, located at the bronchioalveolar duct junction, are hyperproliferative in response to both oncogenic Kras and deletion of Pten, suggesting that they are susceptible to neoplastic transformation (Kim et al., 2005; Tiozzo et al., 2009). In addition, SCA1 can be used to enrich for TPCs in the lung ADC KrasG12D;p53fl/fl (Kras;p53) model (Curtis et al., 2010). In the more proximal lung, nerve growth factor receptor (TNFR superfamily, member 16, Ngfr) is a stem cell marker for the pseudostratified tracheal epithelium in both human and mouse; NGFR expression is specifically observed in the p63+ mouse basal stem cells (Rock et al., 2009). NGFR+ basal cells appear to be the cells of origin in a SOX2-induced model of esophageal SCC, and NGFR has been suggested as a putative marker for human esophageal SCC TPCs (Huang et al., 2009; Liu et al., 2013). Despite these clues as to the molecular phenotype of a potential TPC in SCC, no TPC population able to propagate disease serially has been identified for lung SCC.
Using conditional knockout alleles of both Lkb1 and Pten, we found that biallelic inactivation of these two tumor suppressors in the lung resulted in lung tumors with a purely squamous cell phenotype. These squamous lung tumors were 100% penetrant and recapitulated the genetic, molecular, and microenvironmental aspects of the human disease. With this model, the molecular and genetic mechanisms involved in the pathogenesis of lung squamous tumors, including TPCs, microenvironmental factors, immune tolerance, and potential targets for future therapies, can be explored.
RESULTS
Lkb1fl/fl;Ptenfl/fl Mice Develop Lung SCCs that Recapitulate the Human Disease
In order to examine the possibility that Lkb1 and Pten loss would lead to lung SCC formation, 6- to 8-week-old Lkb1fl/fl, Ptenfl/fl (Lkb1, Pten or LP) mice were administered Adenovirus-Cre (Ad-Cre) via intranasal instillation (Figure 1A). In contrast to other lung-specific genetic mouse models described to date, including KrasG12D (Kras), KrasG12D;p53fl/fl (Kras;p53), KrasG12D;Lkb1fl/fl (Kras;Lkb1), and KrasG12D;p53fl/fl;Lkb1fl/fl (Kras;p53;Lkb1), in which the predominant phenotypes are ADC or mixed adenosquamous cell carcinoma, 100% of the LP mice developed typical lung SCCs with 40 to 50 week latency (Figure 1B; Figures S1A and S1B available online). Small malignant nodules with squamous characteristics were evident at different time points after Ad-Cre infection, ranging from 30 to 40 weeks. Both Lkb1 and Pten were confirmed to be homozygously deleted by PCR on genomic DNA from sorted tumor cells (Figure S1C). The LP tumors were verified as recapitulating human SCC pathology: within the tumor nodules, mature squamous cells growing in a solid configuration with aberrant nuclear morphology (Figure 1Ca), large infiltrates of neutrophils (Figure 1Cb), and keratinized cells or individual cells with markedly dense eosinophilic cytoplasm were all observed (Figure 1Cc). Tumors showed hallmarks of well-differentiated SCC, including invading fibrous stroma with prominent keratinization (keratin pearls) (Figure 1Cd). In some cases, SCC nodules were visible in airways (Figure 1Ce) and at later time points showed lymphovascular invasion (Figure 1Cf). Tumors arose in both the proximal (Figure S1D, yellow arrow) and distal lung (Figures S1D and S1E, green arrows), though many nodules appeared to be surrounded completely by alveolar epithelium. Low-frequency metastatic lesions were visible in the chest wall of these mice (3 of 78) (Figure S1F).
Figure 1. Biallelic Inactivation of Both Lkb1 and Pten in the Mouse Lung Leads to SCC.
(A) Schematic of biallelic inactivation of both Lkb1 and Pten in the mouse lung by Ad-Cre inhalation, followed by tumor dissociation and sorting.
(B) Phenotypic quantification of lung tumor histologies from the indicated conditional mouse models, including KrasG12D (Kras), KrasG12D;p53fl/fl (Kras;p53), KrasG12D;Lkb1fl/fl (Kras;Lkb1), and KrasG12D;p53fl/fl;Lkb1fl/fl (Kras;p53;Lkb1), Lkb1fl/fl;Ptenfl/fl (Lkb1, Pten) Lkb1fl/fl;Ptenfl/fl;p53fl/fl (Lkb1;Pten;p53), which all rely upon Ad-Cre inhalation for tumor initiation (mean ± SEM, n = 5 mice per genotype).
(C) Representative H&E-stained sections derived from tumors arising in the Lkb1;Pten mouse model. Yellow arrows indicate specific features on individual images. (a) Mature squamous cells with aberrant nuclear morphology; (b) large infiltrates of neutrophils in SCC nodules; (c) keratinized cells with markedly dense eosinophilic cytoplasm surrounded by epithelial cells; (d) well-differentiated SCC with keratin pearls; (e) SCC nodules in large airways; (f) squamous-like tumor cell lymphovascular invasion.
Black scale bars represent 200 μm, yellow scale bars represent 50 μm, and the green scale bar represents 25 μm. See also Figure S1.
To confirm the phenotype of the LP tumors, we performed immunohistochemistry (IHC) for markers used clinically to distinguish human lung ADC from lung SCC. TTF1 (also known as NKX2-1) and SOX2 are genomically amplified in lung ADCs and SCCs, respectively, and routinely used as histologic markers (Bass et al., 2009; Weir et al., 2007). In addition, positive staining for the markers p63 and keratin-5/6 (KRT5/6) appears to robustly differentiate SCCs from ADCs (Fatima et al., 2012). Similar to the human SCC samples, the LP tumor nodules displayed high expression of p63, KRT5, and SOX2, while TTF1 staining was negative (Figure 2A). The expression patterns of p63, KRT5, and SOX2 in SCC colocalized with the expression of epithelial cell adhesion molecule (EpCAM) (Figure 2A). In contrast, KrasG12D-driven murine ADC and human ADC tissues were p63, KRT5, and SOX2 negative, while TTF1 staining was strongly positive, confirming their ADC phenotype (Figure 2A). Together these data indicate that LP tumors strongly resemble human SCC by their expression of the classic squamous markers p63, KRT5/6 and SOX2, and hallmarks of squamous differentiation such as keratin deposition.
Figure 2. Lung SCCs in Lkb1fl/fl.
;Ptenfl/fl Mice Closely Recapitulated the Human Disease
(A) Immunohistochemically stained human and mouse ADC and SCC tumors. The SCC canonical markers KRT5, SOX2, and p63 and the ADC canonical marker TTF1 were used to distinguish the tumor types. EpCAM is an epithelial cell marker, and the expression patterns of p63, KRT5, and SOX2 in SCC colocalized with EpCAM expression. Scale bars represent 100 μm for all panels.
(B) Microarray expression profiling of normal lung and SCC tumors from mouse and human (see description in text). Upregulated genes in both mouse and human SCC were enriched for a squamous differentiation signature.
