Abstract
The Janus Kinase (JAK) family members serve essential roles as the intracellular signalling effectors of cytokine receptors. This family, comprising JAK1, JAK2, JAK3 and TYK2, was first described more than 20 years ago, but the complexities underlying their activation, regulation and pleiotropic signalling functions are still being explored. Here, we review the current knowledge of their physiological functions and the causative role of activating and inactivating JAK mutations in human diseases, including haematopoietic malignancies, immunodeficiency and inflammatory diseases. At the molecular level, recent studies have greatly advanced our knowledge of the structures and organisation of the component FERM-SH2, pseudokinase and kinase domains within the JAKs, the mechanism of JAK activation and, in particular, the role of the pseudokinase domain as a suppressor of the adjacent tyrosine kinase domain's catalytic activity. We also review recent advances in our understanding of the mechanisms of negative regulation exerted by the SH2 domain containing proteins, SOCS (Suppressors of Cytokine Signalling) proteins and Lnk. These recent advances highlight the diversity of regulatory mechanisms utilised by the JAK family to maintain signalling fidelity, and suggest alternative therapeutic strategies to complement existing ATP-competitive kinase inhibitors.
Keywords: pseudokinase, kinase, cytokine receptor, Janus kinase
Introduction
The transmission of signals from extracellular stimuli across the plasma membrane via the cytoplasm to the nucleus in eukaryotes principally relies on the post-translational protein modification, phosphorylation. Protein kinases, enzymes that catalyse the transfer of the γ-phosphate from ATP to tyrosine, serine or threonine sidechains in substrate proteins, can thus be thought of “writers” of reversible marks, which function to modify their substrates, intracellular effectors, to regulate their signalling activities. Due to their essential roles in cellular signalling, protein kinases are subjected to many levels of positive and negative regulation to ensure fidelity of signals and restrict signal longevity to guard against aberrant signal activation. Defective kinase activity or regulation are known to underlie proliferative diseases, such as cancers, and consequently protein kinases have come to prominence as therapeutic targets.
Due to their essential roles as signal transducers downstream of cytokine receptor activation, the Janus Kinase (JAK) family of tyrosine kinases have garnered much attention since their discovery more than 20 years ago [1–6]. This family comprises four members: JAK1, JAK2, JAK3 and TYK2. In contrast to receptor tyrosine kinases, such as the c-Kit and Insulin receptors, cytokine receptors lack intrinsic protein kinase domains and consequently rely on the catalytic activities of constitutively-associated Janus kinase (JAK) family of tyrosine kinases to convey signals (Figure 1A). Crucially, cytokine receptors comprise two or more receptor subunits, each associated with a JAK monomer. Upon receptor ligation by a cognate ligand, receptor subunits are either reoriented or oligomerise leading to manoeuvring of receptor-associated JAKs into positions to facilitate their trans-phosphorylation and corresponding activation. Subsequently, activated JAKs phosphorylate tyrosines within the cytoplasmic regions of the receptor with which they are associated, generating docking sites for downstream adaptor and effector (“reader”) proteins that contain phosphotyrosine recognition domains, typified by its SH2 domain, including the signal transducers and activators of transcription (STAT) proteins. Depending on the receptor, and thus the docking sites generated by tyrosine phosphorylation within the cytoplasmic region, any one or more of six STAT family members (STAT1, STAT2, STAT3, STAT4, STAT5 or STAT6) may be recruited via their SH2 domains. STATs exist as preformed dimers [7–9], and by being brought into proximity of the receptor-associated JAK can then be phosphorylated by JAK, leading to a reorientation of subunits within the STAT dimer and translocation into the nucleus where it functions as a transcription factor [10–12]. The resulting transcriptional program dictates whether the cell undergoes proliferation, differentiation, survival or death.
Figure 1.
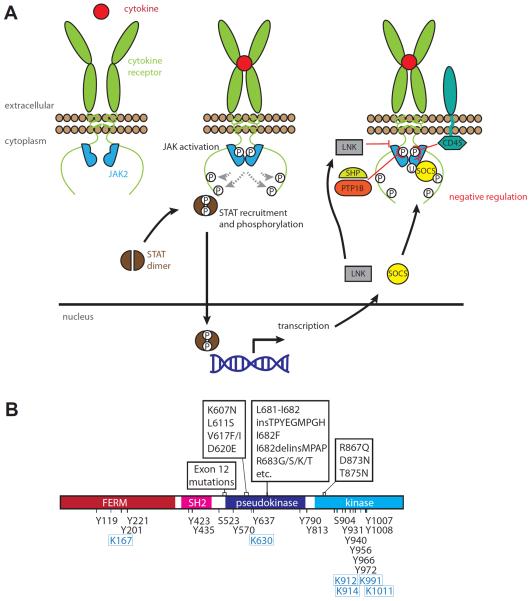
Overview of JAK activation and regulation. (A) Summary of the JAK/STAT signalling pathway and its negative regulation. Ligation of a cytokine receptor leads to a transition of the associated JAK molecules from an inactive (left) to activated state (middle). Activated JAK is characterised by phosphorylation of activation loop residues within its kinase domain (encircled P; middle). Activated JAKs phosphorylate tyrosines within the receptor intracellular region to enable recruitment and phosphorylation of the principal downstream effectors, the STATs. Many layers of negative regulation have been identified (right), including phosphatases (CD45, PTP1B, SHP1 and SHP2) and SH2 domain containing regulators from the LNK and SOCS families. SOCS proteins inhibit JAK signalling by both inhibiting kinase activity and by mediating ubiquitylation (encircled U), leading to proteasomal degradation.
(B) Intrinsic regulatory events annotated on the human JAK2 domain structure. Mutational hotspots associated with myeloproliferative neoplasms are annotated above [36, 37, 39, 41–49, 142, 143]. Residues subject to phosphorylation [65, 144–152] and sumoylation [153] are annotated below in black text and blue boxed text, respectively. In addition to the crucial role of the activation loop tyrosines, 1007 and 1008 [65, 66], Y119, Y221, Y317, Y570, Y637, Y813, Y866, Y913, Y966 and Y972 have been reported to arise from JAK2 auto- or trans-phosphorylation and contribute variously to the modulation of JAK2 activation [146–148, 150, 154–156].
Whilst this pathway is notionally simple, many nuances of the pathway remain mysterious. For instance, how do cytokine receptors dictate that only a subset of JAKs (often only one of the family) associate with the receptor? What are the determinants that govern how a JAK typically signals via only a subset of the STAT family proteins? Additionally, it remains unclear why cytokine receptors would evolve a constitutive association with a protein kinase in preference to containing intrinsic protein kinase domains: does the combinatorial nature of JAK association with receptors facilitate signal pleiotropy or does the separation of receptor and kinase permit the kinase to perform additional functions, such as those proposed for JAK1 and JAK2 in the nucleus [13, 14] and reviewed in [15]? Furthermore, as summarized in Figure 1 and detailed below, regulation of the JAK/STAT signalling pathway is highly nuanced, with signalling flux tuned by layers of regulation by extrinsic regulators, by protein activators and inhibitors, including phosphatases which serve as “erasers” of both activating and inactivating phosphorylation events (Figure 1A,B).
JAKs and their cognate cytokine receptors
While cellular overexpression studies suggested JAKs could signal promiscuously downstream of many cytokine receptors, it is evident from genetic deletion studies that cytokine receptors have clear preferences for the JAK family members they utilize as signalling effectors (Table 1). In light of this, here we have focused our attention on the genetic deletion studies that have illuminated which JAKs couple with which cytokine receptors. The first insights into the specificity of JAKs within each signalling pathway arose from early cell-based genetic screens to identify components of the IFNα/β and IFNγ signalling pathways. Initially, TYK2 was found to complement a defect in IFNα signalling [6]; while subsequent studies implicated JAK2 in IFNγ signalling [16] and JAK1 in IFNα/β and IFNγ signalling [17]. A broader understanding of the relationships between JAKs and cytokine receptors has relied on the generation of knockout mice for each JAK family member and the detailed examination of signalling responses to cytokine stimuli by cells derived from these animals. Although Tyk2−/− and Jak3−/− mice are viable under stress-free conditions and thus readily able to be studied, examination of the signalling pathways mediated by JAK1 and JAK2 have been complicated by the fact that germline deletion of Jak1 or Jak2 causes lethality at postnatal day 1 [18] or E12.5 [19, 20] respectively. Lethality in Jak1−/− mice was attributed to neuronal defects arising from defects in gp130 receptor signalling [18]. The lethality of Jak2 deletion was attributed to an absence of definitive haematopoiesis in knockout mice [19, 20] and the haematopoietic specificity of this phenotype was similarly observed upon conditional deletion of Jak2 in postnatal and adult mice in recent studies [21–23], where defects in the haematopoietic organs, haematopoietic stem cells (HSC), erythropoiesis and thrombopoiesis were most profound. It is remarkable that like JAK1 and TYK2, JAK2 is ubiquitously expressed [18, 20] yet the defects in JAK2 signalling were largely confined to HSC, erythroid and thrombopoietic signalling. In contrast, JAK3 expression is confined to haematopoietic tissues, myeloid and lymphoid cells [4, 24], and deletion of Jak3 principally caused lymphopoietic defects that manifested as severe combined immunodeficiency disease (SCID) in these mice [25, 26]. This phenotype is reminiscent of deletion or deleterious mutation of its cognate receptor, the IL-2, IL-4, IL-7, IL-13, IL-15, IL-21 common-γ (γc) receptor [27–29].
Table 1.
