SUMMARY
We report the case of a patient from Brazil with a bloodstream infection caused by a strain of methicillin-resistant Staphylococcus aureus (MRSA) that was susceptible to vancomycin (designated BR-VSSA) but that acquired the vanA gene cluster during antibiotic therapy and became resistant to vancomycin (designated BR-VRSA). Both strains belong to the sequence type (ST) 8 community-associated genetic lineage that carries the staphylococcal chromosomal cassette mec (SCCmec) type IVa and the S. aureus protein A gene (spa) type t292 and are phylogenetically related to MRSA lineage USA300. A conjugative plasmid of 55,706 bp (pBRZ01) carrying the vanA cluster was identified and readily transferred to other staphylococci. The pBRZ01 plasmid harbors DNA sequences that are typical of the plasmid-associated replication genes rep24 or rep21 described in community-associated MRSA strains from Australia (pWBG745). The presence and dissemination of community-associated MRSA containing vanA could become a serious public health concern.
Since the description in 2002 of the vana gene cluster in mrsa as the mechanism of high-level vancomycin resistance, 13 isolates have been reported in the United States,1,2 with others reported in India3 and Iran.4 Enterococcus faecalis and E. faecium have been implicated as the donors of the vancomycin resistance genes.5,6 The VRSA isolates were recovered from patients with soft-tissue or skin infections, and multilocus sequence typing has shown that 12 U.S. VRSA isolates belong to clonal complex 5 (including ST5, ST85, ST231, and ST371)1 and that the 13th isolate belongs to clonal complex 30. Clonal complex 5 is the most widely disseminated hospital-associated MRSA clonal complex in the United States, with patterns designated as USA100 and USA800 on pulsed-field gel electrophoresis (PFGE).1
Community-associated MRSA emerged during the 1990s and rapidly disseminated across the United States, most often causing severe skin and soft-tissue infections but on occasion causing life-threatening infections such as necrotizing pneumonia.7 Early variants of this community-associated MRSA were initially traced to PFGE pattern USA400, but these variants were rapidly replaced by isolates within the genetic lineage designated USA300-ST8 (clonal complex 8). A variant of USA300-ST8 emerged in South America in 2005 (designated USA300 Latin American Variant [USA300-LV])8 and has spread rapidly, replacing the previously predominant clonal complex 5 hospital-associated clone (known as the Chilean–Cordobes clone).9,10 Here we report on the acquisition of the vanA gene cluster by an invasive bloodstream isolate of community-associated MRSA lineage in a patient at a Brazilian hospital.
CASE REPORT
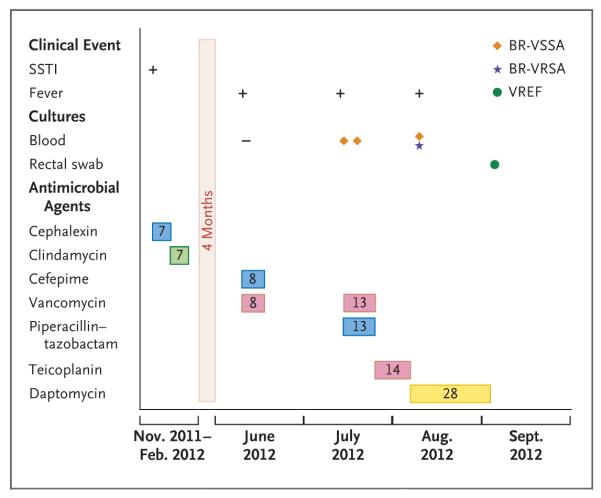
The patient was a 35-year-old man from São Paulo with mycosis fungoides, cocaine addiction, and diabetes mellitus. He was initially admitted to a psychiatric hospital in November 2011 for depression and suicidal ideation. Leg cellulitis developed and was treated with cephalexin and topical gentamicin. The patient was discharged with a prescription for clindamycin but was readmitted in June 2012 because of worsening psychiatric symptoms and recurrent infections of the skin and soft tissues. Vancomycin and cefepime were administered for 8 days; blood cultures obtained during this period were sterile. The patient remained in the hospital to receive chemotherapy with gemcitabine, vinorelbine, and dexamethasone for mycosis fungoides (skin cancer). In July 2012, while the patient remained in the hospital, fever recurred, and treatment with vancomycin and piperacillin–tazobactam was started. On July 16 and 24, blood cultures yielded MRSA isolates that were susceptible to vancomycin, linezolid, and clindamycin (see Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Vancomycin was continued for 13 days (highest vancomycin trough level, 8.7 μg per milliliter), followed by treatment with teicoplanin for an additional 14 days.