(C) Downregulated genes from the analysis in (B) were enriched for a normal lung terminal respiratory unit signature. See also Figure S2.
Next, we compared the transcriptional landscapes of the LP tumors with those found in primary human tumors. To do this, the gene expression profiles of 34 human SCC tumors with either LKB1 or PTEN alterations from the Cancer Genome Atlas (Cancer Genome Atlas Research Network, 2012) were compared with 35 normal human lung tissue samples to generate a list of genotype-specific SCC genes. In parallel, the gene expression profiles of LP tumors from three independent mice were compared with profiles of normal lung from three age-matched LP mice that never received Ad-Cre. In the human comparison, 8,237 genes were significantly differentially expressed in the SCCs versus normal human lung, with a corrected p value (90th percentile false discovery rate) of zero. Compared with normal, 3,658 genes were upregulated in tumors, and 4,579 were downregulated. In the smaller mouse data set, 2,236 genes were differentially expressed, with 916 upregulated and 1,320 downregulated (Figure S2A). Comparison of the mouse and human gene sets yielded 893 genes that were significantly differentially expressed in both human tumors with LKB1 and/or PTEN alterations and LP mouse SCCs (Figure S2B). Among the shared upregulated genes were several known squamous-associated genes, including SOX2, P63, NOTCH3, HRAS, and several keratins (KRT5/KRT6). Gene ontogeny analysis demonstrated enrichment for genes implicated in squamous differentiation (p = 3.62 × 10−10, Figure 2B). In contrast, the shared downregulated genes were enriched for terminal respiratory unit differentiation, consistent with the idea that SCC more closely resembles proximal lung cells than distal epithelia (p = 4.07 × 10−7, Figure 2C).
Lkb1fl/fl;Ptenfl/fl Lung SCCs Display Unique Gene Expression, Metabolism, and Downstream Signaling Pathways
In order to characterize the gene expression profiles specific to the tumor cells within the mouse LP SCCs, we used fluorescence-activated cell sorting (FACS) to enrich for the epithelial cells (CD45−CD31−EpCAM+) from LP SCC tumor nodules, Kras-driven tumor nodules, and normal lung (Figure S3A). We then contrasted the gene expression profiles of these three epithelial cell fractions (Figure 3A, p < 0.001). Remarkably, all of the genes differentially expressed between normal epithelial and LP tumor cells were likewise differentially expressed when comparing Kras tumor with LP tumor cells. This result suggests that Kras tumors retain some gene expression reminiscent of the normal distal lung epithelial cell, from which they likely arise. In contrast, LP tumor cells do not resemble ADC or normal distal lung cells and instead have markers expressed by tracheal basal cells, as discussed below.
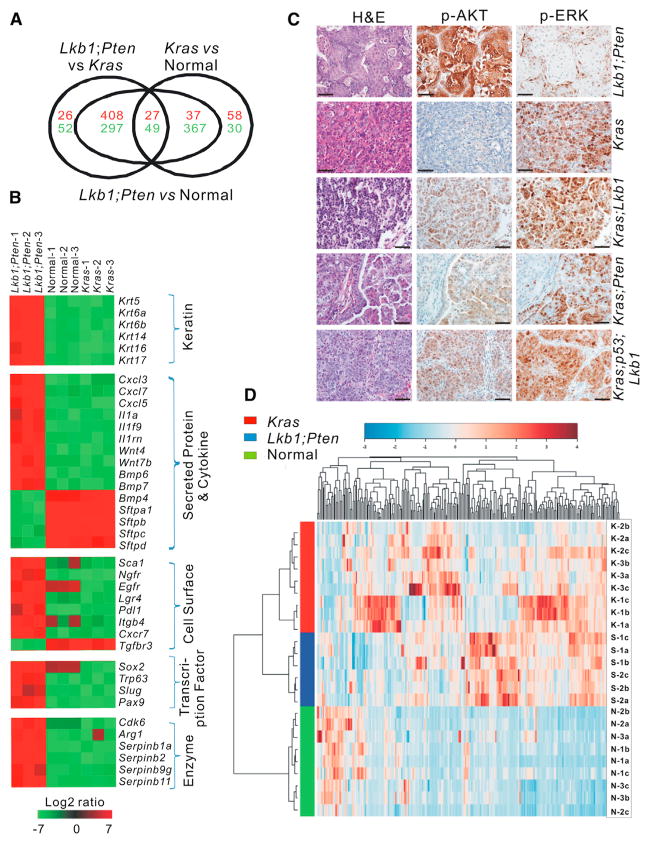
Figure 3. Lkb1fl/fl;Ptenfl/fl Lung SCCs Display Unique Gene Expression, Metabolism, and Downstream Signaling Pathways.
(A) EpCAM+CD31−CD45− cells were isolated by FACS and subject to microarray expression analysis. Euler diagram illustrating the gene expression profiles of epithelial cells from LP SCCs, Kras ADCs, and normal lung tissues.
(B) Heatmap depicting differential expression of selected genes in LP SCCs, Kras ADCs, and normal lung tissues as determined by microarray expression profiling. Red indicates upregulation. and green indicates downregulation.
(C) Immunohistochemical staining for p-AKT and p-ERK on LP, Kras, Kras;Lkb1, Kras;Pten, and Kras;p53;Lkb1 tumor nodules. Scale bars represent 50 μm for all panels.
(D) Hierarchical clustering by Ward method of quantitative metabolomic profiling for LP SCC tumors (S), Kras ADC tumors (K), and normal lung tissues (N).
See also Figure S3 and Tables S1 and S2.
We next focused on the genes that were differentially expressed in the LP tumor cells when compared with both Kras tumor cells and normal lung. In this comparison, 408 genes were upregulated and 297 genes were downregulated, with a log fold change > 1.8 and an adjusted p value < 0.001 (Table S1). Selected genes that can be organized by function or family are illustrated in a heatmap (Figure 3B). Gene sets that were upregulated in LP tumors include the keratin family members, including Krt5, which we observed by IHC, and other squamous keratins such as Krt6a, Krt6b, and Krt14. Also highly upregulated in LP tumors were the transcription factors Sox2 and p63, consistent with our IHC results, and Slug and Pax9. Among the secreted proteins and cytokines produced by these tumors were several Cxcl family members, including Cxcl3, Cxcl7, and Cxcl5, and members of the Wnt, Bmp, and interleukin superfamilies. Several enzymes that were highly expressed in LP cells included Serpin family members and arginase1. Lastly, genes for proteins and receptors known to be localized to the cell membrane that were highly expressed in LP cells included Sca1, Ngfr, Egfr, and Pdl1. Ngfr is of particular interest because it is a known stem cell marker in the tracheal epithelium, and Pdl1 expression suggests a mechanism of immune evasion for LP tumor cells. Genes downregulated in LP tumors included Tgfrb3 and surfactants.
