Abstract
Background
Pancreatic adenocarcinoma is among the most resistant of human cancers, yet specific mechanisms of treatment resistance remain poorly understood. Models to study pancreatic cancer resistance remain limited and should reflect in vivo changes that occur within patient tumors. We sought to identify consistent, differentially expressed genes between treatment of naive pancreatic tumors and those exposed to neoadjuvant therapy using a strict, in vivo direct xenograft model system.
Methods
Over a 42-week period, 12 untreated and treated patient tumors were successfully engrafted into NOD/SCID mice. RNA from each treatment group (5 untreated and 4 treated) was isolated in triplicate and subjected to global gene expression analysis. Consistent gene expression changes with treatment were identified and confirmed using RT-PCR and immunohistochemistry.
Results
Engraftment of untreated patient tumors was more frequent than treated tumors (17 of 21 versus 16 of 49, P = .0002) but without differences in observed time until tumor formation. The histology of patient tumors was recapitulated in direct xenograft tumors. Relative to untreated tumors, treated tumors consistently demonstrated more than a 2-fold reduction in TGFβ-R2 mRNA expression and more than a 5-fold increase in IGFBP3 expression (P < .0218) and were confirmed by immunohistochemistry.
Conclusion
Engraftment of human pancreatic tumors into immunodeficient mice prior to and following neoadjuvant therapy is possible and provides an in vivo platform for comparison of global gene expression patterns. The decreased TGFβ-R2 expression and increased IGFBP3 expression among direct xenograft tumors derived from treated tumors relative to untreated tumors suggests a role in therapy resistance and warrants further study.
Pancreatic cancer remains a lethal disease with only 5% of patients surviving 5 years after diagnosis.1 Numerous clinical trials involving multiple, novel therapeutic agents have failed to demonstrate substantial clinical efficacy.2–5 Principal causes for this lack of efficacy include advanced disease at presentation and resistance to systemic therapy. Limited understanding of the mechanisms contributing to therapeutic resistance stem from a lack of clinically relevant, in vivo model systems critical to investigate mechanisms of resistance to conventional agents for the development of effective therapies.
Studies assessing resistance mechanisms in pancreatic cancer have traditionally used established cell lines or xenograft tumors arising from them after heterotopic or orthotopic implantation in immunodeficient mice.6–11 Such tumors, termed indirect xenografts (cell line intermediary between patient and mouse), have proven useful in identifying specific phenotypes associated with chemoresistance.6 In vitro and in vivo models of resistance in pancreatic cancer based on cultured cell lines may not reflect inherent properties of the patient tumors that contribute to clinical therapeutic resistance. Predictably, success in such preclinical models has not translated into successful clinical strategies.
A more recently popularized strategy described as a method to address perceived limitations of cell lines in the study of drug resistance involves the direct implantation of patient tumors into immunodeficient mice without a cell line intermediary.12–14 Potential advantages of this approach include: (1) the preservation of genetic hallmarks and stromal architecture of the original tumor and (2) the expansion of tumor available for additional in vivo and ex vivo studies, including measurement of therapeutic efficacy. However, a systemic application of these techniques using pancreatic adenocarcinoma tumors after exposure to cytotoxic chemotherapy, biologic therapy, and/or radiation therapy has not been previously reported. Examination of tumor capable of surviving or adapting to neoadjuvant treatment agent, in turn, may offer valuable insight into important resistance mechanisms that render pancreatic cancer such a deadly disease.
Our clinical pancreas cancer program has developed and investigated the use of preoperative therapy in the care of patients with localized pancreatic adenocarcinoma.15–17 Here we report our results establishing heterotopic and orthotopic xenograft tumors from tumors resected from consecutive patients with pancreatic adenocarcinoma at our institution, including the successful establishment of tumors from both treated and untreated patients. Molecular examination of specimens obtained from this unique research platform has identified a consistent pattern of molecular alterations associated with the use of preoperative cytotoxic therapy.
MATERIALS AND METHODS
Surgical Resection, Pathologic Examination, and Tissue Acquisition
In accordance with institutional review board (IRB) approved protocols, all patients with biopsy-proven pancreatic adenocarcinoma undergoing planned pancreatic resection were informed of the acquisition and storage of excess tumor tissue and given the opportunity to either consent or decline participation in research studies. Excess patient tumor was collected only after planned surgical resection and pathologic examination were complete.