A transthoracic echocardiogram showed no evidence of infective endocarditis. On August 15, the day after teicoplanin was discontinued, fever recurred. Blood cultures were positive for two MRSA isolates, one of which was resistant to both vancomycin (minimal inhibitory concentration, >32 μg per milliliter) and teicoplanin (minimal inhibitory concentration, 32 μg per milliliter); the isolate was also resistant to erythromycin, clindamycin, ciprofloxacin, gentamicin, and trimethoprim–sulfamethoxazole (Table S1 in the Supplementary Appendix).
Treatment with daptomycin was started, and the patient was placed in a single room, with a recommendation of contact isolation. A sample from a rectal swab obtained 2 weeks after the initial positive blood culture indicated colonization with vancomycin-resistant E. faecalis (VREF). The fever diminished, and daptomycin was continued for an additional 4 weeks. However, while the patient was receiving daptomycin, oral mucosal lesions developed and his respiratory status and fever worsened. Blood cultures yielded Stenotrophomonas maltophilia; treatment with levofloxacin was started and intravenous catheters were removed. Respiratory secretions yielded extended-spectrum, beta-lactamase–producing Klebsiella pneumoniae and carbapenem-resistant Acinetobacter baumannii. Polymixin B sulfate was added to the treatment regimen. Multisystem organ failure ensued, and blood cultures grew Candida albicans. Because of the patient’s worsening status, care was withdrawn in November 2012. The patient died while receiving meropenem, linezolid, polymyxin B sulfate, and amphotericin B. (A summary of the patient’s course is presented in Fig. 1.)
Figure 1. The Patient’s Clinical Course before and after Isolation of the Vancomycin-Resistant Staphylococcus aureus.
The antibiotics used are depicted in colored rectangles, with beta-lactams in blue (cephalexin, cefepime, and piperacillin–tazobactam), clindamycin in green, glycopeptides in pink (vancomycin and teicoplanin), and daptomycin in yellow; the number in each rectangle corresponds to the number of days of treatment with the antibiotic. The drugs are shown in the order in which they were added to therapy. Dosages were as follows: cephalexin, 500 mg every 6 hours, given orally; clindamycin, 450 mg every 8 hours, given orally; cefepime, 1 g every 8 hours, given intravenously; piperacillin–tazobactam, 3.375 g every 6 hours, given intravenously; vancomycin, 1 g every 12 hours, given intravenously; teicoplanin, 400 mg daily, given intravenously; and daptomycin, 6 mg per kilogram of total body weight daily, given intravenously. The final days of hospitalization are not included. SSTI denotes skin and soft-tissue infection, BR-VSSA vancomycin-susceptible S. aureus, BR-VRSA vancomycin-resistant S. aureus, and VREF vancomycin-resistant Enterococcus faecalis.