We next used gene set enrichment analysis (GSEA) to query the pathways and molecular phenotypes specific to the LP tumors (Subramanian et al., 2007). To do this, we queried a rank-ordered gene list comparing LP EpCAM+ cells to Kras EpCAM+ cells for enrichment of the four known transcriptionally defined subclasses of human lung SCC (Wilkerson et al., 2010). We found that the LP model very closely recapitulates the expression pattern found in the basal subtype of human SCC (Figure S3B, p < 0.0001, normalized enrichment score [NES] = 1.9). Gene sets enriched in the LP tumor cells compared with Kras tumor cells included those positively regulated by AKT1 and mTOR, while a lung-specific KRAS-associated gene set was enriched in the Kras cells (Figure S3C; p < 0.0001 for AKT1 and mTOR, p = 0.012 for KRAS). In addition, compared with tumors driven by Kras, Kras;Lkb1, Kras;Pten, and Kras;p53;Lkb1, the LP tumors had much stronger p-AKT but weaker p-ERK staining (Figure 3C; Figure S3D). Together these data indicate that the oncogenic signaling pathways activated in the LP tumors predominantly involve AKT and mTOR, whereas those in Kras tumors involve downstream mediators of RAS signaling such as MEK and ERK.
To address potential metabolic differences between SCC, ADC, and normal lung, we profiled the metabolites in each tissue. In addition to the transcriptional differences observed among the samples, metabolic profiles of LP tumors, Kras tumors, and normal murine lungs were unique. The metabolic profiles of both Kras and LP tumors clustered completely separately from normal lung. Furthermore, metabolites in Kras tumors and LP tumors segregated the tumor types into two distinct clusters (Figure 3D; Table S2). Among the metabolites most significantly changed in LP cells relative to normal lung were L-arginine (reduced) and creatine (increased) (Figure S3E), which we expected because of the increased expression of arginase1 in these cells, which we confirmed by real-time RT-PCR (Figure S3F, p < 0.001).
Lkb1fl/fl;Ptenfl/fl Lung SCCs Are Enriched for TANs
As noted histologically, the LP SCC lesions contained large neutrophilic infiltrates, suggesting that the immune microenvironment was distinct from the typical TAM-rich microenvironments observed in most mouse Kras- and Kras;p53-driven ADC models. To better understand the role of the inflammatory microenvironment in lung SCC versus Kras ADC, immune cells (CD45+) from LP SCCs and Kras or Kras;p53 ADCs were compared using flow cytometry. In Kras and Kras;p53 tumors, TAMs (alveolar macrophages, CD45+CD11c+CD11b−CD103−) predominated; however, within the LP tumors, the CD45+ population contained significantly fewer macrophages and more TANs (CD45+CD11b+Ly6G+) (Figures 4A and 4B, p < 0.0001). TANs may promote tumorigenesis by stimulating angiogenesis and immunosuppression in the tumor microenvironment (Dumitru et al., 2013). Interestingly, the prevalence of TANs increased with tumor burden, as lung lobes with higher weight (indicative of higher tumor burden) showed substantially more TANs (Figure 4C, p < 0.0001). In contrast, the absolute counts of T cells, B cells, NK cells, and TAMs decreased with increasing lobe weight (Figure S4A), suggesting a selective recruitment and/or proliferation of TANs during SCC tumor progression.
Figure 4. TANs Were the Predominant Inflammatory Cell Population in Lkb1fl/fl.
;Ptenfl/fl SCC Tumors
(A) Representative flow cytometry plots for Kras ADC and LP SCC dissociated tumors. Plots are gated on live single CD45+ cells. In Kras tumors, tumor TAMs (CD45+CD11c+CD11b−CD103−) constituted the majority of CD45+ cells, whereas in LP tumors, TANs (CD45+CD11b+Ly6G+) were predominant. Gating strategies are also described in Experimental Procedures.
(B) Quantification of inflammatory cell populations in Kras tumors (n = 7), Kras;p53 tumors (n = 8), and Lkb1;Pten tumors (n = 7) by flow cytometry (mean ± SEM, p < 0.0001).
(C) Quantification of TANs within right lung lobes from samples with progressively increasing weights (shown in Figure S4A), indicating different tumor burdens: n = 8 for normal lung control (ctrl), n = 5 for mild disease group (tumor plus surrounding tissue weight < 750 mg), n = 5 for severe disease group (tumor plus surrounding tissue weight > 750 mg) (mean ± SEM; mild versus control p = 0.0034, severe versus control p < 0.0001).
(D and E) Representative immunohistochemical staining for MPO, F4/80 (D), and CD163 (E) in SCCs and ADCs; the mouse slides were LP SCC and Kras-driven ADCs. MPO staining indicating neutrophils was positive only in SCC nodules. F4/80 marks macrophages in mice (D), while CD163 marks macrophages in humans (E). Scale bars represent 200 μm for all panels of (D) and 800 μm for all panels of (E).
(F) Representative immunohistochemical staining on Lkb1;Pten;p53 tumors in which distinct areas of ADC and SCC were adjacently located. p63 and MPO staining was restricted to the SCC area. Scale bars represent 200 μm for all panels.
(G) GSEA was used to confirm the major immune cell types within Lkb1;Pten SCCs and Kras ADCs (Abbas et al., 2005; Konuma et al., 2011). See also Figure S4 and Tables S3 and S4.
To further confirm the presence of TANs in LP tumor nodules in situ, we performed staining for myeloperoxidase (Mpo), a marker that is highly expressed by TANs in tumor-bearing mice (Youn et al., 2012). Within the LP SCCs, MPO+ TANs appeared to be specifically localized to squamous lesions surrounded by p63+ epithelial cells (Figure 4D; Figure S4B). Conversely, macrophages, identified by F4/80+ staining in mouse tissue, were distributed widely within or around SCC and ADC lesions (Figure 4D; Figure S4B). Similar patterns were observed in human samples; MPO staining was strongly positive in 13 of 15 human primary SCC samples examined, whereas only 4 of 12 human primary ADCs showed staining (Table S3, p = 0.007); macrophages (CD163+ in human tissue) were scattered in both human ADCs and SCCs (Figure 4E; Figure S4C). These differences in cell infiltrates between ADC and SCC lesions were clearly evident in tumors obtained from the mixed histology p53fl/fl;Ptenfl/fl;Lkb1fl/fl (p53;Lkb1;Pten or PLP) mouse model. In this model, distinct areas of ADC and SCC were sometimes observed in close proximity in the lung. Confirming their histologic identity, the squamous areas in PLP mice expressed high levels of p63, whereas the acinar areas were negative. Importantly, staining for MPO was specific to the SCC area of the tumor, suggesting that TANs are specifically recruited to SCC lesions (Figure 4F). Similarly, the enrichment for TANs specifically in the areas of SCC tumors but not in the adjacent ADC tumors was also observed in the Kras;Lkb1 mouse model, which also has the mixed ADC and SCC histology (Figure S4D).