To minimize time for potential ischemia, a surgical approach was used to preserve the arterial blood supply until just prior to specimen removal.18 The surgical specimen was placed in Tis-U-Sol (Baxter, Deerfield, IL) isotonic solution, and the pancreas was divided along the length of the pancreatic duct. The exposed, inner pancreas was then assessed for the distinct presence of gross tumor tissue and confirmed through immediate pathologic diagnosis. The quantity of tumor available for xenotransplantation was considered sufficient if the acquired sample measured a minimum of 3 × 2 × 1 mm. If a tumor was deemed appropriate for engraftment into mice, a staff pathologist (HW) excised tumor tissue unnecessary for diagnosis or evaluation of tumor margin. In cases of lymph nodes resected along with primary tumor, one-half of the lymph node was submitted for pathologic confirmation of metastatic disease.
Heterotopic Engraftment of Patient Tumor into Immunodeficient Mice
We previously published a protocol detailing the handling and engraftment of patient tumor into immunodeficient mice.13 Briefly, excised patient tumor tissue was immediately placed in chilled (4°C) serum-free RPMI media supplemented with 1% penicillin/streptomycin (antibiotic media) and washed with several 30-mL volumes. Female NOD/SCID mice (NCI, Bethesda) 4–8 weeks old were anesthetized with intraperitoneal injections of a ketamine/ xylazine cocktail (ketamine 100 mg/kg IP and xylazine 20 mg/kg IP). Tumor tissue was then mechanically minced into fragments (~1 mm),3 and 5 tumor fragments were individually placed in a formed tissue pocket. All animals were housed and maintained under guidelines established by the American Association of Laboratory and Animal Care, and animal experiments were performed in accordance with NIH-Animal Care and Use Committee (ACUC) guidelines after MD Anderson Cancer Center IRB approval.
Procurement and Expansion of Direct Xenograft Tumors
Mice were assessed daily for general health and weighed/evaluated weekly for the presence of engrafted tumor(s). The location and date of initial palpation of subcutaneous nodules were recorded, and tumor formation defined as nodules progressing in size to gross tumor. Once tumors reached 1.2 cm in greatest diameter, mice were sacrificed and tumor dissected from mouse subcutaneous tissue under sterile conditions. Approximately one-third of the tumor was placed in a 10% formalin/phosphate-buffered solution for paraffin embedment. A section of newly formed tumor edge measuring 2 mm in thickness was then removed, cut into quadrants, and immediately placed into a prelabeled cryostorage tube and stored in liquid nitrogen for future study. Additional generations of direct xenograft tumors in NOD/SCID mice were generated as outlined.
RNA Isolation, Real-Time PCR Analysis, and Microarray Expression Profiling of Direct Xenograft Tumors
RNA was isolated in triplicate from frozen sections of direct xenograft tumors using an Ambion RNA extraction kit (Ambion, Inc., Austin, TX). Biotin-labeled cRNA was generated using an Illumina RNA amplification kit (Ambion, Inc.) and hybridized to Illumina Human-6v3 chips. Slides were then scanned after washing using BeadStation 500× (Illumina, San Diego, CA) and signal intensities quantified using Beadstudio (Illumina). Quantile normalization was used to normalize the data. Primers for amplification of TGFβ-R2 (Hs00559661m1) and cyclophilin A (Hs03045347gH) were Taq-man primers produced by Applied Biosystems, Foster City, CA. Primers for IGFBP3 were designed using primary design tool (Invitrogen.com: forward: 5′-cagagactcgagcacagcac-3′, reverse: 3′-tggaaatt tggggccagtac-5′; Sigma, St. Louis, MO). Expression of TGFβ-R2 and IGFBP3 mRNA were analyzed by real-time PCR with cyclophilin A serving as an internal control for the quantity of amplifiable RNA in each reaction. The comparative CT method was used to determine relative gene expression levels for each target gene.19
Immunohistochemical Staining of Tumor Specimens
Tumors were fixed in 10% formalin and embedded in paraffin, and 8–10-μm thick sections were affixed to positively charged microscope slides. Slides were rehydrated and steamed in a citrate solution for 90 min and briefly incubated in 3% hydrogen peroxide/PBS solution. Slides were then rinsed in PBS, incubated in a protein solution (Cyto Q immunodiluent buffer, Innovex, Richmond, CA) for 30 min, and then incubated with antibody diluted in protein solution overnight at 4°C. Primary antibodies and dilutions were: anti-cytokeratin 18 (1:500, epitomics, Inc.), anti-TGFβ-R2 (1:400, Millipore, Inc., Billerica, MA), and anti-IGFBP3 (1:50, Abcam, Cambridge, UK). Internal negative controls showed no specific reactivity after staining with secondary antibody. After incubation with anti-rabbit Mach 4 Universal horseradish peroxidase polymer application (Biocare Medical, Inc.), slides were irrigated with PBS and incubated in 3,3-diaminobenzidine (DAB) and counterstained with Gill’s No. 3 hematoxylin.