The phenotypes caused by JAK deficiency
| Gene | Knockout model | Viability | Knockout phenotype | Defective cytokine signalling | Normal cytokine signalling | Reference | |
|---|---|---|---|---|---|---|---|
| Jak1 | Mouse | Germline | Postnatal day 1 lethality | Death attributed to neurological defects from defective gp130 signalling. Small thymi and defective thymocyte production. | IL-3; IL-2, IL-4, IL-7; IL-6, LIF; IFNα; IFNγ; IL-10 | GM-CSF, IL-5; M-CSF; G-CSF | [18] |
|
| |||||||
| Jak2 | Mouse | Germline | Embryonic lethality at E12.5 | Failure of definitive erythropoiesis. | IL-3, IL-5, GM-CSF; IFNγ; EPO; TPO; GH; PRL | IL-6, LIF; IFNα/β; G-CSF; | [19, 20, 158, 159] |
|
| |||||||
| Jak2 | Mouse | Conditional: systemic at in postnatal day 4 (PN4) and 2 month old animals. | Lethal | PN4 lethality due to defective erythropoiesis. Adult mice succumbed to severe anaemia, thrombocytopenia, HSC loss and bone marrow failure. | [21–23, 158] | ||
| Conditional: mammary epithelia | Viable | Impaired mammary gland development and maintenance of functionally differentiated alveolar cells | PRL | [159] | |||
|
| |||||||
| Jak3 | Mouse | Germline | Viable | Develop SCID. Block in B-cell maturation in the bone marrow; smaller thymi and spleen size; defective T cell function | IL-2, IL-4, IL-7, IL-9, IL-13, IL-15, IL-21 | [25, 26, 160] | |
|
| |||||||
| JAK3 | Human | Nonsense mutation | Viable | Diminished T and NK cell numbers; normal B cell numbers. | IL-4 | [161, 162] | |
|
| |||||||
| Tyk2 | Mouse | Germline knockout. B10.Q/J strain devoid of TYK2 protein. | Viable | Macrophages unresponsive to LPS stimulation. Reduced IL-12 mediated T-cell response following viral challenge. | IL-12; IFNα/β and IFNγ (at low doses); IL-23; | IL-3; IL-6, LIF; IL-10; G-CSF, TPO | [30, 163–165] |
|
| |||||||
| Tyk2 | Mouse | Conditional: myeloid, dendritic, T-cell, ubiquitous | Viable | Myeloid deletion reduced MCMV defence. Ubiquitous, but not lineage specific deletions, impaired tumour immunosurveillance | [166] | ||
|
| |||||||
| TYK2 | Human | Nonsense mutation | Viable | Susceptibility to viral, fungal and mycobacteria infection. Atopic dermatitis | IL-12; IFNα/β; IFNγ; IL-6; IL-10; IL-23 | [31] | |
|
| |||||||
| SCID: severe combined immune deficiency; LPS: lipopolysaccharide. | |||||||
Class I cytokines. TPO, EPO, G-CSF, GH, PRL signal via homodimeric receptors (c-MPL, EPOR, G-CSFR, GHR, PRLR, respectively). GM-CSF and IL-5 signal via receptors composed of common-β (βc) and cytokine specific α-subunits; IL-3 via βc or an IL-3 specific β-subunit (βIL-3) and an IL-3 specific α-subunit. IL-2, IL-4, IL-7, IL-9, IL-13, IL-15, IL-21 signal via receptors composed of a common γ-subunit (γc) and cytokine-specific α-subunits. IL-6, IL-11, LIF, CNTF, cardiotrophin-1 and oncostatin-M signal via receptors composed of the shared subunit, gp130, and cytokine-specific α subunits. IL-12 signals via a receptor composed of IL-12β1 and β2 subunits. IL-23 signals via a receptor composed of IL-12β1 and an IL-23-specific α-subunit.
Class II cytokines. IFN-α and IFN-β signal via a receptor composed of Interferon α receptors 1 and 2 (IFNAR1 and IFNAR2). IFNγ signals via a receptor composed of IFNGR1 and IFNGR2. IL-10 signals via a receptor composed of IL-10α and IL-10β subunits. IL-22 signals via a receptor composed of IL-10Rβ and a cytokine-specific α-subunit.
Findings in these mouse knockout cells reinforce the notion that there are obligate relationships between cytokine receptors and their JAK effectors, where a preference exists for which JAK family member serves as the signalling effector of each cytokine receptor. Although these studies have successfully identified obligate relationships between cytokine receptors and JAKs, one shortcoming is that they do not illuminate functional redundancy. This is exemplified by studies of G-CSF receptor signalling, where wild-type foetal liver cells and counterparts lacking Jak1, Jak2 or Tyk2 responded equivalently to G-CSF (Table 1) [18, 20, 30], indicating that in the absence of one JAK family kinase, another family member can fulfil the same signalling function. Relatedly, another confounding factor in interpreting the relationships between JAK family members and cytokine receptors using JAK knockout models is the idea that one JAK family member may function as the upstream activator of a neighbouring heterotypic JAK family member within a cytokine receptor complex. Consequently, where such hierarchies exist, deletion of a JAK family member would be expected to compromise activation of a downstream JAK, which would serve the primary effector role. Broadly, this idea is supported by studies performed on cells derived from JAK-deficient mice. For example, while cells derived from Jak1−/− foetal livers exhibited defective, albeit measurable, signalling upon IL-3 stimulation [18], those from Jak2−/− foetal liver cells exhibited no detectable response to IL-3 [20]. These observations support the idea that the intracellular regions of the IL-3 receptor α and β subunits serve as scaffolds to recruit JAK1 and JAK2 to facilitate JAK1-mediated activation of JAK2 following IL-3 ligation. In such a model, JAK2 serves as the essential signalling effector, which relies on JAK1 for its optimal activation.
JAK mutations in human disease
Mutations in all four JAKs have been associated with human diseases. Inherited mutated JAK alleles lead to inactivated JAK3 and TYK2 in human immune deficiency syndromes (Table 1), while somatic mutations in JAK1, JAK2 and JAK3 result in constitutively active kinases in myeloproliferative diseases and leukaemia/lymphomas.
A single patient with hyper IgE syndrome, multiple infections and impaired responses to interferon and multiple other cytokines (including Th2 polarization) was shown to have a homozygous four nucleotide deletion leading to frameshift mutation and premature termination in the FERM domain resulting in no functional TYK2 (Table 1) [31]. However, a second patient, while also susceptible to viral and other infections, did not display hyper IgE syndrome [32]. There is an interesting discrepancy between the phenotypes of the Tyk2−/− mice and humans lacking TYK2, suggesting that interspecies differences likely exist (Table 1). Like cells derived from Tyk2−/− mice, peripheral blood cells from a TYK2 deficient human exhibited defective IL-12, IL-23 and IFNα/β signalling, but additional severe defects in IL-6 and IL-10 signalling that were not observed in the mouse [31]. These studies may indicate that evolutionary divergence in either the receptor specificity underlying JAK utilization or the capacity for compensatory signalling in different organisms or genetic backgrounds.
Somatic activating mutations in JAK1 have been detected in up to 20% of adult T-cell acute lymphoblastic leukemia (T-cell ALL) and a lower fraction of B-cell ALL with the majority of activating mutations occurring in the pseudokinase domain [33, 34]. Low frequency mutations in JAK1 have also been detected in AML and non-haemopoietic tumours such as hepatocellular adenomas [35].
Somatic activating mutations in JAK2 (most commonly V617F) in the pseudokinase or adjacent domain are found at high frequency in myeloproliferative neoplasms [36–49]. The highest frequency (up to 95%) is in patients with polycythemia vera, with 50–60% in essential thrombocythemia, and idiopathic myelofibrosis [50]. In addition, lower frequencies of the mutation occur in Ph-negative chronic myeloid leukaemia, chronic myelomonocytic leukemia, megakaryocytic AML and juvenile myelomonocytic leukaemia (10–20%) [50]. Other mutations in the JAK2 pseudokinase domain (including point mutations involving R683) have been detected in about 20% of Down Syndrome-associated acute lymphoblastic leukaemia)[47] and other ALLs and AML. A number of JAK2 fusion proteins (usually involving the N-terminal dimerization domains of the fusion partner and the C-terminal kinase domain of JAK2 with or without the adjacent pseudokinase domain) lead to activation of JAK kinase activity and have also been associated with myeloid and lymphoid leukaemias or atypical CMLs. They include the TEL-JAK2 [51, 52], PCM1-JAK2 [53, 54] and BCR-JAK2 fusion proteins [55].
Mutations in JAK3 have been described in 7–14% of patients with severe combined immunodeficiency (SCID) exhibiting a lack of T- and NK cells [56]. At least 34 mutations have been described covering all of the sub-domains of the protein and most result in a lack of functional protein expression. Most patients are compound heterozygotes having inherited a mutated allele from each parent. JAK3 associates with the common γ-chain of the IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 receptors, mutations of which are associated with the most common form of SCID (X-linked) [56] (Table 1). Activating mutations in the FERM or pseudokinase domains of JAK3 have also been associated with acute megakaryocytic leukaemia [57].
The above data point to a clear role for hyperactive JAKs in human hyper-inflammatory/autoimmune diseases and myeloproliferative diseases. Consequently there has been extensive interest by pharmaceutical companies in developing JAK inhibitors to target autoimmune disease/immunosuppression (anti-JAK1, JAK3) and myeloproliferative neoplasms and leukemia/lymphoma (anti-JAK2, JAK1). At least 10 small molecule JAK inhibitors are in current clinical trials for myeloproliferative neoplasms, rheumatoid arthritis, psoriasis and inflammatory bowel disease. All of these compounds target the ATP-binding site of JAKs and so none are absolutely specific for any JAK. Nevertheless Ruxolitinib™ (a JAK1, JAK2 inhibitor) has been approved for use in myelofibrosis and Tofacitinib™ (a JAK1, JAK3 inhibitor) has been approved for use in rheumatoid arthritis. Ruxolitinib™ treatment substantially reduced spleen size and improved quality of life in a significant proportion of patients with myelofibrosis but, surprisingly, treatment was also effective in patients without mutated JAK2 and there was not convincing evidence of reduction in mutated allele burden or progression to AML [58, 59]. Tofacitinib™ was more effective in improving symptoms in rheumatoid arthritis than placebo or existing anti-TNF receptor therapy [60]. While some of these results are promising, it is clear that a better understanding of the specificity of JAK inhibitors is required to draw firm conclusions about optimal therapies for these diseases.