To further explore the differences between CD45+ fractions within SCC and ADC lung tumors, we isolated CD45+EpCAM− cells from LP and Kras tumors and performed microarray analysis. By comparing the gene expression profiles of LP CD45+ cells to LP EpCAM+ cells and Kras CD45+ cells, we constructed a list of 156 genes significantly enriched in LP CD45+ cells (Table S4, adjusted p < 0.025). Among the genes highly expressed by these cells was Ly6G, further confirming the TAN phenotype (Youn et al., 2012). By quantitative PCR, we also confirmed that Mpo, arginase1, and Cxcr2 are enriched in SCC lesions from LP mice, whereas their expression was negligible in Kras-and Kras;p53-tumor derived CD45+ cells (p = 0.005, p < 0.001, and p = 0.0011, respectively; Figure S4E). We then queried immunologic signatures that were enriched in the ranked ordered list of LP versus Kras CD45+ genes. By using two sets of independently derived signatures comparing monocyte/macrophages to neutrophils (Abbas et al., 2005; Konuma et al., 2011), we found a clear enrichment for neutrophil signatures in the LP CD45+ cells, while macrophage signatures were significantly enriched in the Kras CD45+ cells (Figure 4G) (p < 0.001).
Elevated expression of the chemokine receptor Cxcr2 suggests one mechanism through which the LP EpCAM+ cells are able to specifically recruit TANs. Many of the CXC-ligand family members have neutrophil chemoattractant activity (De Filippo et al., 2013) and appeared to be upregulated at the transcriptional level in our EpCAM microarray. Therefore, we assessed the protein concentrations of these cytokines, including CXCL1, CXCL2, CXCL5, and CXCL7, in bronchoalveolar lavage (BAL) fluid from Lkb1;Pten tumor-bearing mice. Compared with levels observed in BAL fluid isolated from normal mice, all these chemokines were significantly elevated in the BAL fluid of LP tumor-bearing mice (Figure S4F, p < 0.002), suggesting a mechanism through which TANs are recruited and stimulated by these tumors. In addition, GCSF, another essential regulator of neutrophil trafficking (Semerad et al., 2002), was also elevated in LP BAL fluid (Figure S4F, p = 0.0002). Together these data confirm that in contrast to murine Kras and Kras;p53 ADC models that contain predominantly macrophages, lung SCCs show accumulation of TANs, indicating that distinct oncogenic drivers in non-small-cell lung cancer sculpt the immune microenvironment in different ways.
Lkb1fl/fl;Ptenfl/fl Lung SCCs Display Hallmarks of Immune Suppression
We next evaluated the types of T cells present in LP SCC tumors by flow cytometry. Compared with T cell populations isolated from normal lung and peritumoral areas, the T cells within LP tumors were significantly enriched for Tregs, as determined by FOXP3 staining (Figure 5A). The ratio of CD8+ T cells to FOXP3+ Tregs within the tumor and surrounding tissues decreased with increasing tumor burden, indicating that the levels of immunosuppression rose with disease progression (Figure 5B, p < 0.0001). The accumulation of Tregs in LP tumors was further confirmed by immunohistochemical staining for FOXP3 in LP nodules (Figure 5C). In addition, T cells in LP tumors highly expressed the negative T cell costimulatory molecule programmed cell death protein 1 (PD-1), and increased percentages of PD-1-positive T cells (both CD4+ and CD8+) correlated with increased tumor burden (Figures 5D and 5E, p < 0.0001). In contrast to the increased PD-1 expression on T cells, lymphocyte-activation gene 3 (LAG3) and T cell immunoglobulin domain and mucin domain 3 (TIM3), two other known immune-modulating proteins, showed modestly increased expression (Figure S5A). Cytokines in BAL fluids were further evaluated, including transforming growth factor β (TGF-β) and interleukin-6 (IL-6). Compared with levels in normal lung, these cytokines were significantly increased in BAL fluids from LP tumor-bearing mice (Figure S5B, p < 0.0009). Previous reports have shown that TGF-β and IL-6 promote tumor growth, regulate Treg cell development, and cause immunosuppression (Flavell et al., 2010).
Figure 5. Lkb1fl/fl;Ptenfl/fl Lung SCCs Display Hallmarks of Immune Suppression.
(A) Representative flow cytometry plots for FOXP3 and CD8 in total CD3+ T cells within LP SCC tumor, uninduced normal lung, and lung surrounding LP SCC tumors.
(B) Ratios of CD8+ T cells to FOXP3+ Tregs as determined with flow cytometry: n = 8 for control lung, n = 5 for mild disease group, and n = 5 for severe disease group (p < 0.0001).
(C) Immunohistochemical staining for FOXP3 confirmed the presence of Tregs in LP SCC nodules. Scale bars represent 50 μm for both panels.
(D and E) Quantification the percentage of PD-1 positive cells within the CD8+ (D) and CD4+ (E) T cell populations: n = 8 for control lung, n = 5 for mild disease group, and n = 5 for severe disease group (p < 0.0001).
(F) Representative immunohistochemical staining for PD-L1 on LP SCC nodules. Scale bars represent 100 μm for both panels.
(G) (a) Percentage of PD-L1 positive cells within the EpCAM+CD45-CD31− fraction from LP SCC as measured by flow cytometry: n = 7 for LP tumors and n = 5 for normal lung (p < 0.0001). (b) Real-time RT-PCR for Pdl1 mRNA levels in the indicated EpCAM+ purified cells from SCC tumors and normal lung tissue: n = 6 for normal lung and n = 5 for LP SCC tumors (p = 0.0013).
Data are presented as mean ± SEM in (B), (D), (E), and (G). See also Figure S5.
Because we observed high levels of Pdl1 in the LP EpCAM+ cells by microarray, and published work suggests that PD-L1 can induce Tregs (Francisco et al., 2009), we further explored the expression of this immunomodulating protein. PD-L1 expression was first observed on LP tumor cells by IHC (Figure 5F). High cell surface expression of PD-L1 on EpCAM+ CD45− cells from LP tumors was confirmed by flow cytometry (Figure 5Ga, p < 0.0001). We further confirmed the Pdl1 expression with real-time PCR on EpCAM+CD45− cells from normal lung and LP tumors and observed 6-fold more Pdl1 mRNA in the LP tumor cells compared with levels in normal distal lung epithelium (Figure 5Gb, p = 0.0013). The increased numbers of Tregs, together with the high levels of PD-1 and PD-L1 on immune and tumor cells, respectively, indicated that immune suppression may play an important role during lung SCC tumorigenesis.