For collagen staining, tissue sections were deparaffinized and hydrated, and nuclei were stained using Weigert’s hematoxylin for 8 min. Picrosirious red solution was generated from Sirius red F3B (C.I. 35782, 0.5 g in 500-mL saturated aqueous solution of picric acid) and slides stained for 60 minutes, washed twice in acidified water, and dehydrated in 100% ethanol. Using brightfield microscopy, nonoverlapping fields from each slide were taken at 2× magnification. After designation of a threshold that best captured stained collagen under a variety of conditions, each image was measured using Nikon software and an average percent collagen area was tabulated for each slide.
Fluorescent Immunohistochemistry
Immediately after resection, tumor was frozen in O.C.T. compound (Sacura, Finetek, Torrance, CA), sectioned, and immersed in cold acetone for 10 minutes. Slides were then incubated in a 4% fish gelatin protein block solution at 21°C. Slides were then incubated with Rabbit anti-human cytokeratin 18 antibody (1:250, Epitomics, Inc. Burlingame, CA) overnight at 4°C. Alexa Fluor 488 goat antirabbit IgG (1:100, A-11008, Invitrogen, Inc., Carlsbad, CA) was then added to slides for 60 min at 21°C. Slides were counterstained with Hoechst dye (1 mg/mL, Invitrogen, Inc.) and visualized with a fluorescence microscope equipped a Hamamatsu C5810 camera (Nikon Microphot-FXA).
Statistical Methods and Data Processing
BRB ArrayTools version 3.7 developed by the National Cancer Institute was used to analyze array data.20 To select genes that are differentially expressed between untreated and treated samples, a class comparison tool using a 2-sample t test was used to calculate the significance of the observations (P < .001 with FDR < 0.1). To evaluate gene expression patterns, each adjusted gene value was used for unsupervised hierarchical clustering with Cluster and TreeView.21 Statistical significance was considered present for P values of < .05. Comparison of tumor engraftment was performed using Fisher exact test and TTF, and differences in the median fold change in expression of TGFβ-R2 and IGFBP3 between treated and untreated tumors was performed using the t test.
RESULTS
Rate of Tumor Collection
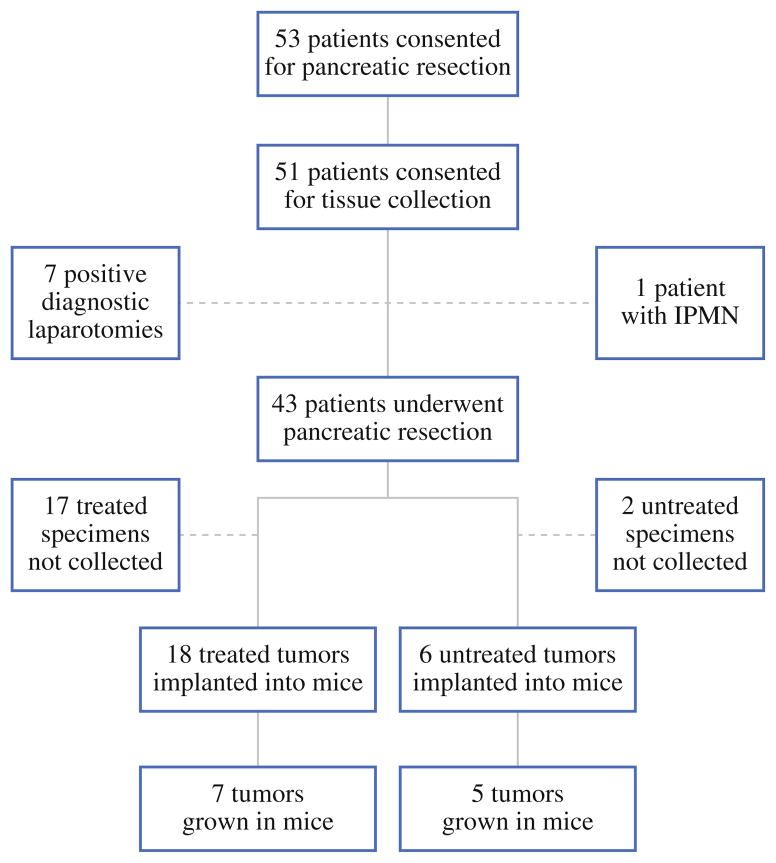
Over a 10-month period, we have systematically implanted pancreatic tumor from 24 different patients into NOD/SCID mice (Fig. 1). During this period, 53 patients consented to pancreatic resection with a preoperative diagnosis of pancreatic adenocarcinoma. The vast majority of patients (51 of 53) consented to pancreatic resection and tumor collection for research and experimental purposes. Among consented patients, 43 ultimately underwent pancreatic resection and represented the true pool from which to harvest tumor for the establishment of xenograft tumors. We collected tumor samples from 24 patients and were unable to collect biospecimen from 19 of 43 patients who ultimately underwent pancreatic resection. Unlike treatment-naive patient tumors that failed to undergo xenotransplantation (2 of 8), patient tumors previously exposed to neoadjuvant therapy more frequently demonstrated loss of tumor volume and definition and were more frequently deemed unsuitable for xenotransplantation (17 of 35). Engraftment of tumor specimen directly into mice has thus far been achieved from 12 of 24 different patient specimens. Among successfully engrafted patient tumor specimens, 5 were derived from untreated patients and the remaining 7 from patients treated with varying regimens of neoadjuvant therapy (Table 1).
