Abstract
Background
Familial hypercholesterolaemia (FH) is an autosomal dominant disease of lipid metabolism, which leads to early coronary heart disease. Mutations in LDLR, APOB and PCSK9 can be detected in 80% of definite FH (DFH) patients. This study aimed to identify novel FH-causing genetic variants in patients with no detectable mutation.
Methods and results
Exomes of 125 unrelated DFH patients were sequenced, as part of the UK10K project. First, analysis of known FH genes identified 23 LDLR and two APOB mutations, and patients with explained causes of FH were excluded from further analysis. Second, common and rare variants in genes associated with low-density lipoprotein cholesterol (LDL-C) levels in genome-wide association study (GWAS) meta-analysis were examined. There was no clear rare variant association in LDL-C GWAS hits; however, there were 29 patients with a high LDL-C SNP score suggestive of polygenic hypercholesterolaemia. Finally, a gene-based burden test for an excess of rare (frequency <0.005) or novel variants in cases versus 1926 controls was performed, with variants with an unlikely functional effect (intronic, synonymous) filtered out.
Conclusions
No major novel locus for FH was detected, with no gene having a functional variant in more than three patients; however, an excess of novel variants was found in 18 genes, of which the strongest candidates included CH25H and INSIG2 (p<4.3×10−4 and p<3.7×10−3, respectively). This suggests that the genetic cause of FH in these unexplained cases is likely to be very heterogeneous, which complicates the diagnostic and novel gene discovery process.
Keywords: Genetics, Lipid Disorders, Diagnosis, Cardiovascular Medicine
Introduction
Familial hypercholesterolaemia (FH (OMIM #143890)) is a genetic disorder, inherited in an autosomal dominant fashion, characterised by the defective plasma clearance of low-density lipoprotein cholesterol (LDL-C) and caused by mutations in three genes: LDLR, APOB and PCSK9.1 A recessive form of FH due to mutations in LDLRAP1 is also known.2 FH is estimated to affect one in 500 individuals3 and if untreated leads to premature coronary heart disease (CHD).4 In the UK, the FH Simon Broome criteria are used for the diagnosis, which classify patients into possible FH, when adults present with total cholesterol >7.5 mmol/L or LDL-C >4.9 mmol/L, and family history of high cholesterol or premature CHD, or the more severe form—definite FH (DFH), when in addition to the above, tendon xanthomas are present in the patient or first or second degree relative.5 The FH mutation detection rate for DFH patients varies between 63% and 87%,6–8 suggesting that there are other genetic causes, located outside of the currently screened regions, which are yet to be identified. The importance of identifying an FH-causing variant, which has clinical utility in providing an unequivocal diagnosis,9 has been emphasised by the National Institute of Health and Care Excellence, which in 2008 recommended cascade testing using DNA information for finding the affected relatives of a patient.10 The risk of early CHD can be significantly reduced by statin treatment,11 and genetic information has been demonstrated to complement the management of treated patients.12
Of FH patients where a mutation can be found, ∼93% occur in the LDLR gene.13 The APOB variant (c.10580G>A, p.(Arg3527Gln)) accounts for ∼5% of UK FH cases,7 8 14 whereas a gain-of-function mutation in PCSK9 (c.1120G>T, p.(Asp374Tyr)) can be found in roughly 1.7% of FH patients.14 In the past few years, several loci have been reported to cosegregate with FH in family linkage studies; however, to date, this has not led to the identification of a specific causal gene.15–17 It is likely that there are novel FH mutations located in unknown genes influencing lipid metabolism and that their discovery may contribute to the identification of novel treatment targets. In order to find novel causes of FH it was agreed that, as part of the UK10K project (http://www.uk10k.org/studies/rarediseases.html), the whole exomes of 125 unrelated DFH patients were sequenced at a high depth. We expected that an FH-causing mutation in a novel gene would be very rare accounting for fewer FH cases than the gain-of-function mutation in PCSK9 (1.7%), since a higher frequency would have made likely its identification in previous studies. We also suspected that a proportion of patients would have polygenic hypercholesterolaemia, due to the combined impact of common LDL-C-raising SNPs.18
Materials and methods
Patients
A total of 125 unrelated patients, diagnosed as DFH using the UK Simon Broome criteria on the basis of the presence or family history of tendon xanthomas, were initially screened and shown to be negative for mutations in known FH genes (LDLR, APOB, PCSK9 and LDLRAP1). All consents and local review board approvals were in accordance with the UK10K project ethical framework. The initial mutation screening methods varied and are summarised in online supplementary table S1.
Controls
The association with FH was tested against consented 1926 UK10K samples with no lipid abnormalities (listed in online supplementary methods), sequenced in parallel, using the same sequence capture and variant calling methods (http://www.uk10k.org/studies/).