Structure and activation of JAK kinases
All four members of the JAK family share a common domain architecture that is distinct from other tyrosine kinases (Figure 1B). The N-terminal half comprises a band 4.1, ezrin, radixin, moesin (FERM) domain and an SH2 domain that mediates JAK association with the cytoplasmic tail of a variety of cytokine receptors [61], while the second half contains a catalytically-defective pseudokinase domain (Jak Homology 2, “JH2”) that is critical for modulating the activity of a C-terminal tyrosine kinase domain (JH1) [62–64]. It is generally accepted that activation of JAKs is initiated by cytokine binding to their cognate receptor dimer, an event that triggers a change of conformation of the receptor dimer and repositioning of the associated JAKs. Such a rearrangement leads to the apposition of the JAK tyrosine kinase domains, transphosphorylation of conserved activation loop tyrosines [65] and a concomitant elevation in catalytic activity [66] (Figure 2A). However the precise molecular mechanisms that drive JAK-receptor specificities and the sequential events that lead to JAK activation within the receptor-JAK complex still remain largely elusive.
Figure 2.
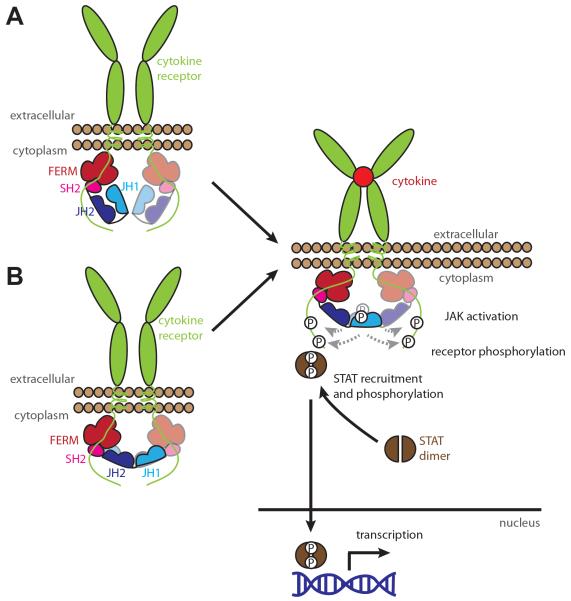
The two prevailing models for regulation of JAK kinase domain catalytic activity by the pseudokinase domain: (A) in cis; (B) in trans. In the in cis inhibition model (A), the pseudokinase domain binds the kinase domain within the same JAK monomer, leading to a suppression in catalytic activity. The in trans model for inhibition (B) involves the binding of the pseudokinase domain from one JAK to the kinase domain of another within a receptor-assembled JAK dimer to suppress the kinase domain's catalytic activity. Activation of JAK in either model involves reorientation of the JAKs to facilitate mutual trans-phosphorylation and thus activation of the JAK kinase domains.
With the overwhelming majority of the activating mutations located in the pseudokinase domain and the adjacent linker region connecting the pseudokinase domain to the SH2 domain (Figure 1B), considerable efforts have been deployed over the past decade to understand how the pseudokinase domain might exerts its inhibitory activity on the catalytic domain. While the recent crystal structures of the pseudokinase domains of JAK1, JAK2 and TYK2 in isolation did not unravel the process of activation of the JAKs, they provided us with some valuable insights [67, 68].
Despite missing key residues that are known to be crucial for catalytic activity, the pseudokinase domain has retained the capability to bind nucleotides in the presence of divalent cations [69]. However, only the JAK2 pseudokinase domain has been shown to possess weak catalytic activity and to autophosphorylate itself in cis on two auto-inhibitory phosphorylation sites, Ser523 and Tyr570 [64]. Those residues are not conserved amongst the other JAKs suggesting a JAK2-specific mechanism of activation. Intriguingly, the residues that compose the “activation loop” in the JAK2, JAK1 and TYK2 pseudokinase domains structures adopt a well-defined conformation reminiscent of inactive protein kinase structures and are predicted to occlude the putative substrate binding site, suggesting that a rearrangement of JAK2 pseudokinase activation loop would need to occur to allow phosphoryl-transfer activity [67]. Although evidently different from JAK1, it remains to be established whether the weak catalytic activity exhibited by the JAK2 pseudokinase domain plays an essential physiological role in vivo. One possibility is that nucleotide binding to these pseudokinase domains solely serves a role in modulating the overall conformation of the JAK. The significant stabilization conferred by ATP binding on purified recombinant JAK1 and JAK2 pseudokinase domains [69] may highlight the necessity to lock a particular conformation for intermolecular interactions to occur. An emerging idea is that nucleotide binding by pseudokinase domains may underlie or be indicative of a “molecular switch” mechanism [69, 70], although the physiological relevance of conformational switching amongst the JAK pseudokinase domains remains to be established.
Interestingly, the crystal structure of the JAK2 pseudokinase domain harbouring the activating mutant, V617F, points toward enhanced structural integrity within this domain underlying pathogenesis. The groups of Silvennoinen and Hubbard proposed that the activating effect of the V617F mutation arises from stabilisation of the αC helix via an aromatic stacking interaction between V617F and two phenylalanines (Phe595 and Phe594) from the αC helix, resulting in an increased length of the αC helix [68]. This is supported by biochemical analyses in which optimal activation of JAK2-V617F required an aromatic amino acid at residue F595 [71]. However, a subsequent structure of the JAK1 pseudokinase domain revealed that the length of the αC region can vary regardless of the presence of the corresponding activating mutation, V658F [67]. In the case of JAK1, Toms, Eck et al. proposed a model in which the V658F mutation in JAK1 promotes activation primarily via rearrangement of the pseudokinase domain and pseudokinase-SH2 linker, again through aromatic stacking interactions. A highly conserved triad Phe-Phe-Val that include Phe575 (Phe537 in JAK2) located in the linker region, Phe636 (Phe595 in JAK2) located in the αC helix and V658 (V617F in JAK2) was shown to be critical for this rearrangement to occur in the JAK1 pseudokinase domain structure [67]. In the structure of the JAK1 pseudokinase domain harbouring the activating V658F mutant, the β4-β5 loop was found to fold toward the αC helix, and the phenyl ring of Phe658 to occupy the site normally occupied by Phe575 in the wild-type structure, pushing away the pseudokinase-SH2 linker region [67]. Nevertheless, it was also shown that this conformation can occur in the wild-type JAK1 pseudokinase domain, suggesting that this region is prone to movement but may be stabilized in an activated conformation by the oncogenic V617F (JAK2)/V658F (JAK1) mutation [67]. Importantly, mutations in the JAK2 pseudokinase-SH2 linker region, such as K539I, were recently identified in polycythemia vera (PV) patients and reported to induce constitutive JAK2 activation [72].
Based on functional studies of activation, two distinct mutational hotspots within the JAK2 pseudokinase domain have been identified. Mutations that map to the N-terminal lobe of the domain, adjacent to V617F (e.g. R588M, N622I, S591L), and in the pseudokinase-SH2 linker region were found to confer factor-independent growth, despite not always exhibiting detectable elevations in catalytic activity [72]. In comparison, JAK2 pseudokinase domain mutations located close to the hinge region on the opposite face (L611S) and at either tip of an adjacent loop that is unique to JAK pseudokinase domains (I682F, R683S and F694L) induced elevated activation loop autophosphorylation within the kinase (JH1) domain and were shown to lead to activation in the absence of erythropoietin (EPO) stimulation [72]. Although two functionally-distinct regulatory interfaces appear to exist on opposite faces of the JAK2 pseudokinase domain, a unifying feature is the observation that cytokine receptor binding is crucial for their transforming potential [73–75].
While it is clear that the JAK pseudokinase domains act as protein interaction modules, whose primary function is to bind and inhibit the tyrosine kinase (JH1) domain activation, there are many conflicting reports on how this inhibition might occur. Two basic models of JAK activation have been proposed, in which the pseudokinase domain binds and inhibits the kinase domain (A) in cis or (B) in trans (Figure 2). Here, we discuss the evidence in support of each model, with particular emphasis on recent advances.
The recent crystal structure of the TYK2 pseudokinase-kinase tandem domains (PDB, 4OLI; [76] supports an inhibition mechanism occurring in cis, with an interacting interface between the pseudokinase domain and the kinase domain predominantly mediated by the N-lobes of each domain (Figure 3A). The pseudokinase domain interacts across the back of the kinase domain's ATP-binding site via an interface comprising residues located in the pseudokinase-SH2 linker region, between the αC helix and β4 strand, and extends out to residues near the unique pseudokinase domain loop in the C-terminal lobe. The interface on the kinase domain that mediates pseudokinase domain interaction is mainly located around the hinge region and the loop opposite to the glycine loop between the β2 and β3 strands. The pseudokinase-SH2 linker mainly interacts with the kinase domain hinge region, but it is yet to be demonstrated if this interaction remains in the full-length context. Importantly, the kinase domain adopts an active-like conformation with the activation loop expelled from the active site, despite the use of a kinase-dead mutant (D1023N), suggesting that the release of the activation loop upon phosphorylation does not participate in the activation process. Instead, the N-terminal back-to-back interaction implies an in cis-autoinhibition mechanism that hinders the flexibility of the kinase domain's hinge region, hence restricting rotation between the N- and C-lobes of the kinase domain and the correct positioning of the αC helix and the catalytic elements that drive catalysis. Nevertheless, it is still unclear how this interplay between kinase and pseudokinase domains has a negative effect on the JAK activity in a receptor complex. The in cis-mechanism of autoinhibition described for TYK2 may represent a snapshot of a part of a process that drives autoinhibition/activation, but it is yet to be demonstrated if it is a conserved mechanism amongst the JAK family.
Figure 3.