FIG. 1.
Flowchart depicting pancreatic tumor acquisition, implantation, and engraftment into immunodeficient mice. Almost half of the surgical specimens obtained from patients who had undergone neoadjuvant therapy were excluded, due largely to poor discernment of tumor. Overall, tumor from 56% of patients that underwent resection was implanted in immunodeficient mice with a 50% engraftment rate
TABLE 1.
Engraftment status and neoadjuvant treatment regimens of pancreatic adenocarcinoma specimens implanted into immunodeficient mice
| Xenograft tumor ID | Growth in mouse | None | XRT | Capecitabine | Gemcitabine | Cisplatin | Oxaliplatin | Cetuximab | Erlotinib | Initial engraftment |
|---|---|---|---|---|---|---|---|---|---|---|
| MDA-PATX1 | + | X | 2/2 | |||||||
| MDA-PATX2 | + | X | 2/4 | |||||||
| MDA-PATX3 | + | X | 1/1 | |||||||
| MDA-PATX4 | + | X | X | X | 2/2 | |||||
| MDA-PATX5 | + | X | 2/2 | |||||||
| MDA-PATX8 | + | X | X | 1/2 | ||||||
| MDA-PATX9 | + | X | X | X | 2/5 | |||||
| MDA-PATX10 | + | X | X | X | X | 4/4 | ||||
| MDA-PATX11 | + | X | 7/7 | |||||||
| MDA-PATX13 | + | X | X | X | X | X | 3/3 | |||
| MDA-PATX16 | + | X | X | 1/5 | ||||||
| MDA-PATX24 | + | X | X | 1/2 | ||||||
| MDA-PATX6 | − | X | X | X | 0/2 | |||||
| MDA-PATX7 | − | X | X | 0/2 | ||||||
| MDA-PATX12 | − | X | 0/1 | |||||||
| MDA-PATX14 | − | X | X | X | X | 0/2 | ||||
| MDA-PATX15 | − | X | X | 0/5 | ||||||
| MDA-PATX17 | − | X | X | X | 0/5 | |||||
| MDA-PATX18 | − | X | X | X | X | 0/4 | ||||
| MDA-PATX19 | − | X | X | 0/3 | ||||||
| MDA-PATX20 | − | X | X | 0/2 | ||||||
| MDA-PATX21 | − | X | X | 0/4 | ||||||
| MDA-PATX22 | − | X | X | 0/5 | ||||||
| MDA-PATX23 | − | X | X | X | X | 0/5 |
Untreated patient tumors engrafted better than treated tumors. Among treated patient tumors, the type or extent of therapy did not appear to affect engraftment of tumor in immunodeficient mice
Engraftment of Tumor from Treated Patients Diminished Relative to Those Untreated
Among the 84 mice into which resected pancreatic tumor were implanted, 33 mice (39%) subsequently developed tumors (Fig. 2a). Engraftment rates among untreated tumors (5 of 6) compare favorably to another published report.14 However, the F1 engraftment rate of tumor derived from treated patients is 40% that of untreated tumors. Irrespective of treatment status, once successful engraftment had been achieved in F1 mice, engraftment of tumor in subsequent generations of mice proved almost universally successful (Fig. 2a). Interestingly, among treated patient specimens that were successfully engrafted in mice, the extent or type of neoadjuvant therapy did not affect the incidence of engraftment (Table 1). Furthermore, the viability of treated tumors as determined by histologic examination of tumor immediately adjacent to implanted tumor did not significantly predict successful tumor engraftment (Fig. 2b).
FIG. 2.
