Abstract
Improving small interfering RNA (siRNA) efficacy in target cell populations remains a challenge to its clinical implementation. Here, we report a chemical modification, consisting of phosphorodithioate (PS2) and 2’-O-Methyl (2’-OMe) MePS2 on one nucleotide that significantly enhances potency and resistance to degradation for various siRNAs. We find enhanced potency stems from an unforeseen increase in siRNA loading to the RNA-induced silencing complex, likely due to the unique interaction mediated by 2’-OMe and PS2. We demonstrate the therapeutic utility of MePS2 siRNAs in chemoresistant ovarian cancer mouse models via targeting GRAM Domain Containing 1B (GRAMD1B), a protein involved in chemoresistance. GRAMD1B silencing is achieved in tumors following MePS2-modified siRNA treatment, leading to a synergistic anti-tumor effect in combination with paclitaxel. Given the previously limited success in enhancing siRNA potency with chemically modified siRNAs, our findings represent an important advance in siRNA design with the potential for application in numerous cancer types.
Introduction
SiRNA-based therapies have shown promise in recent clinical trials as anti-cancer agents1,2. This technology offers tremendous potential in silencing “non-druggable” targets that are not amenable to conventional therapeutics such as small molecules or monoclonal antibodies3,4. However, the lack of stability and low potency at target sites following systemic administration remain the major obstacles to clinical translation of this therapy5,6.
To overcome these problems, efforts have been made to chemically modify the 2’-position of the ribose moiety (e.g. 2’-OMe or 2’-F) or phosphate backbone of the siRNA molecule (e.g. phosphoromonothioate (PS) modification) such that enhanced serum stability7, bioavailability8, and prolonged duration of action9 could be achieved (Reviewed in 10). Importantly, combining PS and 2’-modification impart superior stability over single modifications11,12, leading to the approval of Kynamro, an antisense therapeutic, by the Food and Drug Administration13.
Despite the advance, combination of PS and 2’-modification shows limited enhancement of siRNA potency14. Moreover, synthesis of PS or MePS by standard phosphoramidite methodology also generates a mixture of unresolvable diastereomeric oligomers, which could potentially result in variable biochemical, biophysical, and biological properties. We have recently reported that PS2 modification, which substitutes sulfur atoms for both of the non-bridging phosphate and oxygen atoms, significantly improves serum stability and gene silencing over PS-modified siRNAs15. Its achiral nature at the phosphorous center eliminates the generation of isomeric by-products, thereby permitting batch to batch quality control16,17, a feature that cannot be achieved with the MePS platform. As such, PS2-modified siRNA is a prime candidate for introducing critical new features to further improve siRNA efficacy both in vitro and in vivo.
We hypothesized that the incorporation of two most widely used sugar modifications, 2’-OMe or 2’-F, into a PS2 modified construct could lead to further enhancement of the PS2 siRNA stability and increase their clinical potential. In this study, we show that the novel MePS2 design significantly enhances serum stability and silencing activity of siRNAs. This enhanced potency stems from an unforeseen increase in the loading of siRNAs to the RNA-induced silencing complex (RISC). We further demonstrate the therapeutic utility of MePS2-modified siRNAs by targeting GRAMD1B, a novel gene identified through a whole genome, taxane sensitivity screen. Given that limited success has been achieved thus far toward broadly enhancing siRNA potency with chemically modified siRNAs18-20, MePS2 modification represents a major new direction for enhancing the clinical application of existing RNA interference (RNAi)-based therapies.
Results
Synthesis and activity of chemically modified siRNAs
To investigate the gene silencing activity of MePS2- or FPS2-siRNAs and to gain insight into their regulation of the RNAi pathway, we first synthesized these novel siRNAs via the solid-phase synthesis technique. Given our extensive experience in using unmodified siRNAs to target EPH receptor A2 (EphA2), a tyrosine kinase receptor that promotes tumor progression in cancer21, we chose to initially evaluate our novel siRNAs using EphA2 as a model system. We have previously identified favorable positions of PS2 modification using a siRNA that targets Green Fluorescence Protein (GFP; Supplementary Fig. 1; Supplementary Table 1). The silencing efficacy of selected PS2-modified sequences was further validated using another siRNA sequence15. Informed by these prior observations, we designed MePS2/FPS2-1 through -3 constructs for siEphA2 (Fig. 1, Table 1). We also created MePS2/FPS2-4 sequences, where there are two MePS2/FPS2 groups present at the seed region of the antisense strand. Lastly, counterpart sequences were synthesized with either 2’-OMe/2’-F alone, PS2 alone, or MePS/FPS placed at the same locations.