Exome sequencing and variant calling
The whole exome sequencing was performed and processed at the Wellcome Trust Sanger Institute (Cambridge, UK) as part of the UK10K project (see online supplementary methods). CNVs were called using the ExomeDepth package for R (freely available at the Comprehensive R Archive Network).19
Filtering of the variants
Variants were flagged as rare (frequency<0.5%) and novel (frequency=0) according to their frequency in publicly available databases including 1000 Genomes20 and National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP6500) (http://evs.gs.washington.edu/EVS/). In addition to the frequency filters, a functional flag was added, which prioritised variants that are most likely to affect a protein's function, that is, non-synonymous, stop gain, stop loss, frameshift deletions and insertions, and splice site variants.
Burden test for association
Rare or novel variants were combined in a single gene manner and counted in cases versus 1926 controls (ie, gene by gene). A binomial test was used to assess the excess of functional rare and novel variants in cases in comparison with the controls. p Values lower than 4×10−3 were taken as evidence sufficient to be flagged for follow-up.
Analysis of the variants
Variants within Tier 1 genes (LDLR, APOB, PCSK9, LDLRAP1) were assessed on the basis of their frequency, and manually by looking at their annotations in the UCL FH mutation database.21 Sanger sequencing was used to confirm all called mutations. Samples with known FH mutations and therefore an explained cause were removed from further analysis.
The Tier 2 list (see online supplementary table S2) consisted of genes associated with LDL-C as a lead trait in the largest (at the time) available Global Lipid Genetic Consortium (GLGC) meta-analysis of genome-wide association studies (GWASs).22 Functional rare and novel variants in the Tier 2 genes were compared by the burden test against non-FH controls, as one group (counts in all genes combined) and by each single gene.
LDL-C gene score analysis
The possibility of polygenic hypercholesterolaemia in this cohort was assessed using the LDL-C gene score analysis, recently described.18 Most of the 12 LDL-raising GWAS SNPs are located outside of the coding regions, and thus to obtain these genotypes, methods as in the original publication were used.18 Gene scores were calculated by summing the weights of LDL-raising alleles provided by the GLGC (see online supplementary table S3) and the APOE haplotype was scored as follows: ε2ε2=−0.9, ε2ε3=−0.4, ε2ε4=−0.2, ε3/ε3=0, ε3ε4=0.1 and ε4ε4=0.2.22 Gene scores of a randomly selected subjects from the UK Whitehall II (WHII) study (n=3020) were used as a healthy control comparison group.23 Individuals with a gene score above 1.16, which was the top decile cut-off for the WHII subjects, were considered to have polygenic hypercholesterolaemia. The Welch two sample t test was used to test for an overall difference between the groups.
Results
We first analysed variants in known FH genes (figure 1A) (for gene coverage see online supplementary results). For LDLR, 10 individuals were carrying a missense mutation, five a nonsense mutation, three had small deletions and two individuals had intronic changes known to affect splicing (see online supplementary table S4). Analysis with ExomeDepth for CNVs identified two large duplications and one deletion within the LDLR region (see online supplementary figure S1). For APOB, two individuals carried the known FH mutation, c.10580G>A (p.R3527Q), and several novel and cases-unique APOB variants, distributed across different gene exons, were identified (see online supplementary table S5). These included a recently identified mutation, p.R50W, which cosegregated with the disease.24 Because APOB is highly polymorphic, the overall number of rare variants was not significantly different in comparison with controls. PCSK9 had the lowest mean read depth (18×), with four exons (1, 5, 9 and 10) covered less than 10× due to a high guanine and/or cytosine (GC) content (see online supplementary figure S2). There were no FH-causing variants identified in this gene. There were no homozygous or compound heterozygous calls in the LDLRAP1 gene in any of the samples. One patient was found to be heterozygous for a previously identified frameshift mutation (c.432_433insA (p.(Ala145LysfsX26))).25
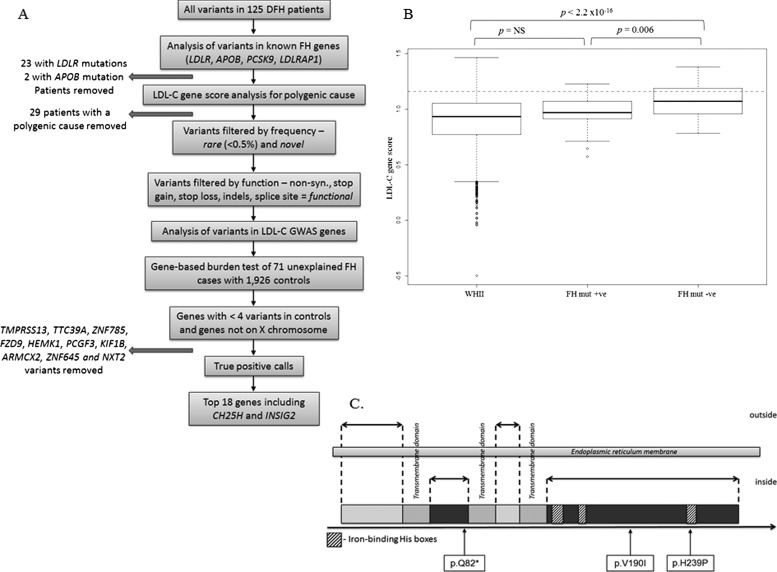
Figure 1.