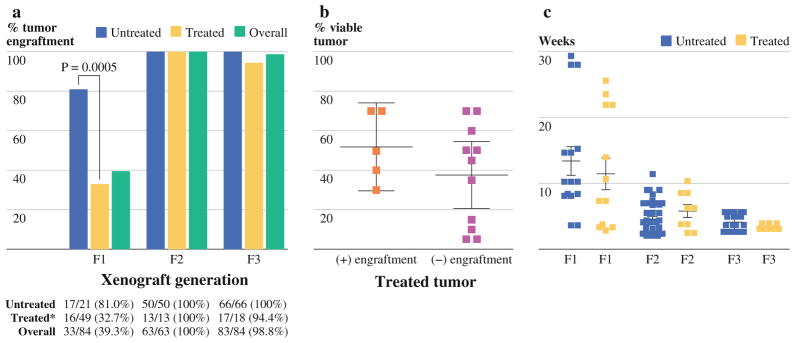
Engraftment of treated and untreated patient specimens into immunodeficient mice. a Specimens derived from patients who have undergone neoadjuvant treatment engraft significantly less than specimens derived from untreated patients. Once grown in immunodeficient mice, direct xenograft tumors derived from either treated or untreated patients may be passaged into subsequent generations of mice (F2, F3) with excellent efficiency. b % viability of treated tumor specimens upon pathologic evaluation does not significantly correlate with engraftment in immunodeficient mice. c The time until tumor formation (TTF) after patient tumor implantation is similar among tumors grown from treated and untreated patient specimens. TTF significantly decreases from the F1 generation to the F3 generation
No Difference in Time Until Tumor Formation (TTF) Observed Between Xenografts Generated From Untreated and Treated Patient Tumors
Despite differences in tumor engraftment among F1 mice, the average time required for tumors to form palpable masses in F1 mice was similar irrespective of neoadjuvant treatment status (Fig. 2c). The average TTF in F1 mice among treatment naïve xenograft tumor was 14.4 weeks (range 3.9–30.1) compared with 12.3 weeks (range 3.6–27.6) for treated xenograft tumors. Likewise, we observed similar variations in TTF within each patient treatment group. Once established in F1 mice, all heterotopic tumors grew significantly faster in subsequent generations of NOD/SCID mice, with the average TTF in untreated and treated F2 mice being 4.9 (range 2.7–12.3 weeks) and 6.2 weeks (2.6–11 weeks), respectively (Fig. 2c). The average TTF among untreated F3 mice was 4.4 weeks (range 2.7–6 weeks) and 3.7 weeks (range 3.4–4.1 weeks), respectively (Fig. 2c).
Histology of Original Patient Tumors Recapitulated in Direct Xenograft Tumors
Direct tumor transfer from patients to mouse hosts results in the production of tumors with strikingly similar histologic features to original patient tumor and include the preservation of tumor glands with cribriform appearance seen in infiltrating ductal adenocarcinoma (Fig. 3a, b). Orthotopic injection of single-cell suspensions obtained from digested xenograft tumors also resulted in tumor gland formation and displayed all histologic features of parental tumors. The histologic appearance of tumors derived from treated samples did not grossly differ from that of untreated tumor and orthotopic xenograft tumors produced from single-cell suspensions in both patient groups displayed similar histology (Fig. 3a). Passage of heterotopic tumor into at least 2 subsequent generations of mice did not affect tumor gland formation or gross histologic features. The human origin of xenograft tumors was confirmed by the presence of human cytokeratin 18 staining (Fig. 3b).
FIG. 3.
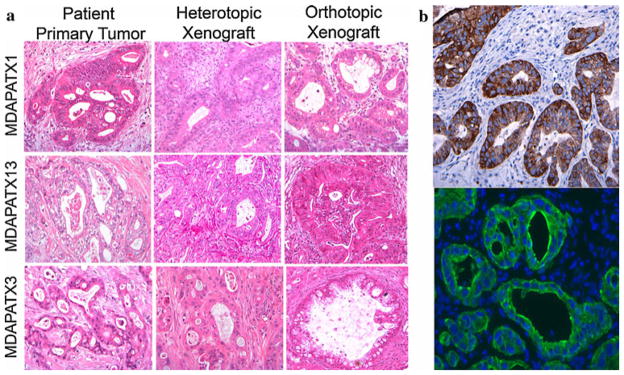