METHODS
BACTERIAL IDENTIFICATION, SUSCEPTIBILITY TESTING, AND MOLECULAR TYPING
S. aureus and VREF isolates recovered from the patient’s blood samples and rectal swabs, respectively, were identified by means of a polymerase-chain-reaction (PCR) assay11,12 and 16S ribosomal RNA sequencing.13 Susceptibility was determined with the use of agar dilution and broth microdilution.14 To assess the genetic background of the isolates, PFGE was performed on two S. aureus isolates recovered from the patient’s blood (BR-VSSA and BR-VRSA) (Fig. 1); on representative MRSA isolates circulating in South American hospitals, including the Latin American variant (USA300-LV) and Chilean–Cordobes clones; and on reference strains from the following pandemic clones: USA300 (NRS482),9 the Brazilian clone (F338), the Iberian clone (NRS385-USA500), the Pediatric clone (NRS387-USA800), and the New York–Japan clone (NRS382-USA100). Typing of BR-VRSA was performed with the use of multilocus sequence typing, SCCmec, and spa. PCR assays were used to detect the genes encoding Panton–Valentine leukocidin (PVL) and the arginine catabolic mobile element, which are often found in MRSA USA300.9
PLASMID CHARACTERIZATION AND BACTERIAL MATINGS
We assessed the transferability of resistance to vancomycin by performing filter matings,15 using BR-VRSA and the VREF isolate from the patient as donors and BR-VSSA, S. aureus RN4220, S. aureus COL, and E. faecalis OG1RF as recipients (Table S2 in the Supplementary Appendix). A donor-to-recipient ratio of 1:10 was used for mating experiments, as described previously.15 Selection was performed on brain–heart infusion (BHI) agar supplemented with vancomycin and fusidic acid at concentrations of 32 μg per milliliter and 25 μg per milliliter, respectively. To determine whether vanA was present on a plasmid, S1 nuclease digestion of total DNA, coupled with PFGE and hybridization with a vanA probe, was performed.16,17 PCR assays targeting pSK41-like6 and Inc18-like plasmids6 and the rep plasmid families18 were performed to classify plasmid replicons. The Biosafety Committee at the University of Texas–Houston approved these experiments.
WHOLE-GENOME SEQUENCING AND PHYLOGENETIC ANALYSIS
BR-VSSA, BR-VRSA, and VREF isolates were evaluated by means of whole-genome sequencing with the use of the Illumina platform. Annotation and assembly were performed as previously described19; details are available at the National Center for Bio technology Information (GenBank Bioproject numbers 205852 [for BR-VSSA], PRJNA183704 [for BR-VRSA], and 205838 [for VREF]). Details of genomic comparisons (including the identification of single-nucleotide polymorphisms [SNPs]) and phylogenetic analysis are described in the Supplementary Appendix.
RESULTS
A NOVEL TRANSFERABLE VAN A PLASMID IN BR-VRSA
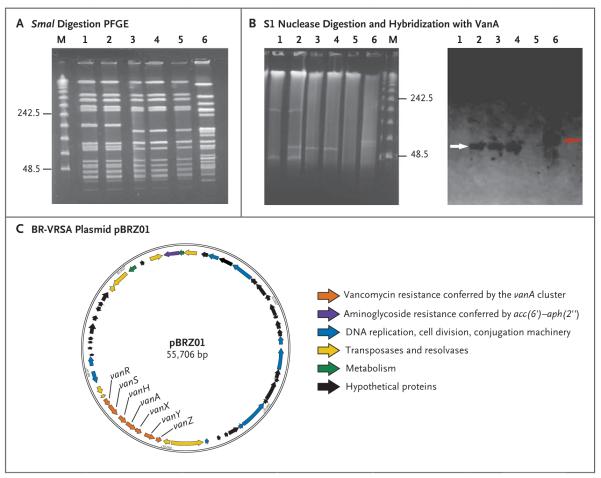
Initial molecular characterization showed that BR-VRSA displayed a PFGE pattern that was indistinguishable from that of BR-VSSA (Fig. 2A), suggesting that BR-VRSA was selected in vivo during the administration of glycopeptides and other antimicrobial drugs. Moreover, S1 nuclease digestion and hybridization showed that BR-VRSA contained a plasmid of approximately 55 kb (not present in BR-VSSA) that carried vanA and aac(6′)–aph(2″), which encode enzymes that confer resistance to vancomycin and gentamicin, respectively (Fig. 2B). Growth rates were similar for both organisms (Fig. S1 in the Supplementary Appendix), suggesting that acquisition of the vanA plasmid did not affect the in vitro fitness of the strain.