Novel familial hypercholesterolaemia (FH) gene discovery pipeline. (A) To increase the chance of detecting true FH-causing variants with a strong effect and reduce the noise, samples with a mutation in LDLR or APOB (apart from novel APOB variants of unknown effect) or those with a high low-density lipoprotein cholesterol (LDL-C) gene score were removed from the analysis. The remaining variants were filtered by their frequency and functional effect and compared against controls. Genes with more than four novel functional variants in controls or genes located on the X chromosome were filtered out to enhance the power of the test. The remaining variants were manually assessed and false positive calls were removed. (B) Comparison of the LDL-C SNPs score among the WHII control population (n=3020), FH mutation positive individuals (n=21) and FH mutation negative individuals (n=83) in a standard boxplot (the minimum, lower quartile, median, upper quartile and maximum). The overall difference between the groups was highly significant (ANOVA, p<2.2×10−16). Dashed line indicates the top decile cut-off for the WHII cohort (=1.16). A gene score was not attainable for 16 samples due to a poor DNA quality and insufficient concentration, which resulted in incomplete genotype data. (C) Schematic representation of the intronless CH25H gene and the localisation of novel variants identified in the FH cohort (in boxes). CH25H encodes an enzyme, cholesterol 25-hydroxylase, known to be spanning the endoplasmic reticulum membrane, with two domains (including the N-terminal) located outside of the membrane (in light grey), three 20 amino acid long transmembrane regions and two domains positioned inside the membrane, which contain three His boxes, essential for the catalytic activity of the enzyme.30
LDL-C gene score analysis
Out of 109 FH samples (21 mutation positive, 88 mutation negative) with sufficient DNA for genotyping for all 12 SNPs, 31 had a gene score above the 1.16 cut-off (figure 1A), within which two samples, in addition to the high gene score, had an LDLR mutation, one in exon 11 (c.1690A>C (p.N564H)) found on the same allele as a 9bp deletion in exon 17 (c.2393_2401del9 (p.L799_V801del)), which has been demonstrated as not fully-penetrant.26 The other was a deletion of a consensus splice site at the 5′ of exon 5, c.695-6_698del, which has not been examined in vivo to confirm its likely effect on splicing.
The mean LDL-C gene score for the FH mutation negative group was 1.08, which was significantly higher than 0.90 for the WHII study (p<2.2×10−16), and 0.96 for the FH mutation positive group (p=0.006) (figure 1B) (for the distribution of scores see online supplementary figure S3). The overall difference between the groups was significant (analysis of variance (ANOVA), p=1.33×10−12). Individuals with a gene score above the top decile cut-off for the WHII subjects (>1.16), were considered to have polygenic hypercholesterolaemia and excluded from further analysis as they were unlikely to carry a single mutation of a strong effect.
GWAS LDL-C genes
We next examined any gene identified through GWAS as being involved in determining levels of LDL-C in healthy individuals.22 A burden test on all functional rare and novel variants in any gene singly or in all Tier 2 genes combined showed no obvious candidate for a novel FH locus (see online supplementary table S6). In addition, there were no loss-of-function variants (ie, premature stop codon formation, loss of a stop codon, frameshift indels, CNVs) observed in these genes in any sample (n=125), or in the 71 with no identified mutation and a low gene score. There was no association of novel functional variants in any gene located within the several loci identified by published family linkage studies (see online supplementary table S7).
Whole exome analysis
In all, 25 samples carrying a mutation in Tier 1 genes and 29 with the LDL-C gene score above 1.16 were removed from further analysis. To interrogate the whole exome, a burden test was performed between 71 cases and 1926 controls. There were 4407 genes with one or more novel functional variant in cases. In order to remove calls less likely to influence the FH phenotype and increase the power of the test, we limited further analysis only to genes where a maximum of four novel functional variants were seen in the controls, based on the expected prevalence of FH of 1 in 500, and therefore any gene with >4 novel functional variants in the controls were excluded (the original gene list is shown in online supplementary table S8). Variants in genes located on the X chromosome were removed from the final list (X chromosome genes shown in online supplementary table S8). The next step involved a visual validation of the quality of calls performed using the Human Genome 19 on the Integrative Genomic Viewer (IGV).27 In order to avoid false negatives, calls that were filtered out due to inadequate quality were reanalysed in genes showing excess of novel variants. An additional loss-of-function variant, a premature stop codon at the position c.244C>T (p.Q81*), was found in the CH25H gene in an FH patient sample with a low LDL-C SNP score. After adjusting for the false positives and false negatives, CH25H remained the top gene (p<4.3×10−4) with three variants in the cases and two in the controls (table 1). To examine the prevalence of nonsense variants in CH25H in public data sources, we analysed the NHLBI ESP database and found one nonsense allele (c.638delT) in 6503 individuals (Minor Allele Frequency (MAF)=0.00008), which was significantly lower than in the FH group (MAF=0.0003, p<1.5×10−3).