Representative histologic sections of patient and direct xenograft tumors. a H&E staining of original patient tumor, direct xenograft tumor (heterotopic), and direct xenograft tumor (orthotopic) from 3 different patients. Pancreatic tumor glands and surrounding stroma present in original patient tumor are recapitulated in all direct xenograft tumors. b Immunohistochemical and immunofluorescent staining for human cytokeratin 18 (CK-18) in direct xenograft tumors (heterotopic) reveals the presence of pancreatic cancer tumor glands without stromal staining, supporting the human origin of tumor in NOD/SCID mice
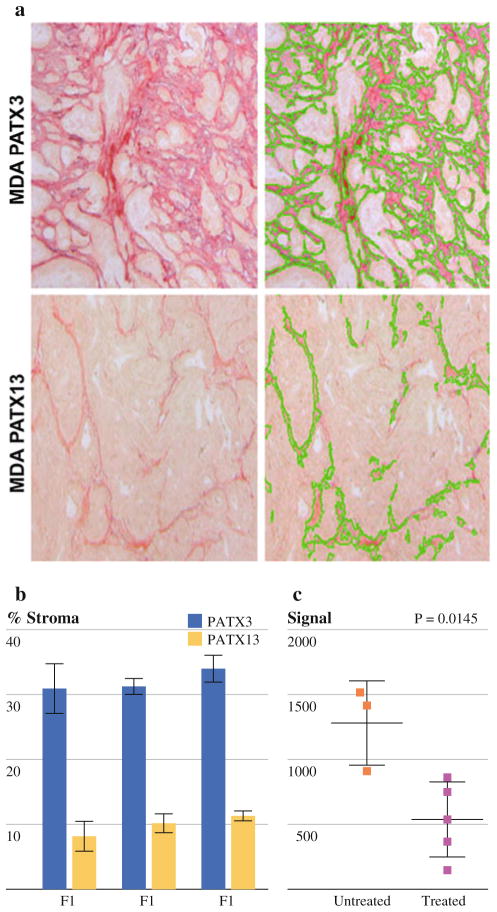
Because of the significant desmoplastic reaction with surrounding stroma in pancreatic cancer, we were interested in evaluating changes in the stromal compartment in engrafted tumors. The percent area of stromal collagen varied between direct xenograft tumors of different lineage (Fig. 4a, b). Direct xenograft tumors from patient 1 (MDA-PATX13) contained ~8%–10% collagen by surface area versus 30%–33% in patient 2 (MDA-PATX1). However, among direct xenograft tumors derived from the same patient, stromal collagen content did not significantly change within the first 3 generations (Fig. 4b). Untreated tumors appeared to form more stroma in direct xenograft tumors relative to treated tumors (P = .0145).
FIG. 4.
Stromal collagen content varies with xenograft lineage. a The % area of collagen within direct xenograft tumors derived from Patient A ranges between 8% and 11% and does not significantly change in generations F1–F3. b Direct xenograft tumors derived from patient B demonstrate % area of collagen between 30% and 33% without significant changes in generations F1–F3
Decreased TGFβ Receptor Type II (TGF β-R2) and Increased Insulinlike Growth Factor Binding Protein 3 (IGFBP3) Expression Identified in Treated Tumors
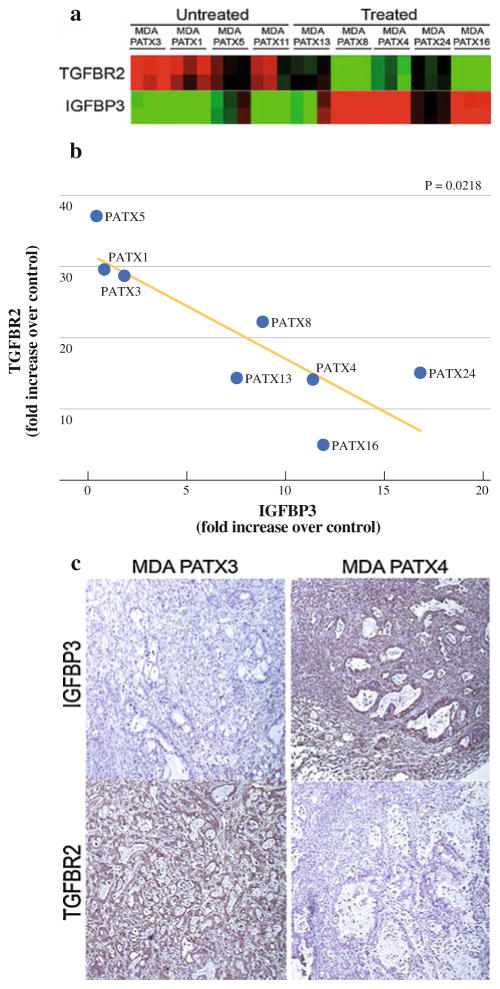
We next sought to identify gene expression changes with treatment through the comparison of global gene expression profiles generated from direct xenograft tumors established from untreated and treated patient specimens. For each patient specimen, RNA was procured from 3 separate F2 mice harboring derived, xenograft tumors. F2 xenograft tumors were not available in triplicate from 3 different patients. Therefore, a total of 9 of 12 direct xenografts underwent genome-wide RNA microarray analysis. Gene expression profiles of direct xenograft tumors derived from untreated (n = 4) patient tumors were compared with expression profiles of xenograft tumors derived from treated (n = 5) patient tumors and consistent differences among each group identified. Examination of expression profiling data identified patterns of expression for TGFβ-R2 and IGFBP3 that were inversely consistent in treated versus untreated specimens (Fig. 5a). To confirm these findings, we performed quantitative RT-PCR analysis of TGFβ-R2 and IGFBP3 gene expression in the treated and untreated samples. We identified a >2-fold decrease in mean TGFβ-R2 expression and a >5 fold increase in IGFBP3 expression among treated xenografts relative to untreated xenografts (Fig. 5b). Immunohistochemical analysis of tumor tissues confirmed decreased protein expression of TGFβ-R2 and a concomitant increase in IGFBP3 expression in samples with identified changes in gene transcript level (Fig. 5c).