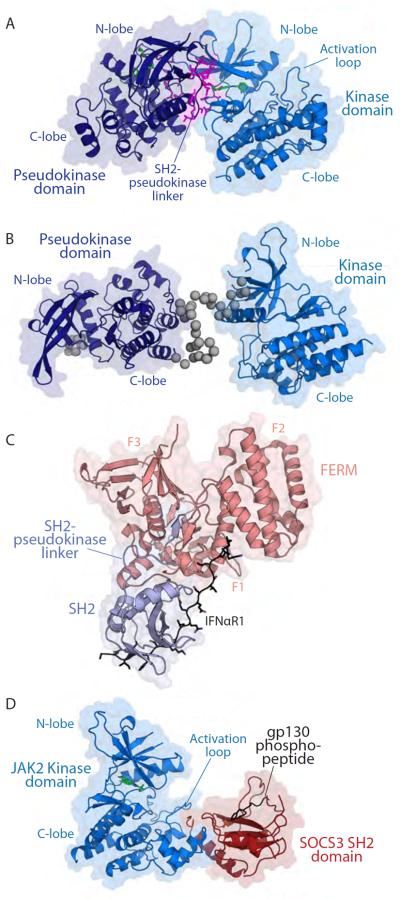
Structures of component domains within JAK family members.
A. Crystal structure of TYK2 pseudokinase-kinase tandem domains (PDB, 4OLI; [76]). The pseudokinase domain is coloured in deep blue and the kinase domain is coloured in marine. Residues involved in the pseudokinase-kinase domain interface are shown as magenta sticks.
B. The solution structure of JAK2 pseudokinase-kinase tandem domains determined by SAXS [77]. Domains are coloured as for TYK2 in panel A, but with grey beads to model the N-terminus and interdomain linker. The component crystal structures of the JAK2 pseudokinase (PDB, 4FVP; [68]) and kinase (PDB, 2B7A; [157]) domains were used to prepare this rigid body model.
C. Crystal structure of TYK2-FERM SH2 domain in complex with IFNAR1 (PDB, 4PO6; [82]). The FERM domain is coloured in salmon with F1, F2 and F3 subdomains labelled. The SH2 domain is coloured pale blue. The IFNAR1 Box 2 peptide is coloured black.
D. Crystal structure of the JAK2 kinase domain (blue) in complex with the SOCS3 SH2 domain (red) and the gp130 phospho-Y757 peptide (black) (PDB, 4GL9; [114]). Small molecule inhibitors are shown as green sticks in panels A and D.
In contrast to the TYK2 pseudokinase-kinase crystal structure, our recent analysis of the JAK2 pseudokinase-kinase tandem domains using small angle X-ray scattering (SAXS) demonstrated that these domains exist in an elongated configuration in solution [77] (Figure 3B). While our data favour a model of inhibition in trans, whereby the pseudokinase domain of one JAK molecule inhibits the activation of the kinase domain of a second JAK molecule, it is important to note that the JAK2 pseudokinase-kinase protein was purified in an activated state with the kinase domain activation loop phosphorylated and presumably expelled from the active site. These data demonstrated that relative orientations of the pseudokinase and kinase domains of JAK2 are not fixed, a finding consistent with earlier cryo-electron microscopy studies of full length JAK1 [78]. If the in cis mechanism of inhibition is applicable to JAK2, this may suggest that only a subtle rearrangement of the catalytic elements around the hinge region is needed to drive pseudokinase domain binding, as seen in the TYK2 crystal structure. Should the in cis autoinhibition model arising from the TYK2 structure similarly operate in JAK2, it is likely that catalytically-dead mutants and/or inhibitors will be required to capture and crystallise the autoinhibited conformation.
Recently, Brooks and colleagues have proposed a model for the activation of JAK2 by the growth hormone receptor (GHR) that also supports an in trans autoinhibition mechanism [79] (Figure 2B). Using a combination of FRET analysis and computational modelling, Brooks et al. found that the GHR exists predominantly as a dimer in vivo held together through interactions of its the transmembrane helices. These helices are parallel in the inactive state and binding of the hormone induces the separation of the membrane proximal region and a rearrangement of the transmembrane helices, leading to the separation of the Box 1 motifs in the cytoplasmic receptor chains and their attached JAK2. Such separation triggered JAK2 activation by the repositioning the inhibitory pseudokinase domains away from their opposing kinase domains, bringing the two kinase domains into close proximity for trans-phosphorylation to occur. This model of activation predicts an interacting interface between the N-terminal lobe of the pseudokinase domain of one JAK2 molecule in the vicinity of the V617F mutation and the activation loop residues of the kinase domain of the other JAK2 molecule. This model is consistent with a scenario earlier proposed by Lee, where differences in proximity of the JAK2 kinase domains were hypothesised to underlie differential and graded signalling responses downstream of different cytokine receptors [80]. Further support for the in trans inhibition model has arisen from the demonstration that activating mutations within the JAK2 pseudokinase domain led to deficits in the inhibition of the kinase domains of other JAK family members [81].
At this juncture, there is supporting evidence for both the in cis and in trans models of inhibition of JAK kinase domains by the adjacent pseudokinase domain. One possibility is that different mechanisms of inhibition and activation operate between different JAK family members (e.g. TYK2 vs JAK2) or that the mode of inhibition/activation is governed by the receptor to which the JAK molecules are tethered. Structural studies of full-length JAKs bound to receptor chains, in both active and inactive states, will be required to definitively address this conundrum and define how the pseudokinase-SH2 linker interface relays signals from an activated receptor to the kinase domain. A further step in this direction has been taken with the recent structure of TYK2 FERM-SH2 domain in complex with IFNAR1 receptor Box 2 motif [82] (Figure 3C). This structure highlights the critical scaffolding function of the SH2 domain in mediating receptor interaction and provides a glimpse of how receptor-JAK specificity can be achieved [82]. Of importance is the formation of an antiparallel β-strand near the SH2 domain–receptor interface between residues located in the linker between the F1 and F2 lobes of the FERM domain and the C-terminus of the SH2 domain. Despite poor sequence conservation within the linker between the F1 and F2 lobes of the FERM domain, a β-strand is predicted at this position for the other JAKs. The residues located within the C-terminal end of the SH2 domain are highly conserved within the JAKs and have been shown to be essential for the interaction of JAK2 with EPOR [72], raising the possibility that this region might play a key role in communicating the event of receptor ligation to the kinase domain, thus leading to activation.
Extrinsic regulation of JAKs
The fidelity of JAK signalling is essential to the health of an organism. Consequently, JAK activity is regulated at many levels, by intrinsic regulatory events, such as post-translational modifications (Figure 1B) and the inhibitory function of the pseudokinase domain (detailed above), in addition to the many extrinsic regulators described below and shown schematically in Figure 1A.
Phosphatases
Like many tyrosine kinases in the cell, JAK activity is specifically inhibited by the action of phosphatases. As well as catalysing dephosphorylation of JAK substrates (for example, STATs), certain phosphatases can act directly on JAK itself, inactivating it by dephosphorylating JAK phosphotyrosines that are critical for its activity, particularly phosphotyrosines in the activation loop. The phosphatases that directly target JAK will be discussed in detail below.
SHP1 and SHP2
SHP1 and SHP2 (PTPN6 and PTPN11 respectively) are ~600 amino acid phosphatases that both consist of a C-terminal phosphatase domain and two N-terminal SH2 domains. SHP1 is primarily expressed in haematopoietic cells and Shp1 knockout and mutant mice display a range of haematopoietic abnormalities [83]. SHP1 directly associates with TYK2, JAK1 [84, 85] and JAK2 [86] as well as a number of different cytokine and haematopoietic growth factor receptors, such as those for IL-3 [87], erythropoietin [88] and IFNα [84] all in a cytokine-dependent fashion. Cells from Shp1−/− mice (also known as motheaten) display significantly enhanced JAK1 autophosphorylation in response to IFNα [84]. Detailed mechanistic insights into this phenotype were recently provided by the co-crystal structure of the SHP1 catalytic domain in complex with a phosphopeptide derived from the JAK1 activation loop [89]. SHP1 is found to be hypermethylated in multiple myeloma, mantle cell lymphoma and follicular lymphoma leading to constitutive STAT3 phosphorylation [90, 91]
In contrast to SHP1, SHP2 is ubiquitously expressed. SHP2 binds JAK1 and JAK2 [92], and genetic knockout leads to increased JAK1 autophosphorylation and enhanced IFNα and IFNγ signalling [93]. These studies support the hypothesis that SHP2 directly dephosphorylates JAKs. In addition, activating mutations in SHP2 have been identified in juvenile myelomonocytic leukemia (JMML), B cell acute lymphoblastic leukemia, and acute myeloid leukemia (AML). SHP2 was the first identified phosphatase proto-oncogene [94]. In humans, mutations in SHP2 cause Noonan syndrome [95].
PTP1B and TCPTP
PTP1B (also known as PTPN1) is a 435 amino acid protein that consists of a phosphatase domain, a C-terminal domain with a proline rich motif that allows binding to SH3 domains, and a terminal targeting sequence that tethers it to the cytoplasmic face of the endoplasmic reticulum. PTP1B has a powerful role in regulating signalling via the insulin receptor, and PTP1B knockout mice are less susceptible to type 2 diabetes. However, in addition to this, PTP1B has also been shown to be an important regulator of JAK/STAT signalling, especially downstream of leptin receptor activation, making it an important metabolic regulator [96, 97]. Substrate trapping mutants of PTP1B have shown both TYK2 and JAK2 (but not JAK1) to be substrates for dephosphorylation [98, 99]. In addition to its effects on leptin signalling, over-expression of PTP1B inhibits IFNα and IFNγ signalling, presumably by dephosphorylation of the conserved tyrosines in the activation loop of TYK2 and JAK2, respectively. Finally, knockdown of PTP1B in a breast cancer cell line led to prolactin hypersensitivity due to enhanced JAK2 (and STAT5) phosphorylation [100].