Table 1.
Summary of genes and their variants which show an excess of novel functional variants in FH cases (n=71) in comparison with controls (n=1926)
| Gene | Ch | Number of variants in cases (n=71) | Number of variants in controls (n=1926) | p Value | ||
|---|---|---|---|---|---|---|
| CH25H | 10 | 3 | 2 | 4.3×10−4 | ||
| Cases | ENST00000371852:exon1:c.G568A:p.V190I; exon1:c.A716C:p.H239P; exon1:c.C244T:p.Q82X | |||||
| Controls | ENST00000371852:exon1:c.T742G:p.C248G; exon1:c.C590A:p.P197Q | |||||
| HSPB7 | 1 | 2 | 0 | 1.3×10−3 | ||
| Cases | 2X ENST00000311890:exon2:c.199+7G>A | |||||
| Controls | None | |||||
| KLRC1 | 12 | 2 | 0 | 1.3×10−3 | ||
| Cases | ENST00000544822:exon5:c.G333C:p.Q111H; exon3:c.C178T:p.H60Y | |||||
| Controls | None | |||||
| MOAP1 | 14 | 3 | 4 | 1.4×10−3 | ||
| Cases | ENST00000556883:exon2:c.C707T:p.A236V; exon2:c.G476C:p.C159S; exon2:c.A182G:p.N61S | |||||
| Controls | ENST00000556883:exon2:c.C655G:p.R219G; exon2:c.C627A:p.S209R; exon2:c.C264G:p.I88M; exon2:c.A919G:p.I307V | |||||
| RBM25 | 14 | 3 | 4 | 1.4×10−3 | ||
| Cases | ENST00000261973:exon6:c.A454T:p.I152F; exon2:c.T50C:p.L17P; exon11:c.C1364A:p.A455D | |||||
| Controls | ENST00000261973:exon7:c.C671T:p.A224V; exon11:c.A1273G:p.R425G; exon18:c.G2392A:p.V798I; exon2:c.T7C:p.F3L | |||||
| ANP32E | 1 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000436748:exon3:c.G227C:p.S76T; ENST00000533654:exon4:c.A434G:p.K145R | |||||
| Controls | ENST00000436748:exon6:c.G629T:p.R210L | |||||
| CABP5 | 19 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000293255:exon4:c.C281A:p.T94N; exon3:c.G201A:p.M67I | |||||
| Controls | ENST00000293255:exon3:c.A169C:p.M57L | |||||
| CELA2B | 1 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000375910:exon6:c.G576A:p.W192X; ENST00000422901:exon3:c.G271A:p.G91R | |||||
| Controls | ENST00000375910:exon7:c.T739C:p.Y247H | |||||
| INSIG2 | 2 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000245787:exon2:c.T89C:p.I30T; exon2:c.C236T:p.T79M | |||||
| Controls | ENST00000245787:exon4:c.G376A:p.D126N | |||||
| KCTD7 | 7 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000275532:exon4:c.G814A:p.V272M; exon4:c.C758T:p.S253L | |||||
| Controls | ENST00000275532:exon4:c.G506A:p.R169Q | |||||
| MRO | 18 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000436348:exon5:c.G578A:p.R193Q; exon5:c.G565A:p.V189I | |||||
| Controls | ENST00000436348:exon3:c.A223G:p.S75G | |||||
| NR2E1 | 6 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000368983:exon1:c.G136A:p.G46S; exon5:c.A634G:p.M212V | |||||
| Controls | ENST00000368983:exon7:c.G1000A:p.V334I | |||||
| PABPC1 | 8 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000318607:exon9:c.A1250C:p.Q417P;exon10:c.G1364A:p.R455H | |||||
| Controls | ENST00000523555:exon3:c.226+3A>G | |||||
| PODXL | 7 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000537928:exon3:c.G821A:p.R274K; exon5:c.A992G:p.H331R | |||||
| Controls | ENST00000537928:exon8:c.C1246G:p.Q416E | |||||
| PUS3 | 11 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000530811:exon1:c.T74C:p.V25A; exon2:c.T824C:p.L275P | |||||
| Controls | ENST00000530811:exon4:c.945-8T>C | |||||
| TXNDC15 | 5 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000511070:exon2:c.C130T:p.R44W; ENST00000507024:exon2:c.G91A:p.A31T | |||||
| Controls | ENST00000358387:exon2:c.G534C:p.E178D | |||||
| WDR89 | 14 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000394942:exon2:c.T821C:p.L274S; exon2:c.A553G:p.M185V | |||||
| Controls | ENST00000394942:exon2:c.A860G:p.D287G | |||||
| ZNF720 | 16 | 2 | 1 | 3.7×10−3 | ||
| Cases | ENST00000398696:exon2:c.T508G:p.L170V; exon2:c.A29G:p.H10R | |||||
| Controls | ENST00000399681:exon6:c.A893G:p.H298R | |||||
Ch, chromosome; FH, familial hypercholesterolaemia.