FIG. 5.
Inverse relationship of TGFβ-R2 and IGFBP3 in direct xenograft tumors derived from treated and untreated patient specimens. a Heatmap of all examined direct xenograft tumors examining the expression of TGFβ-R2 and IGFBP3. b TGFβ-R2 and IGFBP3 are inversely expressed in direct xenograft tumors with reduced TGFβ-R2 expression and increased IGFBP3 expression after therapy (P = .0218). c Protein expression of TGFβ-R2 and IGFBP3 were confirmed by immunohistochemistry. MDA-PATX3 (untreated) demonstrates clear TGFβ-R2 expression and minimal IGFBP3 expression. Conversely, MDA-PATX4 demonstrates minimal TGFβ-R2 expression and abundant IGFBP3 expression
DISCUSSION
In this report, we demonstrate for the first time that systematic engraftment of pancreatic cancer tumors after neoadjuvant therapy is possible in immunodeficient mice from a significant fraction of patient specimens after surgical resection. To efficiently focus our resources and efforts, only tumors from patients with pancreatic adenocarcinoma were engrafted for direct comparison. When maintained in immunodeficient mice, direct xenograft tumors derived from both treated and untreated patient tumors retained key histologic characteristics of pancreatic adenocarcinoma with excellent engraftment efficiency for multiple generations. Engraftment rates of treated tumors are approximately half that of untreated tumor (7 of 18 [39%] versus 5 of 6 [83%]), suggesting that treatment-affected populations of tumor cells arise from this approach. However, one clear limitation of our study is the significant portion of patient tumors that were not able to undergo xenotransplantation because of indiscernible appearance or inadequate size. While such limitations are largely technical and unavoidable, they may nonetheless have introduced selection bias into our study and subsequent analyses.
Interestingly, despite a difference in initial tumor formation, we found no significant difference in the time required to form tumors between treated and treatment naive patient tumors. One obvious explanation for this is the presence of host factors that consistently promote or limit tumor initiation regardless of tumor capabilities. Another explanation may be inefficacious neoadjuvant therapy in tumors that engrafted versus efficacious neoadjuvant therapy in tumors that failed to engraft. Further work is required to distinguish among these possibilities and to fully determine factors that affect tumor initiation in immunodeficient mice.
Most important for this study, the establishment of direct xenograft tumors from treated patient tumors provides an opportunity to evaluate clinically relevant, in vivo products of neoadjuvant treatment in pancreatic cancer. Therapy resistance is a hallmark of pancreatic cancer, and, to our knowledge, our study represents the only expression comparison of treatment naive and treated pancreatic tumors to date. Global comparison of gene expression profiles identified 2 striking differences in treated tumors relative to untreated tumors: a marked increase in IGBFP3 and concomitant decrease in TGFβ-R2 gene and protein expression. TGF-beta signaling has long been recognized as important in the development and progression of pancreatic cancer as well as its downstream effector, Smad4.22–25 Although TGFβ-R2 is inactivated in only a subset of patients with pancreatic cancer, epigenetic changes in TGF-beta signaling or alterations in downstream mediators such as Smad4 (Smad4 is inactivated in ~55% of patient with PDAC) likely promote PDAC progression.23,25 For example, conditional inactivation of TGFβ-R2 in the setting of mutated K-Ras has been shown to act synergistically toward the rapid development of pancreatic cancer in mouse models.26 Likewise, neoadjuvant treatment may attenuate TGFβ-R2 expression and/or alter the tumor/stroma microenvironment and confer survival privileges to select tumor cells with decreased TGFβ-R2 expression. As IGFBP3 has been shown to induce apoptosis through TGF-β1, impaired TGF-beta signaling secondary to TGFβ-R2 inactivation may inhibit apoptosis and potentially increase the expression of IGFBP3 as observed in our expression profiles.27 Other roles of IGFBP3 in resistance are also likely given its role in the induction of apoptosis through IGF-dependent and IGF-independent mechanisms. Overall, TGFβ-R2 and IGFBP3 may be important mediators of therapy resistance with the potential selection of such resistant phenotypes occurring after exposure to modern therapies. Further study is required to identify their potential role(s) in therapy resistance toward therapeutic ends.