A SCA1+NGFR+ Phenotype Is Enriched in Lkb1fl/fl;Ptenfl/fl Lung SCC TPCs
We next wished to determine if LP SCCs contained distinct TPC populations. We decided to begin with analysis for two known stem cell makers, SCA1 and NGFR, which mark BASCs and tracheal basal cells, respectively. We first examined NGFR expression on BASCs and SCA1 expression on basal cells. We found that although nearly 100% of NGFR+ basal cells expressed SCA1, only ~25% of SCA1+ BASCs expressed NGFR (Figure S6A). Within the EpCAM+CD45− cell populations, LP tumor cells showed high expression of SCA1 and NGFR, and LP tumors harbored a unique population of SCA1+NGFR+ cells that constituted an average of 17.5% of the LP EpCAM+ cells. Interestingly this population was nearly absent in both Kras and Kras;p53 ADC models (Figures 6A and 6B, p < 0.0001; Figures S6B and S6C). Ngfr transcript was 30-fold more abundant in LP tumor cells than in normal lung or Kras epithelial fractions (Figure S6D, p < 0.0001). NGFR also specifically stained the LP SCC tumor lesions but was not detectable in Kras tumor lesions by IHC (Figure 6Ca). Likewise, in human primary lung SCCs, NGFR staining was observed in 11 of 13 samples examined, whereas only 2 of 12 human primary ADC sections had detectable NGFR staining (Figure 6Cb; Table S3; p = 0.001). In PLP tumors in which the ADC and SCC lesions were juxtaposed, NGFR staining was specific to the SCC side of the tissue section (Figure 6Cc).
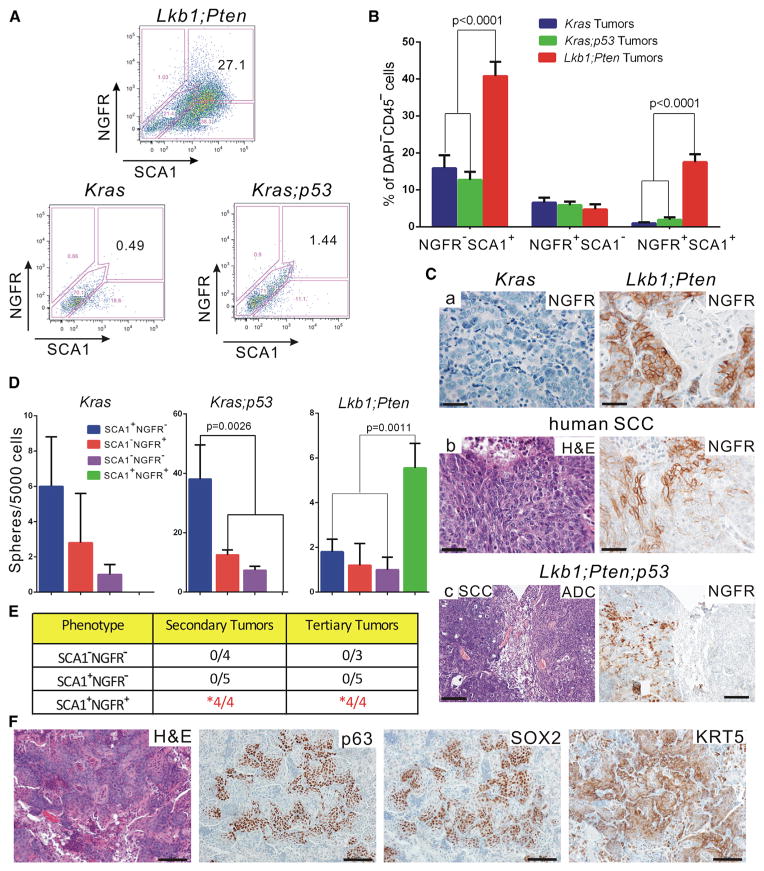
Figure 6. Lkb1fl/fl;Ptenfl/fl Lung SCC Contained SCA1+NGFR+ TPCs that Could Serially Transplant Squamous Disease.
(A) Representative flow cytometry plots for NGFR and SCA1 expression within the indicated EpCAM+CD45−CD31− dissociated tumor cell populations. LP tumor cells showed much higher expression of both SCA1 and NGFR than either the Kras or Kras;p53 tumors.
(B) Quantification of SCA1- and NGFR-expressing cells with the EpCAM+CD45−CD31− population as assessed by flow cytometry. The percentage of SCA1+NGFR+ in LP tumors is much higher than in Kras or Kras;p53 tumors: n = 23 for Kras tumors, n = 25 for Kras;p53 tumors, and n = 34 for Lkb1;Pten tumors (p < 0.0001).
(C) Representative immunohistochemical staining for NGFR on mouse SCC and ADC (a) and human SCC nodules (b). NGFR staining is strongly positive on SCC tumors but negative on ADC tumors. In the Lkb1;Pten;p53 tumors, distinct areas of ADC and SCC were adjacently located. NGFR staining was restricted to the SCC area (c). Scale bars represent 50 μm for (a) and (b) and 200 μm for panel (c).
(D) Quantification of tumorspheres derived from SCA1+NGFR+, SCA1−NGFR+, SCA1+NGFR−, and SCA1−NGFR− FACS purified cells that were cocultured in Matrigel with equal amounts of CD45+CD31+ cells from the same primary tumors. Each fraction was seeded at 5,000 tumor cells/well. The colony-propagating ability of the SCA1+NGFR+ fraction in LP tumors is higher than that of the other fractions (p = 0.0011).
(E) Quantification of tumor propagation ability of FACS-isolated SCA1+NGFR+, SCA1+NGFR−, and SCA1−NGFR− LP tumor cells. The secondary tumors were derived from intratracheal transplantation, with tumor formation latency of ~30 to 40 weeks. The tertiary tumors were derived from intrathoracic injection, with tumor formation latency of ~20 to 30 weeks. Ten thousand sorted cells from each fraction were injected for each fraction and each experiment. Only SCA1+NGFR+ populations could form tumors and be serially transplanted (p = 0.001 for secondary tumors, p = 0.002 for tertiary tumors, Fisher’s exact test).
(F) Representative immunohistochemical staining on tertiary tumors derived from SCA1+NGFR+ LP tumor cells after intrathoracic injection. The tumors retained a squamous histology and were positive for all of the squamous markers examined. Scale bars represent 100 μm for all panels.
Data are presented as mean ± SEM in (B) and (D). See also Figure S6.
We next used FACS to fractionate LP EpCAM+CD31−CD45− tumor cells according to SCA1 and NGFR expression for functional comparison of TPC capacity. First, a surrogate in vitro assay for tumor propagation was used. Four distinct populations, SCA1+NGFR+, SCA1−NGFR+, SCA1+NGFR−, and SCA1−NGFR− cells, were collected and cocultured in Matrigel (BD Biosciences) with CD45+CD31+ “support” cells isolated from the primary tumors. The tumor colony-forming ability of Kras and Kras;p53 sorted tumor fractions was also evaluated in the 3D Matrigel system (Figure S6E). In agreement with previous in vivo results (Curtis et al., 2010) and validating this assay for TPC capacity, SCA1+ cells from Kras;p53 tumors were enriched for tumor colony formation ability (p = 0.0026), but the same was not true for Kras tumors. SCA1+NGFR+ cells from LP tumors formed the most tumor colonies in 3D cultures, suggesting that they are the fraction enriched for tumor propagation (Figure 6D, p = 0.0011). The morphology and histology of the LP tumor colonies were distinct from those found in Kras or Kras;p53 tumor cultures (Figure S6F). By immunofluorescence, the Lkb1;Pten tumor colonies expressed the squamous marker p63 but not the ADC-associated surfactant protein C (SPC), while both the Kras- and Kras;p53-derived colonies expressed SPC (Figure S6F).