Figure 1. Chemical structures of modified siRNAs.
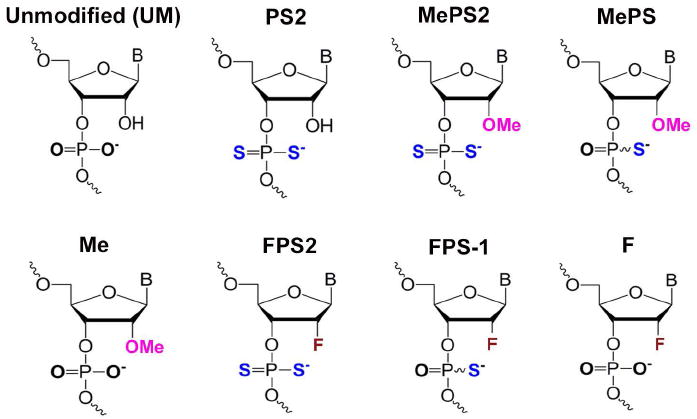
Chemical structures of modified siRNAs used for the initial screen.
Table 1. Chemical structures of modified siRNAs.
Chemical structures of modified siRNAs used for the initial screen. Locations of modification are indicated for each sequence.
| Sense (5’f to 3’f) | Antisense (3’f to 5’f) | |
|---|---|---|
| UM | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| PS2-1 | UGACAUGCCGAUCUACAUS2GS2TT | TTACUGUACGGCUAGAUGUAC |
| PS2-2 | UGAS2CAUGCCGAUS2CUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| PS2-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGS2GS2CUAGAUGUAC |
| PS2-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUS2AGAUS2GUAC |
| MePS2-1 | UGACAUGCCGAUCUACAUMeS2GMeS2TT | TTACUGUACGGCUAGAUGUAC |
| MePS2-2 | UGAMeS2CAUGCCGAUMeS2CUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| MePS2-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGMeS2GMeS2CUAGAUGUAC |
| MePS2-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUMeS2AGAUMeS2GUAC |
| MePS-1 | UGACAUGCCGAUCUACAUMeSGMeSTT | TTACUGUACGGCUAGAUGUAC |
| MePS-2 | UGAMeSCAUGCCGAUMeSCUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| MePS-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGMeSGMeSCUAGAUGUAC |
| MePS-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUMeSAGAUMeSGUAC |
| Me-1 | UGACAUGCCGAUCUACAUMeGMeTT | TTACUGUACGGCUAGAUGUAC |
| Me-2 | UGAMeCAUGCCGAUMeCUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| Me-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGMeGMeCUAGAUGUAC |
| Me-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUMeAGAUMeGUAC |
| FPS2-1 | UGACAUGCCGAUCUACAUFS2GFS2TT | TTACUGUACGGCUAGAUGUAC |
| FPS2-2 | UGAFS2CAUGCCGAUFS2CUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| FPS2-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGFS2GFS2CUAGAUGUAC |
| FPS2-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUFS2AGAUFS2GUAC |
| FPS-1 | UGACAUGCCGAUCUACAUFSGFSTT | TTACUGUACGGCUAGAUGUAC |
| FPS-2 | UGAFSCAUGCCGAUFSCUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| FPS-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGFSGFSCUAGAUGUAC |
| FPS-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUFSAGAUFSGUAC |
| F-1 | UGACAUGCCGAUCUACAUFGFTT | TTACUGUACGGCUAGAUGUAC |
| F-2 | UGAFCAUGCCGAUFCUACAUGTT | TTACUGUACGGCUAGAUGUAC |
| F-3 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGFGFCUAGAUGUAC |
| F-4 | UGACAUGCCGAUCUACAUGTT | TTACUGUACGGCUFAGAUFGUAC |
SKOV3ip1 and HeyA8, both epithelial ovarian cancer cell lines that highly express EphA2, were transfected with synthesized siRNAs in serum-free conditions to test silencing efficacy. Two independent experiments were performed and EphA2 protein quantification was done at 48 h post-transfection. MePS2-1, MePS2-2, Me-4, FPS-1, FPS-4, and F-1 significantly decreased EphA2 levels in SKOV3ip1 compared to unmodified (UM) siRNA and outperformed their counterparts (Fig. 2a). In particular, we found that the MePS2-1 modification silenced EphA2 to a much greater degree compared to UM (6-fold enhancement). It also showed 4, 7.5, and 7-fold enhancement in EphA2 silencing compared to PS2-1, MePS-1, and Me-1 modified sequences, respectively. In contrast to MePS2-1 and MePS2-2, FPS2-1 and FPS2-2 modifications did not increase silencing efficiency of the siRNA. Given the significant improvement in gene silencing observed with MePS2-1, we focused our subsequent effort on Me-modified constructs. In HeyA8 cells, MePS2-1 modified siEphA2 also showed superior silencing activity compared to the UM sequence (80% vs. 50% knockdown, respectively, Fig. 2b). Interestingly, both PS2-1 and PS2-2 modified sequences resulted in significant silencing of EphA2 in this cell line.