Figure 2. Results of Pulsed-Field Gel Electrophoresis (PFGE), S1 Nuclease Digestion of Total DNA, Hybridization, and Plasmid Annotation.
Panel A shows the results of SmaI digestion of total DNA followed by PFGE. Lane M shows the lambda ladder (with the molecular sizes in kilobases shown at left); lane 1, BR-VSSA; lane 2, BR-VRSA (donor); lane 3, transconjugant number 1 Staphylococcus aureus RN4220-RF; lane 4, transconjugant number 2 S. aureus RN4220-RF; lane 5, S. aureus RN4220-RF (recipient); and lane 6, vancomycin-resistant Enterococcus faecalis (vanA). Panel B shows the results of S1 digestion of the total DNA of staphylococcal strains and vancomycin-resistant E. faecalis followed by PFGE (at left) and hybridization with a VanA probe (at right). In the results at left, lane 1 shows BR-VSSA; lane 2, BR-VRSA (donor); lane 3, transconjugant number 1 S. aureus RN4220-RF; lane 4, transconjugant number 2 S. aureus RN4220-RF; lane 5, S. aureus RN4220RF (recipient); lane 6, vancomycin-resistant E. faecalis; and lane M the lambda ladder. In the results at right, the white and red arrows highlight the positive signal for vanA hybridization in staphylococcal and enterococcal strains, respectively. Panel C shows a schematic representation of the annotation of the BR-VRSA plasmid pBRZ01.
Filter-mating assays showed that the rate of transfer of vancomycin resistance from BR-VRSA (donor) to BR-VSSA (recipient) was 2.6×10−4 transconjugants per donor (Table S2 in the Supplementary Appendix). Rates of transfer to other staphylococci (S. aureus RN4220-RF and COL) were 1.65×10−4 and 6×10−5, respectively (Table S2 in the Supplementary Appendix). Experiments with S1 nuclease indicated that the plasmid containing vanA (designated pBRZ01; 55,706 bp) was the only large plasmid that was readily transferred to S. aureus during the mating experiment (Fig. 2B). We were unable to transfer vancomycin resistance to a laboratory strain of E. faecalis (strain OG1RF), which suggests that the plasmid was not able to replicate in enterococci. The transfer of vancomycin resistance from VREF recovered from the patient’s rectal swab was also unsuccessful.
Analysis of the sequence of pBRZ01 (Fig. 2C) indicated that a Tn1546-like element underwent important DNA rearrangements (Fig. S2 in the Supplementary Appendix). An insertion sequence (IS1216) was found at the 5′ end of the Tn1546 variant, with a deletion of 3397 bp eliminating the left inverted repeat, the gene encoding integrase (open reading frame 1), and part of the gene encoding resolvase (open reading frame 2) (Fig. S2 in the Supplementary Appendix). Downstream of vanZ, a deletion of 96 bp, including the right inverted repeat (Fig. S2 in the Supplementary Appendix), was identified, with the insertion of two open reading frames encoding an enterococcal resolvase and transposase (from the Tn3 family), respectively (Fig. S2 in the Supplementary Appendix). The sequence of the vanA cluster (including 614 bp upstream and 222 bp downstream) harbored by the VREF infecting the patient was identical to the sequence in BR-VRSA and was also identical to the sequences from plasmids pWZ7140 and pWZ909, previously identified in E. faecalis.20 These findings support the view that the vanA cluster has an enterococcal origin. Genomic analysis (confirmed by means of PCR assay) of VREF indicated that the repR gene (typical of Inc18 type plasmids previously associated with transfer of the vanA cluster into clonal complex 5 S. aureus) and the traA gene (required for plasmid transfer)6 were absent, which supports the absence of transferability of the enterococcal plasmid and further indicating that VREF was not the direct donor of vanA genes to BR-VRSA.