To determine if SCA1+NGFR+ cells from LP tumors were enriched for TPC characteristics in vivo, three major fractions of the EpCAM+ cells from primary LP tumors, SCA1+NGFR+, SCA1+ NGFR−, and SCA1−NGFR−, were transplanted orthotopically into immunocompromised mouse recipients immediately following FACS purification (Figure S6G). The fraction of SCA1−NGFR+ was not tested, because of its reproducibly small abundance. Of the four mice that received SCA1+NGFR+ cells, all developed typical SCC with p63+, SOX2+, and KRT5+ staining within 30 to 40 weeks (Figures 6E and 6F). To assess the presence of self-renewing TPCs within these tumors, the secondary tumors were dissected, dissociated, and sorted for NGFR and SCA1, and the three major fractions were transplanted for tertiary tumor formation. All mice transplanted with SCA1+NGFR+ developed tertiary SCC within 11 to 27 weeks (Figure 6E, p = 0.001 for secondary, p = 0.002 for tertiary, Fisher’s exact test). All primary, secondary, and tertiary tumors shared the same histological and FACS characteristics (Figure S6H). Together these data demonstrate that LP tumors contained a distinct population of SCA1+NGFR+ TPCs that could transplant disease retaining squamous characteristics.
TPCs Express High Levels of PD-L1
Little is known about how TPCs escape immunologic clearance and clonally expand to form malignant tumor nodules. To address this question, we further quantified the PD-L1 level on LP tumor cell fractions. By gating the four SCA1;NGFR fractions and analyzing the percentage of PD-L1+ cells in each fraction, we found a clear enrichment for PD-L1+ cells within the SCA1+ NGFR+ fraction (Figure 7A). Within a group of seven mice, an average of 69% of SCA1+NGFR+ cells expressed PD-L1 on their cell surface, whereas only 39% of SCA1+NGFR− or 32% of SCA1−NGFR+ cells were PD-L1+ (Figure 7B, p = 0.004). Likewise, by real-time RT-PCR for Pdl1 within the sorted LP tumor fractions, SCA1+NGFR+ cells had 7-fold more Pdl1 mRNA than SCA1−NGFR− cells and about 2-fold more than SCA1+NGFR− or SCA1−NGFR+ cells (Figure 7C, p = 0.035). We also used flow cytometry and real-time RT-PCR to assess PD-L1 levels in BASCs and basal cells. NGFR+ BASCs expressed the most PD-L1 in the distal lung, while PD-L1 expression was uniformly high by flow cytometry in the trachea (Figures S7A and S7B).
Figure 7. SCA1+NGFR+ TPCs in Lkb1fl/fl;Ptenfl/fl Lung SCC Tumor Expressed High Levels of PD-L1.
(A) Representative histogram of PD-L1 expressing cells from a dissociated LP tumor gated on DAPI−EpCAM+CD45−CD31− cells and then for the four indicated fractions of SCA1;NGFR-expressing cells. The unstained control trace in gray is shown for gating. DAPI, 4′,6-diamidino-2-phenylindole.
(B) Quantification of PD-L1 expression level by flow cytometric analysis. PD-L1 expression is higher in SCA1+NGFR+ population than any other population (n = 7 tumors, p = 0.004).
(C) Real-time RT-PCR quantification of Pdl1 mRNA expression in SCA1+NGFR+, SCA1− NGFR+, SCA1+NGFR−, and SCA1−NGFR− sorted populations (n = 7 tumors, p = 0.035).
(D) Representative H&E staining confirming that PDX tumors retained squamous histology. The black scale bar represents 200 μm, and the yellow scale bar represents 2,000 μm.
(E) Quantification of PD-L1 expression level by flow cytometric analysis of PDX samples. Mean fluorescence intensities for PD-L1 antibody on EpCAM+NGFR+ fractions are higher than those for EpCAM+NGFR− fractions. The control is unstained dissociated PDX cells (n = 6 tumors, p = 0.02).
(F) Serial sections of formalin-fixed human SCC tumors stained with H&E, PD-L1, or NGFR. PD-L1 is colocalized to the NGFR+ cells within these tumors. Scale bars represent 100 μm for all panels.
Data are presented as mean ± SEM in (B), (C), and (E). See also Figure S7.
In order to explore the relationship between NGFR and PD-L1 expression in patient tissue, we used human lung SCC tissues that were passaged in immunocompromised mice as patient-derived xenografts (PDXs) (Figure S7C). Hematoxylin and eosin (H&E)-stained sections from the PDX samples showed that the squamous histology of the tumors was retained in the xenograft model (Figure 7D). PDX samples were dissociated and stained with antibodies directed against human CD31, CD45, EpCAM, NGFR, and PD-L1 (Figure S7D). We analyzed the amount of PD-L1 staining on both EpCAM+NGFR+ and EpCAM+NGFR− fractions. Using six different PDX samples, PD-L1 staining was 4.2-fold higher in the NGFR+ fraction relative to the NGFR− fraction of the human EpCAM+ tumor cells (Figure 7E, p = 0.02). We further confirmed this trend with independent human lung SCC tumor samples by staining serial sections for NGFR and PD-L1. Clearly, PD-L1 is colocalized to the NGFR+ cells within these tumors, indicating that the majority of NGFR+ cells coexpress PD-L1 (Figure 7F). Therefore, in lung SCC, PD-L1 is most abundantly expressed on tumor cells that express NGFR, and if these cells are analogous to the NGFR+ cells in mouse tumors, they will also be enriched for TPC activity.
DISCUSSION
Here, we demonstrate that biallelic inactivation of both Lkb1 and Pten in the mouse lung leads to fully penetrant SCC. Compared with lung Kras-driven ADC models, the immune microenvironment of these SCCs was enriched for TANs. Furthermore, we demonstrate that SCA1+NGFR+ tumor cells are enriched for tumor-propagating ability and express high levels of the immune evasion molecule PD-L1. These tumors very closely recapitulate the gene expression profiles of the basal subtype of human lung SCC, indicating that these mice can serve as a valuable model for understanding progression and maintenance of basal lung SCCs. This SCC model will enable the investigation of the molecular mechanisms of human SCC carcinogenesis and will allow further preclinical and coclinical investigation of novel therapies aimed at eradicating lung tumors.