Figure 2. Stability and gene silencing activity of chemically modified siRNA.
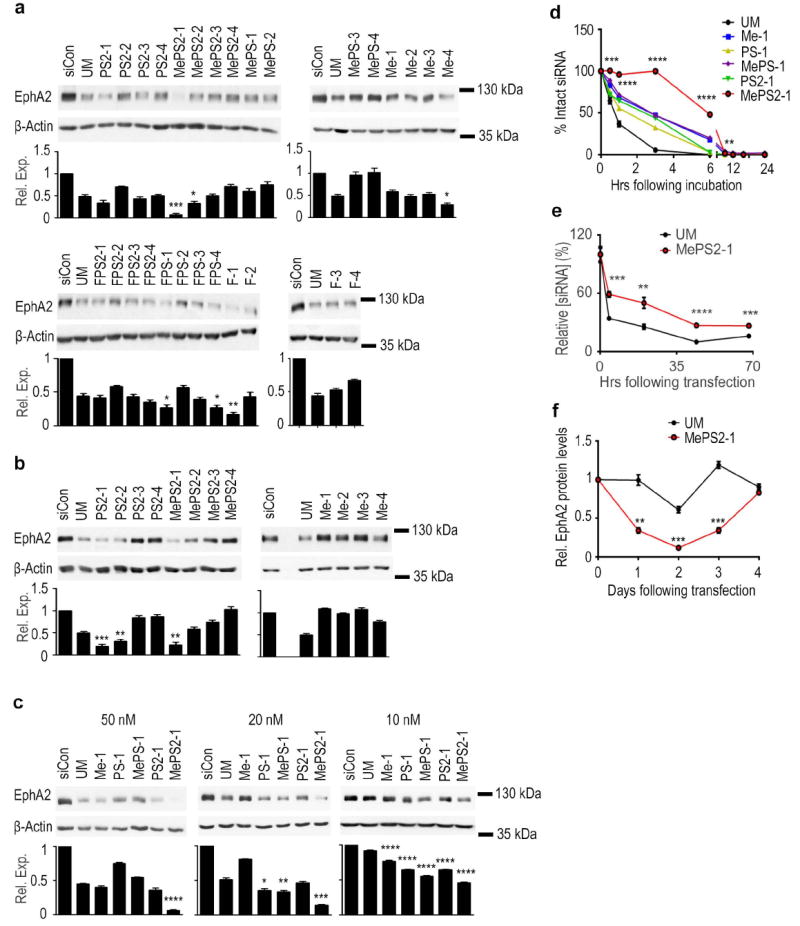
(a) Knockdown of EphA2 in SKOV3ip1 cells using chemically modified siRNAs (100 nM, serum-free condition). (b) Knockdown of EphA2 using selected siRNA sequences in HeyA8 cells (100 nM, serum-free condition). (c) Knockdown of EphA2 using chemically modified siRNAs in SKOV3ip1 cells (10, 20, and 50 nM, 10% FBS-containing media). (a, b, and c) Cells were treated with siRNAs for 4 h and EphA2 protein levels were examined 48 h post-transfection. (d) Stability of chemically modified siRNAs in 10% FBS at 37 °C. (e) Intracellular stability of UM and MePS2-1 modified siEphA2 in SKOV3ip1 cells (50 nM, serum-free condition). (f) Duration of EphA2 knockdown following UM and MePS2-1 modified siEphA2 treatment in SKOV3ip1 cells (50 nM, 10% FBS-containing media). [P-values obtained with Student’s t-test; *, P<0.05; **, P<0.01; ***, P<0.001; or ****, P<0.0001; compared to UM siEphA2 sequence; bars and error bars represent mean values and the corresponding S.E.M.s (n=3)].