CH25H and INSIG2 variants
CH25H codes for cholesterol 25-hydroxylase, known to catalyse the formation of the oxysterol—25-hydroxycholesterol (25-HC) (9). The INSIG2 gene, which also exhibited an excess of novel functional variants in the FH cohort in comparison with the controls (p=3.7×10−3) (table 1), has been demonstrated to regulate the activity of Sterol Regulatory Element-Binding Protein (SREBPs), a family of major lipid metabolism transcription factors, via direct biding of 25-HC.28 Thus, both genes, CH25H and INSIG2, are involved in the same pathway of cholesterol metabolism. There were three heterozygous variants found in CH25H, all confirmed by Sanger sequencing (see online supplementary figure S4), of which one leads to a formation of a premature stop codon at residue 81, predicted to have a damaging effect on the protein; the second affects a well-conserved residue across species, c.568G>A (p.V190I); and the third, c.716A>C (p.H239P), alters one of the crucial residues of the His Box 3 domain, known to play a crucial role, together with His Boxes 1 and 2, in the catalytic activity of CH25H29 (figure 1C). Two novel functional variants were found in the control cohort, both being non-synonymous (p.P197Q and p.C248G). The p.P197Q is located in a conserved region of the protein; however, it is predicted as tolerated/benign/neutral by SIFT/PolyPhen/Mutation Taster. The p.C248G variant affects a residue that is not conserved.30
Sanger sequencing also confirmed two novel functional variants in the INSIG2 gene called in the cases, both non-synonymous changes (see online supplementary figure S5). A mutation prediction report generated by Project HOPE31 highlighted that the c.89T>C (p.I30T) variant will cause an empty space in the core of INSIG2 because of the size differences between the wild type Isoleucine and the smaller mutant—Threonine. The other variant, c.236C>T (p.T79M), located in the transmembrane domain of INSIG2, is predicted to have an effect on the hydrophobic interactions within the core of the protein or with the membrane lipids, because the mutant Methionine is more hydrophobic than the wild type Threonine. One rare missense variant was found in INSIG2 in the controls (p.D126N), which was predicted as tolerated/probably damaging/disease causing (by SIFT/PolyPhen/Mutation Taster).
Discussion
In this study, we have identified 25 mutations in known FH genes (23 in LDLR and two in APOB), which were missed by the current screening protocol. Because the sequencing coverage of the PCSK9 gene was lower than for LDLR and APOB, we cannot rule out that there may have been undetected mutations in this gene also. This finding confirmed that LDLR locus is highly heterogeneous and mutations within this gene account for the majority of FH causes. The issue of genetic misdiagnosis and the need for an update of current screening methods have been previously discussed.32 In addition to the known FH mutations, we identified six novel APOB variants, distributed across different exons, in five patients, which included the recently examined p.R50W variant.33 The pathogenicity of these variants remains to be tested. Most of the current mutation screening strategies for FH are focused on a selected region of exon 26 of APOB, because of its established function;34 however, the whole exome sequencing enabled us to analyse the entire coding sequence of the gene, by which we found novel variants unique to the FH cohort.
Polygenic hypercholesterolaemia
The cumulative effect of common LDL-raising alleles in genes identified by GWAS was shown to be the likely cause of high LDL-C in a significant proportion (27%) of the examined patients. A gene score above the top decile for a healthy population cut-off (1.16) was also observed in two patients with considerably mild LDLR mutations, which demonstrates that common polymorphisms can contribute to the presentation of an individual carrying a mild effect FH mutation with LDL-C levels above the diagnostic threshold.
GWAS LDL-C genes
Since common variants in the LDL-C-associated GWAS genes were found to be important in the FH pathogenesis, we looked for evidence that rare variants in these genes were causing FH. Rare and novel functional variants in genes associated with LDL-C levels in the GWAS meta-analysis were not significantly over-represented in the FH cohort, when compared with controls. This suggests that rare variants that have a major effect on function in these genes known to have common LDL-C variants of modest effect are unlikely to be a common cause of FH.
CH25H and INSIG2 variants
Genes CH25H and INSIG2 are the strongest candidates for novel FH loci among the final 18 genes, showing an excess of novel functional variants, based on the available reports on functions of the proteins for which they code. CH25H encodes 25-cholesterol hydroxylase, which catalyses the formation of 25-HC from cholesterol. The gene is located in close proximity to the LIPA gene in which mutations were recently found in patients with autosomal recessive FH phenotype.35 It has been demonstrated that both cholesterol and 25-HC can regulate the function of SREBP, a transcription factor known to regulate the expression of several key players in the lipid metabolism.36 37 It is known that the regulation of SREBP activity depends on binding of 25-HC to INSIG2, encoded by the INSIG2 gene.28 The recently updated GLGC GWAS study with >180 000 individuals has identified an association at the genome-wide level of LDL-C with an INSIG2 gene variant (rs10490626, MAF=0.08).38
The CH25H variants identified in this study have not been observed in 1000 Genomes, 6500ESP and 69CG or the 1926 control exomes. We therefore decided to sequence the gene in an additional cohort of 150 mutation negative FH patients with a low gene score, but no additional amino acid changes were identified.