TCPTP (T-cell protein tyrosine phosphatase, PTPN2) is a 415 or 387 amino acid phosphatase that, depending on processing, is similar in sequence to PTP1B. Like PTP1B, TCPTP is found tethered to the ER however the shorter form is also found in the nucleus. TCPTP has been shown to dephosphorylate JAK1 and JAK3 and TCPTP-deficient lymphocytes are hyper-responsive to IL-2 and IFNα and γ [101]. Inactivation of TCPTP occurs in a small percentage of T-ALL cases [102] and these mutations can cooperate with JAK1-activating mutations to give rise to disease [103]
CD45
Unlike the aforementioned phosphatases, CD45 (PTPRC) is a receptor tyrosine phosphatase. It consists of a large extracellular region that includes three fibronectin (FnIII) domains, a transmembrane domain and a cytoplasmic domain that contains two individual phosphatase domains. The major evidence for its role in regulation of JAK/STAT signalling was discovered by Irie-Sasaki et al., who showed that CD45 dephosphorylates all four JAKs. In particular phosphotyrosines in the activation loops of the JAK catalytic domains were found to be CD45 substrates. Consistent with this, CD45−/− cells are hyper-responsive to IL-3 and EPO and show elevated levels of phospho-JAK2 [104]. Loss-of-function mutations of CD45 have been found in a small proportion of T-ALL patients and can be found in combination with activating mutations in JAK1 [105].
SH2 domain containing proteins
The trans-phosphorylation of JAKs, especially at the di-tyrosine motif found in their activation loop is a key step in their activation and is required to initiate downstream signalling pathways [66]. Consequently, physiological inhibitors of JAK signalling often contain an SH2 domain that allows them to target JAK-phosphotyrosines to induce its dephosphorylation, degradation or inhibition. Such inhibitors include the SOCS and LNK families of SH2 domain containing proteins, in addition to the phosphatases, SHP1 and SHP2 (described above). The SOCS and LNK family proteins will now be discussed in detail.
SOCS family
The SOCS family members were initially discovered on the basis of their ability to bind JAK [106] and inhibit cytokine signalling [107, 108]. For the majority of the SOCS family, SOCS protein expression is induced by JAK/STAT signalling and they then act to inhibit the cytokine signalling cascade, forming a negative feedback loop [106–108]
The human genome encodes eight SOCS proteins [109]. Each member of the family consists of an N-terminal domain, a central SH2 domain and a C-terminal SOCS box domain. The latter is the defining characteristic of the SOCS family but is also found in a wider protein superfamily that, like SOCS proteins themselves, function as E3 ubiquitin ligases to promote the ubiquitination (and subsequent proteasomal degradation) of protein substrates. The majority of the SOCS family inhibit cytokine signalling by inducing the degradation of JAK-associated cytokine receptors once they are activated. This results in receptor (and possibly JAK) turnover. However, SOCS1 and SOCS3, alone of the SOCS proteins, can directly inhibit the catalytic activity of JAKs and these will now be discussed further.
SOCS1 and SOCS3
Extensive mutagenesis and characterisation of SOCS1 was used by the laboratory of Yoshimura to define a short region of the protein, upstream of its SH2 domain, that enabled SOCS1 to directly inhibit the catalytic activity of JAK [110]. This short motif was termed the Kinase Inhibitory Region (KIR). Further analysis by Yoshimura's group showed that SOCS3 was also capable of inhibiting JAK catalytic activity via the same KIR motif [111], a trait peculiar to SOCS1 and SOCS3.
Recent structural and mechanistic analyses have revealed the molecular basis of JAK inhibition by SOCS3 (and therefore by analogy, SOCS1). The KIR of SOCS3 is unstructured in the absence of JAK [112, 113], but upon binding to JAK, adopts an extended, β-strand-like conformation that sits in the substrate binding groove of the kinase [114] (Figure 3D). This partially occludes the substrate-binding site and prevents JAK from interacting with substrates, thus inhibiting its ability to initiate downstream signalling. SOCS3 binds JAK2 with approximately micromolar affinity and this is reflected in a similar IC50 in in vitro kinase assays. SOCS1 appears to have the more potent KIR, as replacing the SOCS3 KIR with that of SOCS1 allows 10-fold more potent inhibition of JAK2 kinase activity [115].
SOCS3 can inhibit JAK1, JAK2 and TYK2 via its kinase inhibitory region, but not JAK3 [115]. This is due to the absence of an evolutionarily conserved “GQM” sequence in JAK3 that is present in all vertebrate forms of JAK1, JAK2 and TYK2, where it lines one edge of the substrate binding groove. Despite the ability to inhibit three of the four JAKs, knockout studies have shown that SOCS3 shows specificity, in vivo, for IL-6 family cytokines [116–119], G-CSF [120, 121] and leptin [122]. These cytokines all signal via receptors that contain binding sites for the SOCS3 SH2 domain [123] and this allows SOCS3 to bind to these receptors and their associated JAK1, JAK2 and TYK2 molecules simultaneously [114], indicating that it is specific JAK:receptor dimers that are the true high-affinity targets for SOCS3 action. To date there is no structural data regarding SOCS1, but high sequence conservation with SOCS3 in the KIR and the JAK binding surfaces suggest it may inhibit JAK via the same mechanism. Differences in the SH2 domain sequence, especially around the phosphotyrosine binding groove, indicate that the SH2 domain of SOCS1 will target different sites to SOCS3, potentially explaining why SOCS1 (but not SOCS3) is a potent inhibitor of interferon [124, 125] and IL-2 family [126] cytokine signalling.
LNK
LNK (also known as SH2B3) [127], together with the proteins APS and SH2B comprise a small family of signalling adapter proteins that contain both an SH2 and a PH (Pleckstrin homology) domain [128]. Whilst the SH2 domain of these molecules binds to specific JAK phosphotyrosines, the PH domains bind to phosphoinositides and direct these proteins to the cell membrane. Each member of the family also contains a phenylalanine zipper dimerisation motif and a proline rich domain.
Of the three members of the SH2B family, LNK has a well-characterised negative regulatory role in JAK2-based signalling. SH2B and APS, in contrast, appear to activate JAK signalling in certain contexts. These differential effects are intriguing given that the SH2 domains of LNK, APS and SH2B all target the same site, phosphotyrosine 813 of JAK2, which resides within the SH2-pseudokinase domain linker sequence [129]. It is also remarkable that an analogous motif is present in JAK3 (but not JAK1 or TYK2), yet neither SH2B nor APS impact JAK3 activity or tyrosyl phosphorylation [130]. Thus, it appears that the SH2B family proteins specifically modulate JAK2 activation, although the molecular basis for this specificity is currently poorly understood.
Consistent with a negative regulatory role in haematopoietic JAK/STAT signalling, Lnk−/− mice [131] [132] show increased numbers of HSCs [133] and haematopoietic cells showed increased responsiveness to a number of cytokines including IL-7 [132], EPO [134] and TPO [135, 136],. The effects of LNK action on TPO signalling have been especially well characterised; LNK expression is induced by TPO [137] and LNK and TPO play opposing roles in HSC expansion as revealed by Lnk and Tpo double knockout mice [135]. Overexpression of LNK inhibits megakaryocyte development in mice [138], again consistent with a role in regulating TPO/JAK2 signalling.
Whilst the molecular details of LNK inhibition of TPO signalling have not been fully elucidated, LNK is known to interact directly with JAK2. In particular, the SH2 domain of LNK binds to phospho-Y813 of JAK2 [139, 140], which is located in the short linker sequence between the kinase and pseudokinase domains. The PH domain is also required for efficient regulation of TPO signalling as point mutations in this domain are found in human cases of V617F-negative MPNs [141]. This mutation led to TPO hyper-responsiveness, as did mutating the SH2 domain, as seen by aberrant growth as well as enhanced phospho-JAK2 and phospho-STAT in Ba/F3(MPL) presumably due to mislocalisation of LNK from the plasma membrane into the cytoplasm.
Summary
Although the four members of the JAK family were first cloned more than 20 years ago, it is clear that our understanding of their physiology, molecular mechanisms of activation and negative regulation are far from complete. Guided by very recent molecular level studies, our fundamental understanding of JAK activation and negative regulation have vastly improved. Importantly, this work has opened up a number of novel possibilities for therapeutic development to counter diseases of JAK hyperactivation, including myeloproliferative neoplasms, leukaemias and inflammatory disease. For example, targeting the receptor:JAK interaction or emulating the negative regulation by the JAK pseudokinase domain or exogenous regulators, such as the SOCS proteins, with small molecules may offer a way forward to potent and highly specific JAK modulators to complement ongoing efforts to develop conventional ATP-mimetic kinase inhibitors.
Acknowledgments
FUNDING The authors acknowledge fellowship support from the Australian Research Council (to JJB and JMM) and NHMRC (to NAN); scholarship support from the Leukaemia Foundation and Australian Stem Cell Centre (to LV); grant support from NHMRC (1011804) and the National Institutes of Health, USA (CA22556); with additional support from the Victorian State Government Operational Infrastructure Scheme and NHMRC IRIISS grant (361646).