Given the promise of the MePS2-1 modification in both of the cell lines examined, we next investigated the impact of different modifications at the same position on gene silencing and serum stability (Supplementary Fig. 2, Fig. 2c,d). To mimic the condition in the pre-clinical or clinical setting, sequences were introduced into SKOV3ip1 cells in media containing 10% Fetal Bovine Serum (FBS). EphA2 protein levels were examined 48 h post-transfection. MePS2-1 displayed a significant improvement over all other sequences tested, including PS2-1 (Fig. 2c, Supplementary Fig. 3). Of note, further modifications at different positions of the MePS2-1 siEphA2 sequence resulted in reduced silencing efficiency (Supplementary Fig. 4). The improved gene-silencing as the result of MePS2-1 modification was also observed when another siRNA sequence was used [parathymosin (PTMS) targeted siRNA, Supplementary Fig. 5a]. SiRNA stability assays show that the majority of UM siRNA was broken down within 20 min, with near-complete degradation at 3 h (Fig. 2d). Overall improvements in serum stability were seen for all modified sequences in comparison to UM, but MePS2-1 displayed the highest sequence stability, with a large portion of the input molecules remaining intact at 3 h. An even greater enhancement in stability was observed with PTMS-targeted siRNA (Supplementary Fig. 5b).
We next wondered if the enhanced stability after modification accounted for these constructs’ increased silencing efficacy. To test this, we employed stem loop polymerase chain reaction (PCR) technique to quantify intact siRNA within the cell following transfection (Fig. 2e, Supplementary Fig. 5c). Increased intracellular siRNA stability via MePS2 modification was confirmed, but not to a degree that would explain its dramatic increase in siRNA potency. Our conclusion was that MePS2-1 showed increased potency independent of improvements in stability. This was consistent with the finding that the duration of silencing was not significantly prolonged for MePS2-1 sequence compared to the UM counterpart (Fig. 2f).
Biophysical properties of MePS2-1 siRNA
We next examined the mechanism by which MePS2-1 sequence enhances the gene silencing effect. First, we examined thermodynamic stability [measured by RNA duplex melting temperature (Tm)] since the destabilization of the 5’-end of duplex by introducing modified units into 3’-end of the sense strand is crucial in creating favorable thermodynamic asymmetry for siRNA activity15,22,23. Measuring the Tm of MePS2-1 and its counterpart sequences showed MePS2-1 to have the largest decrease in Tm (1.24 °C, ΔTm, P<0.05) when compared to UM siRNA, suggesting duplex unwinding may have a role in its enhanced RNAi activity (Table 2). Importantly, in contrast to 2’-OMe modification alone which increased Tm, its addition at the site of PS2 modification did not elevate Tm.
Table 2.
Thermostability of chemically modified siRNAs
| SiRNA sequence | Average Tm (°C) | Std. Dev. (°C) | ΔTm (°C) |
|---|---|---|---|
| UM | 76.51 | 0.64 | N/A |
| Me-1 | 76.68 | 0.50 | 0.17 |
| PS-1 | 75.91 | 0.65 | -0.60 |
| MePS-1 | 76.37 | 0.08 | -0.14 |
| PS2-1 | 75.82 | 0.50 | -0.69 |
| MePS2-1 | 75.27 | 0.07 | -1.24 |
We also hypothesized that the MePS2 modification may potentially alter helix conformation between the target and guide strand by changing local structure, which can affect silencing activity24,25. However, circular dichroism (CD) spectra of all the modified sequences tested were similar to the spectrum of UM duplexes and are consistent with the typical A-type structure (a maximum of the positive Cotton effect at 268 nm and a crossover point at 240 nm) (Fig. 3a).
Figure 3. Biophysical properties of MePS2-1 siRNA.