A detailed literature search and gene ontology analysis of the remaining 16 most significant genes did not reveal any clear association with lipid metabolism. We suspect that the majority of these associations are false positives, and that increasing the number of DFH cases would help to reduce the number of chance signals. It is also possible that some of the top genes are indeed affecting the plasma clearance of LDL-C; however, their biology is yet to be understood.
There are a number of limitations to our study. An alternative study design would be to use Next Generation Sequencing (NGS) of relatives (or trios) from selected families with clear autosomal dominant hypercholesterolaemia. The UK10K study only allowed for 125 subjects with FH to be included, and we calculated that, if we selected 125 singleton no-mutation patients with a clinical diagnosis of DFH, we would expect four to carry a shared mutated locus leading to the defective plasma clearance of LDL cholesterol. The power of the study is clearly dependent on the number of singletons included, with the idea that any identified candidate locus would be sequenced in the family members of the affected proband. While a group of singletons may be genetically heterogeneous, the use of the ‘burden’ analysis and not a single-variant test means that heterogeneity should not reduce power to detect a novel FH-causing gene. Another limitation is that we did not have lipid profile information for individuals in the control comparison cohort, only their rare disease phenotype status, which did not overlap with FH pathogenesis. The possibility that the control cohort includes FH-affected individuals was considered. Assuming that the prevalence of FH is 1/500, we would expect by chance to find ∼4 individuals in this cohort carrying an FH-causing mutation. We have analysed variants in LDLR, APOB and PCSK9 in the control cohort and identified three LDLR and two APOB mutations as incidental findings, which was similar to the expected FH frequency of one in 500. We have also allowed for this prevalence in the control comparison cohort by using a frequency cut-off of four novel gene functional variants in controls, in case any of the novel variants identified in FH cases were also present in the controls. A final limitation is that it is possible that some of the identified variants in the 18 genes in table 1 may be technical false positives, since only for the CH25H and INSIG2 genes were all variants confirmed by Sanger Sequencing, However, to be as certain as possible using bioinformatics that the variants we observed are not false positives, for all these variants we included a visual validation of the quality of calls performed using the Human Genome 19 on the IGV.27
In summary, in 125 DFH unrelated patients without an identified mutation by conventional screening methods, analyses identified 25 disease-causing variants in already known FH loci, as well as six previously unreported APOB variants in five patients. LDL-C gene score analysis found that 31 (29 mutation negative) patients had an SNP score in the top decile of the general population and therefore had a definite polygenic aetiology, and an additional five had a potential functional variant in CH25H or INSIG2. This means that the explanation for the FH phenotype is still lacking in 50% of the patients, suggesting that some causal variants may have been missed at different stages of the data processing or analysis. The variant calling pipeline used for this study was carefully optimised for the majority of the exome regions, though some calls in poorly covered regions could be missed. There is a possibility that there are genetic causes located outside of the protein coding region, affecting protein expression, posttranscriptional stability or altering gene splicing. Also, it is possible that the LDL-C gene score cut-off of 1.16 for polygenic hypercholesterolaemia is too stringent. Thus, using the 9th decile cut-off of 1.08, in which a 41% of WHII individuals had LDL-C above the 4.9 mmol/L (mean LDL-C=4.68±1.05 mmol/L) FH diagnostic level, could be more appropriate. By doing so, the phenotype would be explained in an additional nine mutation-negative patients. A polygenic explanation in additional subjects might also be achieved if SNPs in recently identified LDL-C-raising loci38 were included in the score. Finally, because the burden test results are dependent on the number of associated variants and variants diluting the signal, it is possible that novel FH mutations are located in a highly polymorphic gene, in which it is difficult to pick up the true mutation.
Thus, overall, no major novel locus for FH was detected, with no gene having a functional variant in more than three patients. This suggests that the genetic cause of FH in these unexplained cases is likely to be very heterogeneous, which complicates the novel gene discovery and diagnostic process.
Supplementary Material
Acknowledgments
This study makes use of data generated by the UK10K Consortium. A full list of the investigators who contributed to the generation of the data is available online (http://www.UK10K.org). Funding for UK10K was provided by the Wellcome Trust under award WT091310.
Footnotes
Collaborators: UK10K Consortium (http://www.uk10k.org).