Although lung ADC and SCC occur at relatively equal frequencies worldwide, developing a genetic model of lung SCC has been challenging. The Ad-Cre inhalation method may specifically target more distal lung progenitors, thus selecting for tumor cells of origin that predispose toward an ADC phenotype. Several studies have targeted deletion of squamous tumor suppressors, such as Pten, or activation of squamous oncogenes, such as Sox2, but despite these efforts, only partial SCC differentiation was observed in either model (Lu et al., 2010; Malkoski et al., 2013). Here, we demonstrate that deletion of both Pten and Lkb1, via the traditional Ad-Cre inhalation system, is able to produce lung tumors of purely squamous phenotype. Lkb1;Pten tumor lesions appeared to grow into the distal lung, suggesting that if basal cells are the cells of origin, they are able to migrate more distally to propagate disease. When we examined the expression of stem cell markers in normal lung tissue, 100% of basal cells were SCA1+, while ~25% of BASCs were NGFR+. These data suggest an alternative possibility that a rare subset of NGFR+ BASCs could serve as distal cells of origin for these tumors. Surfactant protein C Cre-recombinase and club cell secretory protein Cre-recombinase both failed to produce tumors when used with the Lkb1;Pten alleles (data not shown), indicating that distal lung cells may not be the primary targets of oncogenic transformation in this model. Further examination of the cells of origin for these tumors, including the use of basal cell-specific Cre strains or repetitive injury that targets particular cell populations, will help us understand which lung cells can serve as precursors for these squamous tumors.
As expected, the loss of both Lkb1 and Pten in these tumors activated the AKT and mTOR pathways, likely driving cellular proliferation and tumorigenesis. The deletion of these genes was also associated with the upregulation of specific cytokines and other immune-modulating proteins, leading to a unique tumor microenvironment. Compared with Kras tumor cells, the LP EpCAM+ cells expressed very high levels of the chemokines CXCL3 and CXCL5, and the BAL fluid contained elevated CXCL1, CXCL2, CXCL5, and CXCL7. The CXC chemokine family controls the migration and adhesion of monocytes and neutrophils, mediating its effects on target cells by interacting with CXCR2 (Põld et al., 2004). CXCL5 is also known as epithelial-derived neutrophil-activating peptide 78, and its expression is associated with PI3K/AKT and Raf/MEK/ERK activation (Hsu et al., 2013). Recent findings in tumor-bearing mice and cancer patients indicate that the increased metabolism of L-arginine by TANs producing arginase1 can inhibit T cell lymphocyte responses (Raber et al., 2012), and it is likely that this mechanism is in play in the LP tumors. Furthermore, we observed strong MPO staining in patient SCC tissues, suggesting that activated TANs are a key component of SCC in both mouse and human.
In addition to expression of the TAN-attracting cytokines, the LP EpCAM+ cells expressed high levels of the immune evasion molecule PD-L1. Recently, there has been much excitement surrounding the potential of targeting molecules such as PD-L1 to “reawaken” the immune system and cause tumor destruction. In the phase 1 study of nivolumab, a fully humanized monoclonal antibody to PD-1, PD-L1 expression was determined by IHC in pretreatment tumor biopsies of various tumor types (n = 42). Thirty-six percent of patients whose tumors showed PD-L1 expression achieved objective response to nivolumab treatment (9 of 25), while none of the 17 patients with PD-L1-negative tumors showed any objective response, although some achieved prolonged stable disease (Topalian et al., 2012). These data indicate that PD-L1 expression might influence response to anti-PD-1 antibody therapy. With the accumulation of clinical data, the actual correlation between PD-L1 expression and response to anti-PD-1 therapy should become clearer.
Intriguingly, the TPCs within our SCC model showed enrichment for PD-L1 expression, suggesting that TPCs have unique immune evasion properties. Strikingly, we found that the SCA1+NGFR1+ cell population had enhanced tumor-propagating activity compared with other tumor cell populations. Notably, our studies compared three tumor cell populations from murine SCC, but the SCA1−NGFR+ population could not be assessed because of low abundance. Thus, there could be additional TPCs to characterize in Lkb1;Pten SCCs. Furthermore, although PDX analyses showed that NGFR+ human SCC cells are enriched for PD-L1, we have not yet established the identity of TPCs in human SCC with a functional assay. SCA1 as a TPC marker is not useful for human cell studies, and markers in addition to NGFR may be required to enrich for propagating activity from primary patient SCCs. It is also currently unclear how universal the immune evasion ability of TPCs we found will be in human SCC or in other tissues. It will be crucial in future studies to investigate TPCs in other tumor types and characterize their PD-L1 expression to further explore this intriguing link between immune escape and tumor propagation. In the future, it may be possible to first debulk the tumor with a more generally targeted inhibitor or surgery and then prevent tumor recurrence and/or metastasis through administration of anti-PD-1 therapy to target TPCs. Together these data demonstrate the potential of immunotherapy for the treatment of lung SCC and lay the groundwork for further investigation into the response of both cancer cells and the immune microenvironment to such treatments.
EXPERIMENTAL PROCEDURES
Mouse Cohorts and Human Samples
All mice were housed in a biosafety level 2 lab at Dana-Farber Cancer Institute. All care and treatment of experimental animals were in accordance with Harvard Medical School/Dana-Farber Cancer Institute institutional animal care and use committee guidelines. Detailed mouse cohort information can be found in Supplemental Experimental Procedures. Patient slides were provided by the Pathology Department of Brigham and Women’s Hospital. The human samples were obtained from consented subjects at the Dana-Farber Cancer Institute/Harvard Cancer Center under institutional review board-approved protocol 02-180. Frozen PDX tissues were purchased from the Van Andel Institute.
Flow Cytometry Analysis and Sorting
Tumors were dissected from the lungs of primary mice, and tumor tissue was prepared as previously described (Curtis et al., 2010). Single-cell suspensions were stained using rat-antimouse antibodies. Detailed antibody information and gating strategy can be found in Supplemental Experimental Procedures.
3D Culture and Tumor Transplants
FACS-sorted mouse cells were resuspended in MTEC/Plus containing 20 ng/ml EGF and FGF2, mixed 1:1 with growth factor-reduced Matrigel, and pipetted into a 12-well 0.4 μm Transwell insert (Falcon). MTEC/Plus medium (700 μl) was added to the lower chamber and refreshed every other day. Intratracheal transplantation was performed as previously described (Curtis et al., 2010). Intrathoracic injections were performed as previously described (Jongsma et al., 2008).
Metabolomics Profiles Analysis
Metabolite extraction and targeted mass spectrometry analysis for metabolomics profiles were conducted as reported previously (Yuan et al., 2012). Briefly, the frozen tumors were smashed in cold 80% high-performance liquid chromatography-grade methanol on dry ice twice, and then the extractions were Speed Vac/lyophilized to a pellet using no heat. The data were normalized and analyzed with MetaboAnalyst 2.0 (Xia et al., 2012). In hierarchical cluster analysis, each sample begins as a separate cluster, and the algorithm proceeds to combine them until all samples belong to one cluster. Clustering results are shown as heatmap (distance measure using Pearson and clustering algorithm using Ward).
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism (GraphPad Software). All numerical data are presented as mean ± SEM. Grouped analysis was performed using two-way ANOVA. Column analysis was using one-way ANOVA or Student’s t test. A p value < 0.05 was considered statistically significant.
Supplementary Material
Significance.