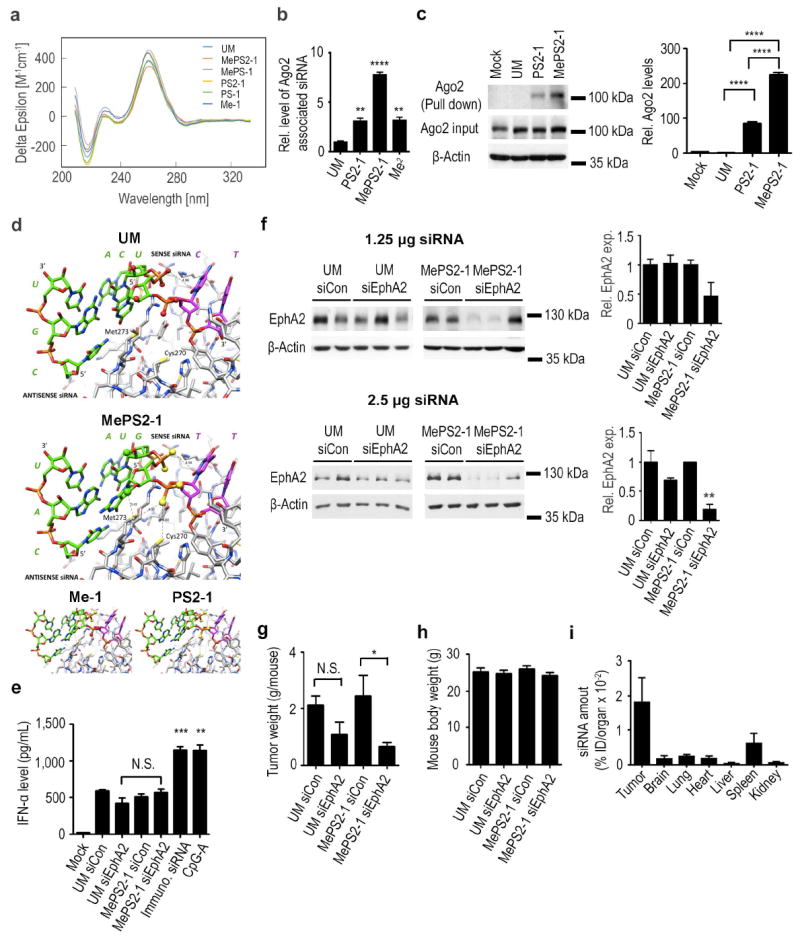
(a) Circular dichroism spectra of Me-1, PS-1, MePS-1, PS2-1, and MePS2-1 modified siEphA2s. (b) Intracellular binding of UM, PS2-1, MePS2-1, and Me2 modified siEphA2 to Ago2 protein in SKOV3ip1 cells (50 nM, serum-free condition). (c) Ago2 association with UM, PS2-1, and MePS2-1 biotin-labeled siRNAs. Apart from mock-treated (no treatment control) sample, each sample contains the same amount of siRNA (quantified using stem loop PCR). Transfection was performed using 50 nM biotin-labeled siRNA in serum-free condition. The bottom two panels show the effect of transfection on total Ago2 level in cells. (d) Computational modeling of the siRNA:PAZ interaction. UM, MePS2-1, Me-1, and PS2-1 modified siRNAs were modeled, all shown in the same orientation. (e) Induction of IFN-α by UM and MePS2-1 modified siEphA2 in C57BL/6-derived dendritic cells (75 nM, serum-free condition). A high IFN-α-inducing siRNA sequence and CpG2216 were used as positive controls. (f) Knockdown of EphA2 protein in tumors following a single dose of siRNA-DOPC treatment in SKOV3ip1 ovarian cancer mouse model (1.25 and 2.5 μg per dose). Effect of UM-siEphA2-DOPC and MePS2-1-siEphA2-DOPC on tumor burden (g) and body weight (h) in SKOV3ip1 orthotopic ovarian cancer mouse model following 4 weeks of siRNA treatment (n=10). (i) Biodistribution of MePS2-1-siEphA2-DOPC. SiRNA levels were measured in various organs at 24 h post intraperitoneal administration. Stemloop PCR technique was employed to assess intact siRNA levels. The total amount of siRNA in each organ was measured (n=4) and was expressed as percentage of injected dose (ID). [P-values obtained with Student’s t-test; (b) **, P<0.01; ****, P<0.0001; compared to UM; (c) ****, P<0.0001; compared to respective controls; (e) **, P<0.01; ***, P<0.001; compared to UM siCon; (f, g) *, P<0.05 or **, P<0.01; compared to the corresponding control groups; bars and error bars represent mean values and the corresponding S.E.M.s (n= 2-3)].