Contributors: All authors listed on the manuscript fulfil the criteria for authorship. Specific contributions are: MF: writing of the manuscript, data analysis. VP: data analysis, CNVs calling. KWL: LDL-C gene score genotyping. RAW: sample preparation. HAWN, MS, SB, SC, OSD, CAG, RAH, FK, RD, EL, NL, DRN, HS, FMVB: patient clinical diagnosis, selection and study samples providers. UK10K: exome sequencing and data production. SEH: data analysis, project supervision, guarantor of the publication.
Funding: SEH holds a Chair funded by the British Heart Foundation, and SEH and RAW are supported by the BHF (PG08/008) and by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. MF is funded by an MRC CASE award with Gen-Probe Life Sciences, and VP is partially funded by a MRC research grant (G1001158). HAWN is a NIHR Senior Investigator.
Competing interests: None.
Ethics approval: Ethical Governance Framework UK10K. National Research Ethics Service, Cambridgeshire 2 Research Committee (http://www.uk10k.org/ethics.html).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Marks D, Thorogood M, Neil HA, Humphries SE. A review on the diagnosis, natural history, and treatment of familial hypercholesterolaemia. Atherosclerosis 2003;168:1–14 [DOI] [PubMed] [Google Scholar]
- 2.Ciccarese M, Pacifico A, Tonolo G, Pintus P, Nikoshkov A, Zuliani G, Fellin R, Luthman H, Maioli M. A new locus for autosomal recessive hypercholesterolemia maps to human chromosome 15q25-q26. Am J Hum Genet 2000;66:453–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neil HA, Hammond T, Huxley R, Matthews DR, Humphries SE. Extent of underdiagnosis of familial hypercholesterolaemia in routine practice: prospective registry study. BMJ 2000;321:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slack J. Risks of ischaemic heart-disease in familial hyperlipoproteinaemic states. Lancet 1969;2:1380–2 [DOI] [PubMed] [Google Scholar]
- 5.[No authors listed] Mortality in treated heterozygous familial hypercholesterolaemia: implications for clinical management. Scientific Steering Committee on behalf of the Simon Broome Register Group. Atherosclerosis 1999;142:105–12 [PubMed] [Google Scholar]
- 6.Taylor A, Patel K, Tsedeke J, Humphries SE, Norbury G. Mutation screening in patients for familial hypercholesterolaemia (ADH). Clini Genet 2010;77:97–9 [DOI] [PubMed] [Google Scholar]
- 7.Futema M, Whittall RA, Kiley A, Steel LK, Cooper JA, Badmus E, Leigh SE, Karpe F, Neil HA. Analysis of the frequency and spectrum of mutations recognised to cause familial hypercholesterolaemia in routine clinical practice in a UK specialist hospital lipid clinic. Atherosclerosis 2013;229:161–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham CA, McIlhatton BP, Kirk CW, Beattie ED, Lyttle K, Hart P, Neely RD, Young IS, Nicholls DP. Genetic screening protocol for familial hypercholesterolemia which includes splicing defects gives an improved mutation detection rate. Atherosclerosis 2005;182:331–40 [DOI] [PubMed] [Google Scholar]
- 9.Humphries SE, Norbury G, Leigh S, Hadfield SG, Nair D. What is the clinical utility of DNA testing in patients with familial hypercholesterolaemia? Curr Opin Lipidol 2008;19:362–8 [DOI] [PubMed] [Google Scholar]
- 10.Wierzbicki AS, Humphries SE, Minhas R. Familial hypercholesterolaemia: summary of NICE guidance. BMJ 2008;337:a1095. [DOI] [PubMed] [Google Scholar]
- 11.Neil A, Cooper J, Betteridge J, Capps N, McDowell I, Durrington P, Seed M, Humphries SE. Reductions in all-cause, cancer, and coronary mortality in statin-treated patients with heterozygous familial hypercholesterolaemia: a prospective registry study. Eur Heart J 2008;29:2625–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphries SE, Whittall RA, Hubbart CS, Maplebeck S, Cooper JA, Soutar AK, Naoumova R, Thompson GR, Seed M, Durrington PN, Miller JP, Betteridge DJ, Neil HA. Genetic causes of familial hypercholesterolaemia in patients in the UK: relation to plasma lipid levels and coronary heart disease risk. J Med Genet 2006;43:943–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usifo E, Leigh SE, Whittall RA, Lench N, Taylor A, Yeats C, Orengo CA, Martin AC, Celli J, Humphries SE. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet 2012;76:387–401 [DOI] [PubMed] [Google Scholar]
- 14.Taylor A, Wang D, Patel K, Whittall R, Wood G, Farrer M, Neely RD, Fairgrieve S, Nair D, Barbir M, Jones JL, Egan S, Everdale R, Lolin Y, Hughes E, Cooper JA, Hadfield SG, Norbury G, Humphries SE. Mutation detection rate and spectrum in familial hypercholesterolaemia patients in the UK pilot cascade project. Clini Genet 2010;77:572–80 [DOI] [PubMed] [Google Scholar]