REFERENCES
- 1.Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zürcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Molecular and cellular biology. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 4.Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rane SG, Reddy EP. JAK3: a novel JAK kinase associated with terminal differentiation of hematopoietic cells. Oncogene. 1994;9:2415–2423. [PubMed] [Google Scholar]
- 6.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. The Journal of biological chemistry. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 8.Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Analysis of Stat3 (signal transducer and activator of transcription 3) dimerization by fluorescence resonance energy transfer in living cells. Biochem J. 2004;377:289–297. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt M, Domoszlai T, Kleshchanok D, Lehmann S, Schmitt A, Poli V, Richtering W, Muller-Newen G. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci. 2011;124:900–909. doi: 10.1242/jcs.072520. [DOI] [PubMed] [Google Scholar]
- 10.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 12.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 13.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobie PE, Ronsin B, Silvennoinen O, Haldosen LA, Norstedt G, Morel G. Constitutive nuclear localization of Janus kinases 1 and 2. Endocrinology. 1996;137:4037–4045. doi: 10.1210/endo.137.9.8756581. [DOI] [PubMed] [Google Scholar]
- 15.Zouein FA, Duhe RJ, Booz GW. JAKs go nuclear: emerging role of nuclear JAK1 and JAK2 in gene expression and cell growth. Growth factors. 2011;29:245–252. doi: 10.3109/08977194.2011.614949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, Rogers NC, Schindler C, Stark GR, Ihle JN, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 17.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur AG, Barbieri G, Witthuhn BA, Schindler C, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and - gamma signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 18.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr., Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 19.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 20.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 21.Park SO, Wamsley HL, Bae K, Hu Z, Li X, Choe SW, Slayton WB, Oh SP, Wagner KU, Sayeski PP. Conditional deletion of Jak2 reveals an essential role in hematopoiesis throughout mouse ontogeny: implications for Jak2 inhibition in humans. PloS one. 2013;8:e59675. doi: 10.1371/journal.pone.0059675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grisouard J, Hao-Shen H, Dirnhofer S, Wagner KU, Skoda RC. Selective deletion of Jak2 in adult mouse hematopoietic cells leads to lethal anemia and thrombocytopenia. Haematologica. 2014;99:e52–54. doi: 10.3324/haematol.2013.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akada H, Akada S, Hutchison RE, Sakamoto K, Wagner KU, Mohi G. Critical role of Jak2 in the maintenance and function of adult hematopoietic stem cells. Stem cells. 2014 doi: 10.1002/stem.1711. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musso T, Johnston JA, Linnekin D, Varesio L, Rowe TK, O'Shea JJ, McVicar DW. Regulation of JAK3 expression in human monocytes: phosphorylation in response to interleukins 2, 4, and 7. J Exp Med. 1995;181:1425–1431. doi: 10.1084/jem.181.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 26.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 27.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 28.DiSanto JP, Muller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 30.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 31.Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, Takada H, Hara T, Kawamura N, Ariga T, Kaneko H, Kondo N, Tsuge I, Yachie A, Sakiyama Y, Iwata T, Bessho F, Ohishi T, Joh K, Imai K, Kogawa K, Shinohara M, Fujieda M, Wakiguchi H, Pasic S, Abinun M, Ochs HD, Renner ED, Jansson A, Belohradsky BH, Metin A, Shimizu N, Mizutani S, Miyawaki T, Nonoyama S, Karasuyama H. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Kilic SS, Hacimustafaoglu M, Boisson-Dupuis S, Kreins AY, Grant AV, Abel L, Casanova JL. A patient with tyrosine kinase 2 deficiency without hyper-IgE syndrome. The Journal of pediatrics. 2012;160:1055–1057. doi: 10.1016/j.jpeds.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, Ariola C, Fodale V, Clappier E, Paoloni F, Martinelli S, Fragale A, Sanchez M, Tavolaro S, Messina M, Cazzaniga G, Camera A, Pizzolo G, Tornesello A, Vignetti M, Battistini A, Cave H, Gelb BD, Renauld JC, Biondi A, Constantinescu SN, Foa R, Tartaglia M. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. The Journal of experimental medicine. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ, Yoo NJ, Lee SH. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14:3716–3721. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 35.Pilati C, Letouze E, Nault JC, Imbeaud S, Boulai A, Calderaro J, Poussin K, Franconi A, Couchy G, Morcrette G, Mallet M, Taouji S, Balabaud C, Terris B, Canal F, Paradis V, Scoazec JY, de Muret A, Guettier C, Bioulac-Sage P, Chevet E, Calvo F, Zucman-Rossi J. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428–441. doi: 10.1016/j.ccr.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR, Project CG. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 37.James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 38.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 39.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Fröhling S, Döhner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ. Identification of an acquired JAK2 mutation in polycythemia vera. The Journal of biological chemistry. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kratz CP, Böll S, Kontny U, Schrappe M, Niemeyer CM, Stanulla M. Mutational screen reveals a novel JAK2 mutation, L611S, in a child with acute lymphoblastic leukemia. Leukemia. 2006;20:381–383. doi: 10.1038/sj.leu.2404060. [DOI] [PubMed] [Google Scholar]
- 42.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF, Green AR. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JW, Kim YG, Soung YH, Han KJ, Kim SY, Rhim HS, Min WS, Nam SW, Park WS, Lee JY, Yoo NJ, Lee SH. The JAK2 V617F mutation in de novo acute myelogenous leukemias. Oncogene. 2006;25:1434–1436. doi: 10.1038/sj.onc.1209163. [DOI] [PubMed] [Google Scholar]
- 44.Grunebach F, Bross-Bach U, Kanz L, Brossart P. Detection of a new JAK2 D620E mutation in addition to V617F in a patient with polycythemia vera. Leukemia. 2006;20:2210–2211. doi: 10.1038/sj.leu.2404419. [DOI] [PubMed] [Google Scholar]
- 45.Schnittger S, Bacher U, Kern W, Schröder M, Haferlach T, Schoch C. Report on two novel nucleotide exchanges in the JAK2 pseudokinase domain: D620E and E627E. Leukemia. 2006;20:2195–2197. doi: 10.1038/sj.leu.2404325. [DOI] [PubMed] [Google Scholar]
- 46.Bercovich D, Ganmore I, Scott LM, Wainreb G, Birger Y, Elimelech A, Shochat C, Cazzaniga G, Biondi A, Basso G, Cario G, Schrappe M, Stanulla M, Strehl S, Haas OA, Mann G, Binder V, Borkhardt A, Kempski H, Trka J, Bielorei B, Avigad S, Stark B, Smith O, Dastugue N, Bourquin JP, Tal NB, Green AR, Izraeli S. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 47.Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, Devidas M, Atlas SR, Chen IM, Clifford RJ, Gerhard DS, Carroll WL, Reaman GH, Smith M, Downing JR, Hunger SP, Willman CL. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2009;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W, Kantarjian H, Zhang X, Yeh C, Zhang ZJ, Verstovsek S, Albitar M. Mutation Profile of JAK2 Transcripts in Patients with Chronic Myeloproliferative Neoplasias. J Mol Diagn. 2010;11:49–53. doi: 10.2353/jmoldx.2009.080114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passamonti F, Elena C, Schnittger S, Skoda R, Green A, Girodon F, Kiladjian J, Mcmullin M, Ruggeri M, Besses C, Vannucchi A, Lippert E, Gisslinger H, Rumi E, Lehmann T, Ortmann C, Pietra D, Pascutto C, Haferlach T, Cazzola M. Molecular and clinical features of the myeloproliferative neoplasm associated with JAK2 exon 12 mutations. Blood. 2011;117:2813–2816. doi: 10.1182/blood-2010-11-316810. [DOI] [PubMed] [Google Scholar]
- 50.Godfrey AL, Green AR. Genotype-phenotype interactions in the myeloproliferative neoplasms. Hematology/oncology clinics of North America. 2012;26:993–1015. doi: 10.1016/j.hoc.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 52.Peeters P, Raynaud SD, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 53.Bousquet M, Quelen C, De Mas V, Duchayne E, Roquefeuil B, Delsol G, Laurent G, Dastugue N, Brousset P. The t(8;9)(p22;p24) translocation in atypical chronic myeloid leukaemia yields a new PCM1-JAK2 fusion gene. Oncogene. 2005;24:7248–7252. doi: 10.1038/sj.onc.1208850. [DOI] [PubMed] [Google Scholar]
- 54.Reiter A, Walz C, Watmore A, Schoch C, Blau I, Schlegelberger B, Berger U, Telford N, Aruliah S, Yin JA, Vanstraelen D, Barker HF, Taylor PC, O'Driscoll A, Benedetti F, Rudolph C, Kolb HJ, Hochhaus A, Hehlmann R, Chase A, Cross NC. The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer research. 2005;65:2662–2667. doi: 10.1158/0008-5472.CAN-04-4263. [DOI] [PubMed] [Google Scholar]
- 55.Griesinger F, Hennig H, Hillmer F, Podleschny M, Steffens R, Pies A, Wormann B, Haase D, Bohlander SK. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes, chromosomes & cancer. 2005;44:329–333. doi: 10.1002/gcc.20235. [DOI] [PubMed] [Google Scholar]
- 56.O'Shea JJ, Husa M, Li D, Hofmann SR, Watford W, Roberts JL, Buckley RH, Changelian P, Candotti F. Jak3 and the pathogenesis of severe combined immunodeficiency. Molecular immunology. 2004;41:727–737. doi: 10.1016/j.molimm.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Walters DK, Mercher T, Gu TL, O'Hare T, Tyner JW, Loriaux M, Goss VL, Lee KA, Eide CA, Wong MJ, Stoffregen EP, McGreevey L, Nardone J, Moore SA, Crispino J, Boggon TJ, Heinrich MC, Deininger MW, Polakiewicz RD, Gilliland DG, Druker BJ. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, Estrov Z, Fridman JS, Bradley EC, Erickson-Viitanen S, Vaddi K, Levy R, Tefferi A. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, Cervantes F, Vannucchi AM, Barbui T, Barosi G. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Liang F, Yin X, Xiao X, Shi P, Wei D, Yao L, Wang Q, Chen Y. Tofacitinib for acute rheumatoid arthritis patients who have had an inadequate response to disease-modifying antirheumatic drug (DMARD): a systematic review and meta-analysis. Clin Rheumatol. 2014;33:165–173. doi: 10.1007/s10067-013-2452-7. [DOI] [PubMed] [Google Scholar]