Comparison of pBRZ01 with pWBG745 (38,204 bp), a plasmid identified in community-associated MRSA isolates from remote areas of Australia (clonal complex 5), showed 99% identity across 21,164 nucleotides.21 In addition, a sequence analysis indicated that the plasmid containing vanA is not related to the pSK41-like staphylococcal or Inc18-like enterococcal plasmids previously associated with vanA-carrying plasmids from VRSA isolates identified in the United States and belonging to clonal complex 5.1,6 Instead, the vanA plasmid harbors sequences that are typical of the recently designated rep24 family18 or rep21 family22 found in pWBG745. (Other genetic determinants conferring antibiotic resistance that are present in the BR-VRSA genome are shown in Table S3 in the Supplementary Appendix.)
ST8 COMMUNITY-ASSOCIATED GENETIC BACKGROUND IN BR-VRSA
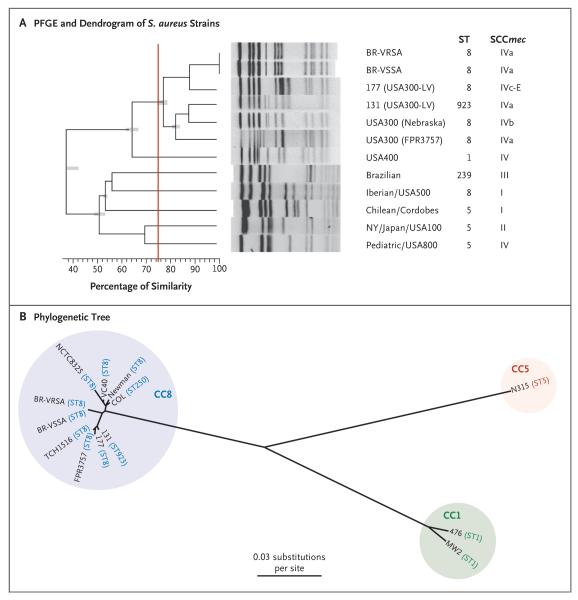
PFGE indicated that BR-VRSA is genetically related to MRSA USA300 (Fig. 3A), and multilocus sequence typing showed that BR-VRSA belongs to ST8 (as do USA300 and USA300-LV), harbors SCCmec type IVa, and is spa type t292. Unlike USA300 and USA300-LV, BR-VRSA lacked genes encoding PVL. Neither the arcA gene nor the entire arginine catabolic mobile element was present in BR-VRSA, as has also been noted for MRSA strains belonging to the USA300-LV lineage. BR-VRSA and BR-VSSA harbor an intact bsa operon (for bacteriocin production) that is characteristic of community-associated MRSA; VRSA strains from clonal complex 5 lack this operon.1 Whole-genome, SNP-based phylogenetic analysis (performed with either the whole genome [Fig. 3B] or the core genome [Fig. S3 in the Supplementary Appendix]) showed that there is a close relationship between BR-VSSA and BR-VRSA and between each of these variants and other ST8 genomes, including USA300 and USA300-LV. Genomic comparisons of BR-VSSA and BR-VRSA revealed only 288 SNPs (134 in the core genome) with a score indicating high quality (defined as a score of q>20) differentiating the two strains, whereas a comparison of each of these strains with the USA300 strain TCH1516 showed a difference of 1637 SNPs (965 core SNPs) for BR-VSSA and 1757 SNPs (1005 core SNPs) for BR-VRSA, suggesting that BR-VSSA is more similar to the progenitor of BR-VRSA. (The SNP score is a measure of the number of reads from a sequencing that agree with regard to a particular SNP.)