Although many of the molecular alterations underlying human lung SCC were recently described, lung SCC has been challenging to model in the mouse, and there are few in vivo models for the development of novel cancer therapeutics for this disease. The SCC model described herein is an important tool for investigating the molecular mechanisms of SCC carcinogenesis, including the identity of TPCs and the involvement of the immune microenvironment in tumor progression. This model also provides a platform to study new therapeutic opportunities for lung SCC through the targeting of a TPC-specific interaction with the immune cell microenvironment.
Acknowledgments
We thank the Boston Children’s Hospital flow cytometry facility and members of the C.F.K. and Zon Labs for helpful discussions. This work was supported in part by the United Against Lung Cancer and the Susan Spooner Research Fund (to K.-K.W.); American Cancer Society Post-Doctoral Fellowship PF-12-151-01-DMC (to C.M.F.); a Margaret A. Cunningham Immune Mechanisms in Cancer Research Fellowship Award (to S.K.); American Cancer Society Research Scholar grant RSG-08-082-01-MGO, the V Foundation for Cancer Research, a Basil O’Conner March of Dimes Starter Award, the Harvard Stem Cell Institute, and the Lung Cancer Research Foundation (to C.F.K.); the Cancer Prevention Research Institute of Texas (RP100550); and grants HL090136, HL100402, CA122794, CA140594, CA163896, CA166480, P0CA154303, CA098101, CA141576, CA137181, P0CA120964, CA143083, and CA163677 from the NIH.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession number for the microarray data reported in this paper is GSE54353.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2014.03.033.
AUTHOUR CONTRIBUTIONS
C.X. and C.M.F. worked closely on the paper. C.X. and C.M.F. contributed to the experimental design and performed the majority of the experiments. C.X. was responsible primarily for the mouse model establishment and characterization, microenvironment, and immune marker identification. C.M.F. worked primarily on validating the TPC markers in vitro and in vivo, the bioinformatic analysis, and tumor microenvironment characterization. C.X. and C.M.F. contributed to manuscript writing.
References
- Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- Adeegbe DO, Nishikawa H. Natural and induced T regulatory cells in cancer. Front Immunol. 2013;4:190. doi: 10.3389/fimmu.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK, Kim CF. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, Gunzer M, Roers A, Hogg N. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–4937. doi: 10.1182/blood-2013-02-486217. [DOI] [PubMed] [Google Scholar]
- Dumitru CA, Moses K, Trellakis S, Lang S, Brandau S. Neutrophils and granulocytic myeloid-derived suppressor cells: immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol Immunother. 2012;61:1155–1167. doi: 10.1007/s00262-012-1294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru CA, Lang S, Brandau S. Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol. 2013;23:141–148. doi: 10.1016/j.semcancer.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Fatima N, Cohen C, Lawson D, Siddiqui MT. Combined double CK5/P63 stain: useful adjunct test for diagnosing pulmonary squamous cell carcinoma. Diagn Cytopathol. 2012;40:943–948. doi: 10.1002/dc.21678. [DOI] [PubMed] [Google Scholar]
- Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limón P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YL, Hou MF, Kuo PL, Huang YF, Tsai EM. Breast tumor-associated osteoblast-derived CXCL5 increases cancer progression by ERK/MSK1/Elk-1/Snail signaling pathway. Oncogene. 2013;32:4436–4447. doi: 10.1038/onc.2012.444. [DOI] [PubMed] [Google Scholar]
- Huang SD, Yuan Y, Liu XH, Gong DJ, Bai CG, Wang F, Luo JH, Xu ZY. Self-renewal and chemotherapy resistance of p75NTR positive cells in esophageal squamous cell carcinomas. BMC Cancer. 2009;9:9. doi: 10.1186/1471-2407-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Jongsma J, van Montfort E, Vooijs M, Zevenhoven J, Krimpenfort P, van der Valk M, van de Vijver M, Berns A. A conditional mouse model for malignant mesothelioma. Cancer Cell. 2008;13:261–271. doi: 10.1016/j.ccr.2008.01.030. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Konuma T, Nakamura S, Miyagi S, Negishi M, Chiba T, Oguro H, Yuan J, Mochizuki-Kashio M, Ichikawa H, Miyoshi H, et al. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp Hematol. 2011;39:697–709.e5. doi: 10.1016/j.exphem.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, Kim CF. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkoski SP, Cleaver TG, Thompson JJ, Sutton WP, Haeger SM, Rodriguez KJ, Lu SL, Merrick D, Wang XJ. Role of PTEN in basal cell derived lung carcinogenesis. Mol Carcinog. 2013 doi: 10.1002/mc.22030. Published online April 26, 2013 http://dx.doi.org/10.1002/mc.22030. [DOI] [PMC free article] [PubMed]
- Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- Nalbandian A, Yan BS, Pichugin A, Bronson RT, Kramnik I. Lung carcinogenesis induced by chronic tuberculosis infection: the experimental model and genetic control. Oncogene. 2009;28:1928–1938. doi: 10.1038/onc.2009.32. [DOI] [PubMed] [Google Scholar]
- Põld M, Zhu LX, Sharma S, Burdick MD, Lin Y, Lee PP, Põld A, Luo J, Krysan K, Dohadwala M, et al. Cyclooxygenase-2-dependent expression of angiogenic CXC chemokines ENA-78/CXC ligand (CXCL) 5 and interleukin-8/CXCL8 in human non-small cell lung cancer. Cancer Res. 2004;64:1853–1860. doi: 10.1158/0008-5472.can-03-3262. [DOI] [PubMed] [Google Scholar]
- Raber P, Ochoa AC, Rodríguez PC. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest. 2012;41:614–634. doi: 10.3109/08820139.2012.680634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG, Ladanyi M. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002;17:413–423. doi: 10.1016/s1074-7613(02)00424-7. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Tiozzo C, De Langhe S, Yu M, Londhe VA, Carraro G, Li M, Li C, Xing Y, Anderson S, Borok Z, et al. Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am J Respir Crit Care Med. 2009;180:701–712. doi: 10.1164/rccm.200901-0100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trellakis S, Bruderek K, Dumitru CA, Gholaman H, Gu X, Bankfalvi A, Scherag A, Hütte J, Dominas N, Lehnerdt GF, et al. Polymorphonuclear granulocytes in human head and neck cancer: enhanced inflammatory activity, modulation by cancer cells and expansion in advanced disease. Int J Cancer. 2011;129:2183–2193. doi: 10.1002/ijc.25892. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DR, Schissel DJ, Howrey RP, Aeed PA. Tumor-elicited polymorphonuclear cells, in contrast to “normal” circulating polymorphonuclear cells, stimulate invasive and metastatic potentials of rat mammary adenocarcinoma cells. Proc Natl Acad Sci U S A. 1989;86:5859–5863. doi: 10.1073/pnas.86.15.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40(Web Server issue):W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, Back T, Song NY, Datla M, Sun Z, et al. The pivotal role of IKKα in the development of spontaneous lung squamous cell carcinomas. Cancer Cell. 2013;23:527–540. doi: 10.1016/j.ccr.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JI, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.