- 15.Cenarro A, Garcia-Otin AL, Tejedor MT, Solanas M, Jarauta E, Junquera C, Ros E, Mozas P, Puzo J, Pocovi M, Civeira F. A presumptive new locus for autosomal dominant hypercholesterolemia mapping to 8q24.22. Clini Genet 2011;79:475–81 [DOI] [PubMed] [Google Scholar]
- 16.Marques-Pinheiro A, Marduel M, Rabes JP, Devillers M, Villeger L, Allard D, Weissenbach J, Guerin M, Zair Y, Erlich D, Junien C, Munnich A, Krempf M, Abifadel M, Jais JP, Boileau C, Varret M. A fourth locus for autosomal dominant hypercholesterolemia maps at 16q22.1. Eur J Hum Genet 2010;18:1236–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li X, Zhang YB, Zhang F, Sun L, Lin J, Wang DM, Wang LY. Genome-wide linkage scan of a pedigree with familial hypercholesterolemia suggests susceptibility loci on chromosomes 3q25-26 and 21q22. PLoS ONE 2011;6:e24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talmud PJ, Shah S, Whittall R, Futema M, Howard P, Cooper JA, Harrison SC, Li K, Drenos F, Karpe F, Neil HA, Descamps OS, Langenberg C, Lench N, Kivimaki M, Whittaker J, Hingorani AD, Kumari M, Humphries SE. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet 2013;381:1293–301 [DOI] [PubMed] [Google Scholar]
- 19.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, Kumararatne D, Doffinger R, Nejentsev S. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012;28:2747–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usifo E, Leigh SE, Whittall RA, Lench N, Taylor A, Yeats C, Orengo CA, Martin AC, Celli J, Humphries SE. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet 2012;76:387–401 [DOI] [PubMed] [Google Scholar]
- 22.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marmot MG, Smith GD, Stansfeld S, Patel C, North F, Head J, White I, Brunner E, Feeney A. Health inequalities among British civil servants: the Whitehall II study. Lancet 1991;337:1387–93 [DOI] [PubMed] [Google Scholar]
- 24.Thomas ER, Atanur SS, Norsworthy PJ, Encheva V, Snijders AP, Game L, Vandrovcova J, Siddiq A, Seed M, Soutar AK, Aitman TJ. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol Genet Genomic Med 2013;1:155–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, Barnes R, Cohen JC, Hobbs HH. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 2001;292:1394–8 [DOI] [PubMed] [Google Scholar]
- 26.Mozas P, Cenarro A, Civeira F, Castillo S, Ros E, Pocovi M. Mutation analysis in 36 unrelated Spanish subjects with familial hypercholesterolemia: identification of 3 novel mutations in the LDL receptor gene. Hum Mutat 2000;15:483–4 [DOI] [PubMed] [Google Scholar]
- 27.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA 2007;104:6511–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox BG, Shanklin J, Ai J, Loehr TM, Sanders-Loehr J. Resonance Raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry 1994;33:12776–86 [DOI] [PubMed] [Google Scholar]
- 30.Holmes RS, Vandeberg JL, Cox LA. Genomics and proteomics of vertebrate cholesterol ester lipase (LIPA) and cholesterol 25-hydroxylase (CH25H). 3 Biotech 2011;1:99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC bioinformatics 2010;11:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Futema M, Plagnol V, Whittall RA, Neil HA, Humphries SE. Use of targeted exome sequencing as a diagnostic tool for Familial Hypercholesterolaemia. J Med Genet 2012;49:644–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas ERA, Atanur SS, Norsworthy PJ, Encheva V, Snijders AP, Game L, Vandrovcova J, Siddiq A, Seed M, Soutar AK, Aitman TJ. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol Genet Genomic Med 2013;1:155–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boren J, Lee I, Zhu W, Arnold K, Taylor S, Innerarity TL. Identification of the low density lipoprotein receptor-binding site in apolipoprotein B100 and the modulation of its binding activity by the carboxyl terminus in familial defective apo-B100. J Clin Invest 1998;101:1084–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stitziel NO, Fouchier SW, Sjouke B, Peloso GM, Moscoso AM, Auer PL, Goel A, Gigante B, Barnes TA, Melander O, Orho-Melander M, Duga S, Sivapalaratnam S, Nikpay M, Martinelli N, Girelli D, Jackson RD, Kooperberg C, Lange LA, Ardissino D, McPherson R, Farrall M, Watkins H, Reilly MP, Rader DJ, de Faire U, Schunkert H, Erdmann J, Samani NJ, Charnas L, Altshuler D, Gabriel S, Kastelein JJ, Defesche JC, Nederveen AJ, Kathiresan S, Hovingh GK. Exome sequencing and directed clinical phenotyping diagnose cholesterol ester storage disease presenting as autosomal recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol 2013;33:2909–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem 2004;279:52772–80 [DOI] [PubMed] [Google Scholar]
- 37.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab 2012;16:414–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.