Figure 3. Phylogenetic Comparisons of BR-VRSA and BR-VSSA with Representatives of Other Methicillin-Resistant Staphylococcus aureus (MRSA) Clones.
Panel A shows a dendrogram of the pandemic MRSA clones, BR-VRSA and BR-VSSA, generated with PFGE and the use of GelCompar II software, version 6.5 (Applied Maths). Patterns were clustered by means of UPGMA (unweighted pair group method with arithmetic mean), with the use of the Dice similarity coefficient and an optimization of 0.50% and a tolerance of 1.0%. PFGE types, or clusters, were identified on the basis of a similarity of 75% or higher (indicated by the vertical red line). ST denotes sequence type. The phylogenetic tree shown in Panel B is based on whole-genome single-nucleotide polymorphisms (SNPs) and was generated with the use of the maximum-likelihood optimality criterion. Branch lengths are proportional to the number of evolutionary changes (substitutions per site). All nodes have 100% bootstrap support. Sequence types (STs) and clonal complexes (CCs) of the S. aureus strains are indicated.
DISCUSSION
The acquisition of high-level vancomycin resistance by S. aureus has been deemed a major clinical and epidemiologic threat. However, since the first characterization of a strain of VRSA in the United States,5,23 only a few other isolates have been reported. All the VRSA isolates from the United States described to date have been isolated from infections of the skin and soft-tissue or from colonizers of the skin. This case documents a bloodstream infection caused by a VRSA strain in a patient with a serious skin condition that probably predisposed the patient to bacterial colonization. The patient was treated repeatedly with beta-lactams and glycopeptides, and our analyses suggest that the MRSA isolate had also acquired the vanA plasmid during therapy. The genetic characterization and genomic analyses also suggest that the vanA gene cluster present in BR-VRSA may have originated from an enterococcal donor. The DNA sequence of the vanA gene cluster (and partial flanking sequences) of BR-VRSA is identical to the sequence present in the genome of VREF that was recovered from a swab of the patient’s rectum, suggesting that VREF may have been the donor. However, the presence of a different plasmid in this isolate and the absence of transferability of the vanA cluster from VREF to staphylococci or enterococci suggest that acquisition occurred through different intermediaries. Indeed, the Tn1546-like element in BR-VRSA was altered, indicating that potentially important genetic rearrangements of Tn154624 had occurred. The presence of the insertion sequences flanking the vanA gene cluster (Fig. S2 in the Supplementary Appendix) may also provide the truncated Tn1546-derived element with mobility.
The results of our genomic and phylogenetic analyses (with more SNP changes detected on the branches leading to BR-VRSA than on those leading to BR-VSSA) suggest that BR-VRSA is derived from BR-VSSA or a strain closely resembling BR-VSSA. However, since BR-VSSA did not contain pBRZ01, the origin of this plasmid is unknown. The two MRSA isolates recovered (but then discarded) from the patient’s blood before the isolation of BR-VSSA and BR-VRSA (Fig. 1, and Table S1 in the Supplementary Appendix) were gentamicin-resistant and vancomycin-susceptible, and it is tempting to speculate that they may have been carriers of a version of pBRZ01 that did not contain the vanA gene cluster.