- 61.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Molecular and cellular biology. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. The Journal of biological chemistry. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 64.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nature structural & molecular biology. 2011;18:971–976. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Molecular and cellular biology. 1997;17:2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chatti K, Farrar WL, Duhé RJ. Tyrosine phosphorylation of the Janus kinase 2 activation loop is essential for a high-activity catalytic state but dispensable for a basal catalytic state. Biochemistry. 2004;43:4272–4283. doi: 10.1021/bi036109b. [DOI] [PubMed] [Google Scholar]
- 67.Toms AV, Deshpande A, McNally R, Jeong Y, Rogers JM, Kim CU, Gruner SM, Ficarro SB, Marto JA, Sattler M, Griffin JD, Eck MJ. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol. 2013;20:1221–1223. doi: 10.1038/nsmb.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nature structural & molecular biology. 2012;19:754–759. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J. 2014;457:323–334. doi: 10.1042/BJ20131174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, Young SN, Varghese LN, Tannahill GM, Hatchell EC, Majewski IJ, Okamoto T, Dobson RCJ, Hilton DJ, Babon JJ, Nicola NA, Strasser A, Silke J, Alexander WS. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 71.Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN. JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PloS one. 2010;5:e11157. doi: 10.1371/journal.pone.0011157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao L, Dong H, Zhang C, Kinch L, Osawa M, Iacovino M, Grishin N, Kyba M, Huang L. A JAK2 Interdomain Linker Relays Epo Receptor Engagement Signals to Kinase Activation. Journal of Biological Chemistry. 2009;284:26988–26998. doi: 10.1074/jbc.M109.011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu X, Huang LJ, Lodish HF. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. The Journal of biological chemistry. 2008;283:5258–5266. doi: 10.1074/jbc.M707125200. [DOI] [PubMed] [Google Scholar]
- 74.Wernig G, Gonneville JR, Crowley BJ, Rodrigues MS, Reddy MM, Hudon HE, Walz C, Reiter A, Podar K, Royer Y, Constantinescu SN, Tomasson MH, Griffin JD, Gilliland DG, Sattler M. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood. 2008;111:3751–3759. doi: 10.1182/blood-2007-07-102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorantla SP, Dechow TN, Grundler R, Illert AL, Zum Büschenfelde CM, Kremer M, Peschel C, Duyster J. Oncogenic JAK2V617F requires an intact SH2-like domain for constitutive activation and induction of a myeloproliferative disease in mice. Blood. 2010;116:4600–4611. doi: 10.1182/blood-2009-07-236133. [DOI] [PubMed] [Google Scholar]
- 76.Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1401180111. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varghese LN, Ungureanu D, Liau NP, Young SN, Laktyushin A, Hammaren H, Lucet IS, Nicola NA, Silvennoinen O, Babon JJ, Murphy JM. Mechanistic insights into activation and SOCS3-mediated inhibition of myeloproliferative neoplasm-associated JAK2 mutants from biochemical and structural analyses. Biochem J. 2014;458:395–405. doi: 10.1042/BJ20131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lupardus PJ, Skiniotis G, Rice AJ, Thomas C, Fischer S, Walz T, Garcia KC. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Ralpha cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure. 2011;19:45–55. doi: 10.1016/j.str.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344:1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 80.Lee TS. On the regulation and activation of JAK2: a novel hypothetical model. Mol Cancer Res. 2013;11:811–814. doi: 10.1158/1541-7786.MCR-12-0555. [DOI] [PubMed] [Google Scholar]
- 81.Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, Liu F, Saunders LM, Mullally A, Abdel-Wahab O, Leung L, Weinstein A, Marubayashi S, Goel A, Gönen M, Estrov Z, Ebert BL, Chiosis G, Nimer SD, Bernstein BE, Verstovsek S, Levine RL. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shultz LD, Rajan TV, Greiner DL. Severe defects in immunity and hematopoiesis caused by SHP-1 protein-tyrosine-phosphatase deficiency. Trends Biotechnol. 1997;15:302–307. doi: 10.1016/S0167-7799(97)01060-3. [DOI] [PubMed] [Google Scholar]
- 84.David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yetter A, Uddin S, Krolewski JJ, Jiao H, Yi T, Platanias LC. Association of the interferon-dependent tyrosine kinase Tyk-2 with the hematopoietic cell phosphatase. J Biol Chem. 1995;270:18179–18182. doi: 10.1074/jbc.270.31.18179. [DOI] [PubMed] [Google Scholar]
- 86.Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, Yi T. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. 1996;16:6985–6992. doi: 10.1128/mcb.16.12.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bone H, Dechert U, Jirik F, Schrader JW, Welham MJ. SHP1 and SHP2 protein-tyrosine phosphatases associate with betac after interleukin-3-induced receptor tyrosine phosphorylation. Identification of potential binding sites and substrates. J Biol Chem. 1997;272:14470–14476. doi: 10.1074/jbc.272.22.14470. [DOI] [PubMed] [Google Scholar]
- 88.Tauchi T, Damen JE, Toyama K, Feng GS, Broxmeyer HE, Krystal G. Tyrosine 425 within the activated erythropoietin receptor binds Syp, reduces the erythropoietin required for Syp tyrosine phosphorylation, and promotes mitogenesis. Blood. 1996;87:4495–4501. [PubMed] [Google Scholar]
- 89.Alicea-Velazquez NL, Jakoncic J, Boggon TJ. Structure-guided studies of the SHP-1/JAK1 interaction provide new insights into phosphatase catalytic domain substrate recognition. Journal of structural biology. 2013;181:243–251. doi: 10.1016/j.jsb.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chim CS, Kwong YL, Fung TK, Liang R. SOCS1 and SHP1 hypermethylation in multiple myeloma: Potential implications for epigenetic activation of the Jak/STAT pathway. Blood. 2003;102:369b–369b. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- 91.Chim CS, Wong KY, Loong F, Srivastava G. SOCS1 and SHP1 hypermethylation in mantle cell lymphoma and follicular lymphoma: implications for epigenetic activation of the Jak/STAT pathway. Leukemia. 2004;18:356–358. doi: 10.1038/sj.leu.2403216. [DOI] [PubMed] [Google Scholar]
- 92.Yin T, Shen R, Feng GS, Yang YC. Molecular characterization of specific interactions between SHP-2 phosphatase and JAK tyrosine kinases. The Journal of biological chemistry. 1997;272:1032–1037. doi: 10.1074/jbc.272.2.1032. [DOI] [PubMed] [Google Scholar]
- 93.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Molecular and Cellular Biology. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–867. doi: 10.1182/blood-2006-07-028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nature Genetics. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 96.Zabolotny JM, Bence-Hanulec KK, Stricker-Krongrad A, Haj F, Wang Y, Minokoshi Y, Kim YB, Elmquist JK, Tartaglia LA, Kahn BB, Neel BG. PTP1B regulates leptin signal transduction in vivo. Developmental cell. 2002;2:489–495. doi: 10.1016/s1534-5807(02)00148-x. [DOI] [PubMed] [Google Scholar]
- 97.Lund IK, Hansen JA, Andersen HS, Moller NPH, Billestrup N. Mechanism of protein tyrosine phosphatase 1B-mediated inhibition of leptin signalling. Journal of Molecular Endocrinology. 2005;34:339–351. doi: 10.1677/jme.1.01694. [DOI] [PubMed] [Google Scholar]
- 98.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 99.Cheng A, Uetani N, Simoncic PD, Chaubey VP, Lee-Loy A, McGlade CJ, Kennedy BP, Tremblay ML. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Developmental cell. 2002;2:497–503. doi: 10.1016/s1534-5807(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 100.Johnson KJ, Peck AR, Liu C, Tran TH, Utama FE, Sjolund AB, Schaber JD, Witkiewicz AK, Rui H. PTP1B suppresses prolactin activation of Stat5 in breast cancer cells. Am J Pathol. 2010;177:2971–2983. doi: 10.2353/ajpath.2010.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Current Biology. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 102.Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, Van Roosbroeck K, Ferrando AA, Langerak AW, Meijerink JPP, Sigaux F, Haferlach T, Wlodarska I, Vandenberghe P, Soulier J, Cools J. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nature Genetics. 2010;42:530–U584. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kleppe M, Soulier J, Asnafi V, Mentens N, Hornakova T, Knoops L, Constantinescu S, Sigaux F, Meijerink JP, Vandenberghe P, Tartaglia M, Foa R, Macintyre E, Haferlach T, Cools J. PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood. 2011;117:7090–7098. doi: 10.1182/blood-2010-10-314286. [DOI] [PubMed] [Google Scholar]
- 104.Irie-Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, Welstead G, Griffiths E, Krawczyk C, Richardson CD, Aitken K, Iscove N, Koretzky G, Johnson P, Liu P, Rothstein DM, Penninger JM. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 105.Porcu M, Kleppe M, Gianfelici V, Geerdens E, De Keersmaecker K, Tartaglia M, Foa R, Soulier J, Cauwelier B, Uyttebroeck A, Macintyre E, Vandenberghe P, Asnafi V, Cools J. Mutation of the receptor tyrosine phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood. 2012;119:4476–4479. doi: 10.1182/blood-2011-09-379958. [DOI] [PubMed] [Google Scholar]
- 106.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 107.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 108.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 109.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes to Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 112.Babon JJ, Yao S, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. FEBS J. 2005;272:6120–6130. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 113.Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 114.Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nature structural & molecular biology. 2013;20:469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WS. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature Immunology. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 118.Kinjyo I, Inoue H, Hamano S, Fukuyama S, Yoshimura T, Koga K, Takaki H, Himeno K, Takaesu G, Kobayashi T, Yoshimura A. Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-beta 1. Journal of Experimental Medicine. 2006;203:1021–1031. doi: 10.1084/jem.20052333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ogata H, Chinen T, Yoshida T, Kinjyo I, Takaesu G, Shiraishi H, Iida M, Kobayashi T, Yoshimura A. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta 1 production. Oncogene. 2006;25:2520–2530. doi: 10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 120.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nature Immunology. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 121.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 122.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Medicine. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 123.Babon JJ, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012;30:207–219. doi: 10.3109/08977194.2012.687375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, Nicola NA, Hertzog PJ, Metcalf D, Hilton DJ. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 125.Bullen DV, Darwiche R, Metcalf D, Handman E, Alexander WS. Neutralization of interferon-gamma in neonatal SOCS1−/− mice prevents fatty degeneration of the liver but not subsequent fatal inflammatory disease. Immunology. 2001;104:92–98. doi: 10.1046/j.0019-2805.2001.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ. JAB/SOCS1/SSI-1 is an interleukin-2-induced inhibitor of IL-2 signaling. Blood. 2001;97:221–226. doi: 10.1182/blood.v97.1.221. [DOI] [PubMed] [Google Scholar]