Acknowledgments
Supported by the Various Donor Fund for Pancreatic Cancer Research, The Viragh Family Foundation (JBF, JA), Gillson Longenbaugh Foundation (GEG), NIH 5P20 CA101936 (JLA, DM, JBF, GEG), NIH T32 09599 (MK), and IRB No. LAB07-0854.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Berlin JD, Catalano P, Thomas JP, Kugler JW, Haller DG, Benson AB., III Phase III study of gemcitabine in combination with fluorouracil versus gemcitabine alone in patients with advanced pancreatic carcinoma: Eastern Cooperative Oncology Group Trial E2297. J Clin Oncol. 2002;20:3270–5. doi: 10.1200/JCO.2002.11.149. [DOI] [PubMed] [Google Scholar]
- 3.Heinemann V, Quietzsch D, Gieseler F, Gonnermann M, Schönekäs H, Rost A, et al. Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J Clin Oncol. 2006;24:3946–52. doi: 10.1200/JCO.2005.05.1490. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann R, Bodoky G, Ruhstaller T, Glimelius B, Bajetta E, Schüller J, et al. Gemcitabine plus capecitabine compared with gemcitabine alone in advanced pancreatic cancer: a randomized, multicenter, phase III trial of the Swiss Group for Clinical Cancer Research and the Central European Cooperative Oncology Group. J Clin Oncol. 2007;25:2212–7. doi: 10.1200/JCO.2006.09.0886. [DOI] [PubMed] [Google Scholar]
- 5.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–6. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 6.Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14:3629–37. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 7.Pan X, Arumugam T, Yamamoto T, Levin PA, Ramachandran V, Ji B, et al. Nuclear factor-kappaB p65/relA silencing induces apoptosis and increases gemcitabine effectiveness in a subset of pancreatic cancer cells. Clin Cancer Res. 2008;14:8143–51. doi: 10.1158/1078-0432.CCR-08-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanbolooki S, Nawrocki ST, Arumugam T, Andtbacka R, Pino MS, Kurzrock R, et al. Nuclear factor-kappaB maintains TRAIL resistance in human pancreatic cancer cells. Mol Cancer Ther. 2006;5:2251–60. doi: 10.1158/1535-7163.MCT-06-0075. [DOI] [PubMed] [Google Scholar]
- 9.Nawrocki ST, Carew JS, Pino MS, Highshaw RA, Dunner K, Jr, Huang P, et al. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res. 2005;65:11658–66. doi: 10.1158/0008-5472.CAN-05-2370. [DOI] [PubMed] [Google Scholar]
- 10.Duxbury MS, Ito H, Benoit E, Waseem T, Ashley SW, Whang EE. RNA interference demonstrates a novel role for integrin-linked kinase as a determinant of pancreatic adenocarcinoma cell gemcitabine chemoresistance. Clin Cancer Res. 2005;11:3433–8. doi: 10.1158/1078-0432.CCR-04-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Inhibition of SRC tyrosine kinase impairs inherent and acquired gemcitabine resistance in human pancreatic adenocarcinoma cells. Clin Cancer Res. 2004;10:2307–18. doi: 10.1158/1078-0432.ccr-1183-3. [DOI] [PubMed] [Google Scholar]
- 12.Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc Natl Acad Sci USA. 1992;89:5645–9. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4:1670–80. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–61. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 15.Katz MH, Varadhachary GR, Fleming JB, Wolff RA, Lee JE, Pisters PW, et al. Serum CA 19-9 as a marker of resectability and survival in patients with potentially resectable pancreatic cancer treated with neoadjuvant chemoradiation. Ann Surg Oncol. 17:1794–801. doi: 10.1245/s10434-010-0943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans DB, Varadhachary GR, Crane CH, Sun CC, Lee JE, Pisters PW, et al. Preoperative gemcitabine-based chemoradiation for patients with resectable adenocarcinoma of the pancreatic head. J Clin Oncol. 2008;26:3496–502. doi: 10.1200/JCO.2007.15.8634. [DOI] [PubMed] [Google Scholar]
- 17.Varadhachary GR, Wolff RA, Crane CH, Sun CC, Lee JE, Pisters PW, et al. Preoperative gemcitabine and cisplatin followed by gemcitabine-based chemoradiation for resectable adenocarcinoma of the pancreatic head. J Clin Oncol. 2008;26:3487–95. doi: 10.1200/JCO.2007.15.8642. [DOI] [PubMed] [Google Scholar]
- 18.Hwang RF, Wang H, Lara A, Gomez H, Chang T, Sieffert N, et al. Development of an integrated biospecimen bank and multidisciplinary clinical database for pancreatic cancer. Ann Surg Oncol. 2008;15:1356–66. doi: 10.1245/s10434-008-9833-1. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–55. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 21.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–32. [PubMed] [Google Scholar]
- 23.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–4. [PubMed] [Google Scholar]
- 24.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–3. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 25.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–60. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajah R, Valentinis B, Cohen P. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272:12181–8. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]