Apart from the type of infection, several factors that may have important public health implications distinguish this case from previously reported cases of VRSA infection. First, this case is an example of the acquisition of a vanA gene cluster that occurred independently of the hospital-associated MRSA clonal complex 5 lineage. The molecular and genomic data indicate that the genetic background of the VRSA strain is closely related to community-associated MRSA strains disseminated in several parts of the world (USA300 ST8 harboring SCCmec type IVa). An ST8 community-associated variant of the MRSA USA300 clone (USA300-LV), which has been reported in the northern part of South America,8,9,25 has almost replaced the common hospital-associated clone (ST5 Chilean–Cordobes clone).9,10,25 However, the genetic background of BR-VRSA differs from that of USA300-LV and from that of the prototypical USA300 in that it lacks the genes encoding PVL, a genetic marker of community-associated MRSA strains. This difference suggests that BR-VRSA is a novel variant of ST8 community-associated MRSA, closely related to the USA300 genetic lineage and capable of producing severe, invasive bloodstream infections. This observation is supported by our finding that BR-VRSA, unlike previously reported VRSA strains, harbors an intact bsa operon,1 a genetic marker of community-associated MRSA strains.26 In addition, characterization and sequencing of pBRZ01 indicate that rep24 or rep21 family plasmids recently described in community-associated MRSA (belonging to clonal complex 5)21 in Australia have been acquired by the MRSA ST8 genetic lineage and are capable of capturing the vanA cluster on a transferable plasmid. Finally, because USA300-like strains spread rapidly and replace other MRSA lineages with such efficiency, our findings raise the possibility that vancomycin resistance may be disseminated to MRSA clonal complex 8 isolates in both the hospital and the community.
Supplementary Material
Acknowledgments
Supported by grants from the National Institute of Allergy and Infectious Diseases (R01 AI093749, to Dr. Arias) and the National Institutes of Health (1U54 HG004968, to Dr. Weinstock).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Kos VN, Desjardins CA, Griggs A, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most com monly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;22:3(3):e00112–12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limbago BM, Kallen AJ, Zhu W, et al. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J Clin Microbiol. 2014;52:998–1002. doi: 10.1128/JCM.02187-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia) J Med Microbiol. 2008;57:72–9. doi: 10.1099/jmm.0.47144-0. [DOI] [PubMed] [Google Scholar]
- 4.Azimian A, Havaei SA, Fazeli H, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50:3581–5. doi: 10.1128/JCM.01727-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigel LM, Clewell DB, Gill SR, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–71. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 6.Zhu W, Clark N, Patel JB. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother. 2013;57:212–9. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias CA, Rincon S, Chowdhury S, et al. MRSA USA300 clone and VREF — a U.S.–Colombian connection? N Engl J Med. 2008;359:2177–9. doi: 10.1056/NEJMc0804021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes J, Rincón S, Díaz L, et al. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis. 2009;49:1861–7. doi: 10.1086/648426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Noriega E, Seas C, Guzmán-Blanco M, et al. Evolution of methicillin-resistant Staphylococcus aureus clones in Latin America. Int J Infect Dis. 2010;14:e560–6. doi: 10.1016/j.ijid.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–7. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; J Clin Microbiol. 1995;33:1434. doi: 10.1128/jcm.33.5.1434-1434.1995. Erratum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo PC, Leung AS, Leung KW, Yuen KY. Identification of slide coagulase positive, tube coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol Pathol. 2001;54:244–7. doi: 10.1136/mp.54.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement (CLSI document no. M100-S21) Clinical and Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 15.Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J Clin Microbiol. 2002;40:3326–33. doi: 10.1128/JCM.40.9.3326-3333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;53:4240–6. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226:235–40. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 18.Lozano C, García-Migura L, Aspiroz C, Zarazaga M, Torres C, Aarestrup FM. Expansion of a plasmid classification system for Gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl Environ Microbiol. 2012;78:5948–55. doi: 10.1128/AEM.00870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arias CA, Panesso D, McGrath DM, et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365:892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, Murray PR, Huskins WC, et al. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:4314–20. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien FG, Coombs GW, Pearman JW, et al. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Anti microb Chemother. 2009;64:684–93. doi: 10.1093/jac/dkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy AJ, Lindsay JA. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012;12:104. doi: 10.1186/1471-2180-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang S, Sievert DM, Hageman JC, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–7. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 24.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–27. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiménez JN, Ocampo AM, Vanegas JM, et al. CC8 MRSA strains harboring SCCmec type IVc are predominant in Colombian hospitals. PLoS One. 2012;7(6):e38576. doi: 10.1371/journal.pone.0038576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.