- 127.Huang X, Li Y, Tanaka K, Moore KG, Hayashi JI. Cloning and characterization of Lnk, a signal transduction protein that links T-cell receptor activation signal to phospholipase C gamma 1, Grb2, and phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 1995;92:11618–11622. doi: 10.1073/pnas.92.25.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 129.Kurzer JH, Saharinen P, Silvennoinen O, Carter-Su C. Binding of SH2-B family members within a potential negative regulatory region maintains JAK2 in an active state. Mol Cell Biol. 2006;26:6381–6394. doi: 10.1128/MCB.00570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O'Brien KB, O'Shea JJ, Carter-Su C. SH2-B family members differentially regulate JAK family tyrosine kinases. The Journal of biological chemistry. 2002;277:8673–8681. doi: 10.1074/jbc.M109165200. [DOI] [PubMed] [Google Scholar]
- 131.Takaki S, Sauer K, Iritani BM, Chien S, Ebihara Y, Tsuji K, Takatsu K, Perlmutter RM. Control of B cell production by the adaptor protein lnk. Definition Of a conserved family of signal-modulating proteins. Immunity. 2000;13:599–609. doi: 10.1016/s1074-7613(00)00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Velazquez L, Cheng AM, Fleming HE, Furlonger C, Vesely S, Bernstein A, Paige CJ, Pawson T. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med. 2002;195:1599–1611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ema H, Sudo K, Seita J, Matsubara A, Morita Y, Osawa M, Takatsu K, Takaki S, Nakauchi H. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Developmental cell. 2005;8:907–914. doi: 10.1016/j.devcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 134.Tong W, Zhang J, Lodish HF. Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood. 2005;105:4604–4612. doi: 10.1182/blood-2004-10-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buza-Vidas N, Antonchuk J, Qian H, Mansson R, Luc S, Zandi S, Anderson K, Takaki S, Nygren JM, Jensen CT, Jacobsen SE. Cytokines regulate postnatal hematopoietic stem cell expansion: opposing roles of thrombopoietin and LNK. Genes Dev. 2006;20:2018–2023. doi: 10.1101/gad.385606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seita J, Ema H, Ooehara J, Yamazaki S, Tadokoro Y, Yamasaki A, Eto K, Takaki S, Takatsu K, Nakauchi H. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci U S A. 2007;104:2349–2354. doi: 10.1073/pnas.0606238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baran-Marszak F, Magdoud H, Desterke C, Alvarado A, Roger C, Harel S, Mazoyer E, Cassinat B, Chevret S, Tonetti C, Giraudier S, Fenaux P, Cymbalista F, Varin-Blank N, Le Bousse-Kerdiles MC, Kiladjian JJ, Velazquez L. Expression level and differential JAK2-V617F-binding of the adaptor protein Lnk regulates JAK2-mediated signals in myeloproliferative neoplasms. Blood. 2010;116:5961–5971. doi: 10.1182/blood-2009-12-256768. [DOI] [PubMed] [Google Scholar]
- 138.Tong W, Lodish HF. Lnk inhibits Tpo-mpl signaling and Tpo-mediated megakaryocytopoiesis. J Exp Med. 2004;200:569–580. doi: 10.1084/jem.20040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bersenev A, Wu C, Balcerek J, Tong W. Lnk controls mouse hematopoietic stem cell self-renewal and quiescence through direct interactions with JAK2. J Clin Invest. 2008;118:2832–2844. doi: 10.1172/JCI35808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Louria-Hayon I, Frelin C, Ruston J, Gish G, Jin J, Kofler MM, Lambert JP, Adissu HA, Milyavsky M, Herrington R, Minden MD, Dick JE, Gingras AC, Iscove NN, Pawson T. Lnk adaptor suppresses radiation resistance and radiation-induced B-cell malignancies by inhibiting IL-11 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20599–20604. doi: 10.1073/pnas.1319665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, Jr., Merker JD, Zehnder JL, Nolan GP, Gotlib J. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang JH, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, Gerhard DS, Loh ML, Reaman GH, Relling MV, Camitta BM, Bowman WP, Smith MA, Willman CL, Downing JR, Hunger SP. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loh ML, Zhang J, Harvey RC, Roberts K, Payne-Turner D, Kang H, Wu G, Chen X, Becksfort J, Edmonson M, Buetow KH, Carroll WL, Chen IM, Wood B, Borowitz MJ, Devidas M, Gerhard DS, Bowman P, Larsen E, Winick N, Raetz E, Smith M, Downing JR, Willman CL, Mullighan CG, Hunger SP. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children's Oncology Group TARGET Project. Blood. 2013;121:485–488. doi: 10.1182/blood-2012-04-422691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsuda T, Feng J, Witthuhn BA, Sekine Y, Ihle JN. Determination of the transphosphorylation sites of Jak2 kinase. Biochemical and biophysical research communications. 2004;325:586–594. doi: 10.1016/j.bbrc.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 145.Godeny MD, Sayyah J, VonDerLinden D, Johns M, Ostrov DA, Caldwell-Busby J, Sayeski PP. The N-terminal SH2 domain of the tyrosine phosphatase, SHP-2, is essential for Jak2-dependent signaling via the angiotensin II type AT1 receptor. Cellular signalling. 2007;19:600–609. doi: 10.1016/j.cellsig.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 146.Feener EP, Rosario F, Dunn SL, Stancheva Z, Myers MG. Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol Cell Biol. 2004;24:4968–4978. doi: 10.1128/MCB.24.11.4968-4978.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Argetsinger LS, Kouadio JL, Steen H, Stensballe A, Jensen ON, Carter-Su C. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol Cell Biol. 2004;24:4955–4967. doi: 10.1128/MCB.24.11.4955-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Robertson SA, Koleva RI, Argetsinger LS, Carter-Su C, Marto JA, Feener EP, Myers MG., Jr. Regulation of Jak2 function by phosphorylation of Tyr317 and Tyr637 during cytokine signaling. Mol Cell Biol. 2009;29:3367–3378. doi: 10.1128/MCB.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 150.Kurzer JH, Argetsinger LS, Zhou YJ, Kouadio JL, O'Shea JJ, Carter-Su C. Tyrosine 813 is a site of JAK2 autophosphorylation critical for activation of JAK2 by SH2-B beta. Mol Cell Biol. 2004;24:4557–4570. doi: 10.1128/MCB.24.10.4557-4570.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ishida-Takahashi R, Rosario F, Gong Y, Kopp K, Stancheva Z, Chen X, Feener EP, Myers MG., Jr. Phosphorylation of Jak2 on Ser(523) inhibits Jak2-dependent leptin receptor signaling. Molecular and cellular biology. 2006;26:4063–4073. doi: 10.1128/MCB.01589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mazurkiewicz-Munoz AM, Argetsinger LS, Kouadio JL, Stensballe A, Jensen ON, Cline JM, Carter-Su C. Phosphorylation of JAK2 at serine 523: a negative regulator of JAK2 that is stimulated by growth hormone and epidermal growth factor. Molecular and cellular biology. 2006;26:4052–4062. doi: 10.1128/MCB.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sedek M, Strous GJ. SUMOylation is a regulator of the translocation of Jak2 between nucleus and cytosol. The Biochemical journal. 2013;453:231–239. doi: 10.1042/BJ20121375. [DOI] [PubMed] [Google Scholar]
- 154.Funakoshi-Tago M, Pelletier S, Matsuda T, Parganas E, Ihle JN. Receptor specific downregulation of cytokine signaling by autophosphorylation in the FERM domain of Jak2. EMBO J. 2006;25:4763–4772. doi: 10.1038/sj.emboj.7601365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Funakoshi-Tago M, Tago K, Kasahara T, Parganas E, Ihle JN. Negative regulation of Jak2 by its auto-phosphorylation at tyrosine 913 via the Epo signaling pathway. Cell Signal. 2008;20:1995–2001. doi: 10.1016/j.cellsig.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 156.Argetsinger LS, Stuckey JA, Robertson SA, Koleva RI, Cline JM, Marto JA, Myers MG, Jr., Carter-Su C. Tyrosines 868, 966, and 972 in the kinase domain of JAK2 are autophosphorylated and required for maximal JAK2 kinase activity. Molecular endocrinology. 2010;24:1062–1076. doi: 10.1210/me.2009-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, Rossjohn J. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 158.Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, Wagner KU. Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis. 2004;40:52–57. doi: 10.1002/gene.20063. [DOI] [PubMed] [Google Scholar]
- 159.Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, Rui H. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Molecular and cellular biology. 2004;24:5510–5520. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 161.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O'Shea JJ, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 162.Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, O'Shea JJ, Leonard WJ. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 163.Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, Shibata S, Asano Y, Gondo H, Sekiguchi K, Nakayama K, Nakayama T, Okamura T, Okamura S, Niho Y, Nakayama K. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 164.Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Muller M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 165.Shaw MH, Boyartchuk V, Wong S, Karaghiosoff M, Ragimbeau J, Pellegrini S, Muller M, Dietrich WF, Yap GS. A natural mutation in the Tyk2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11594–11599. doi: 10.1073/pnas.1930781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vielnascher RM, Hainzl E, Leitner NR, Rammerstorfer M, Popp D, Witalisz A, Rom R, Karaghiosoff M, Kolbe T, Muller S, Rulicke T, Lassnig C, Strobl B, Muller M. Conditional ablation of TYK2 in immunity to viral infection and tumor surveillance. Transgenic research. 2014;23:519–529. doi: 10.1007/s11248-014-9795-y. [DOI] [PubMed] [Google Scholar]