Our next hypothesis was that the thiol groups on MePS2-1’s phosphate backbone may interact with metal ions or amino acid residues in RISC to increase the sequence’s affinity for the complex26,27. We tested this hypothesis by transfecting UM, PS2-1, and MePS2-1 into SKOV3ip1 cells, then immunoprecipitating argonaute-2 (Ago2). SiEphA2 with two 2’-OMe residues at the 5’-end of the sense strand (Me2-siEphA2)28 was used as a positive control. From a panel of 10 different microRNAs (miR) tested, hsa-miR-484 was found to be both unaffected by siEphA2 and present at a high level in SKOV3ip1 cells, thus we chose it as our normalizing control (Supplementary Fig. 6). We found that Ago2-associated MePS2-1 and PS2-1 sequences were enriched 7-fold and 3-fold compared to UM siRNA, respectively (Fig. 3b). In contrast to UM siRNA where the level of associated Ago2 protein was below the limit of detection via Western blotting, a high level of siRNA-associated Ago2 was detected for both PS2-1 and MePS2-1 siRNAs in the biotin-siRNA pull-down study (Fig. 3c). In both of these experiments, there was a significant increase in Ago2 binding for MePS2-1 compared to PS2-1 sequence (>2-fold). The ability of MePS2-1 modification to enhance loading of siRNAs into RISC was also demonstrated using another siRNA sequence (Supplementary Fig. 5d). Of note, the enhanced binding of MePS2-1 siRNAs to RISC had minimal effect on miR biogenesis (Supplementary Fig. 7).
We further investigated the impact of MePS2-1 modification on strand loading. SKOV3ip1 cells were transfected with siEphA2 sense (S) and antisense (AS) strand-specific luciferase reporters. MePS2-1(S) decreased the luciferase signal to a much greater degree (2.2-fold enhancement) compared to UM siRNA in cells transfected with the AS reporter construct (Supplementary Fig. 8a). As expected, MePS2-1(AS) did not reduce the luciferase signal significantly when compared to control siRNA (siCon) treatment. Conversely, in cells transfected with the S reporter construct, MePS2-1(AS) further improved the silencing ability of the siRNAs when compared to the UM sequence, while no silencing was observed with MePS2-1(S) (Supplementary Fig. 8b). We further compared the silencing efficacy of MePS2-1(S) with Me2(S)-siRNA. In cells transfected with the AS reporter construct, Me2(S) siRNAs decreased the luciferase signal to a greater extent compared to UM, as expected (Supplementary Fig. 8a). This extent of silencing was comparable to that achieved by MePS2-1(S). The addition of MePS2-1(S) modification to Me2(S)-siRNA (Me2MePS2-1) did not significantly alter its silencing efficiency. These results are consistent with the level of EphA2 mRNA knockdown observed with these siRNAs (Supplementary Fig. 8c).
We next used computational modeling to examine the potential effects of the MePS2 modification on the interaction of siRNAs with RISC, focusing on the PAZ domain that recognizes 3’-overhanging nucleotides of siRNA duplexes29. In the crystal structure, the siRNA 5’-(CGUGACU)-d(CU)-3’ forms a duplex with one G-G and two U-C mismatch pairs and 3’-terminal d(CU) overhangs. Modeling entailed changing the penultimate 3’-dC to T, converting the terminal RNA U-C pair in the native duplex to G:C, and changing the adjacent G:C pair to A:U. Methyl groups were also added to the sugar 2’-oxygen atoms of U and G in the antiperiplanar orientation (torsion angle C3’-C2’-O2’-Cmethyl30) Inspection of the model of MePS2-1 bound to PAZ indicates that the combination of the 2’-OMe and PS2 modifications will likely benefit the interaction between siRNA and PAZ (Fig. 3d). Thus, the proximities of the 2’-OMe moiety of G and Cε of M273 as well as the PS2 moiety of T and both Cε of Met273 and the sulfhydryl group of C270 are consistent with favorable hydrophobic interactions that are absent in the complex with the UM siRNA and are superior to those for the Me-1 and PS2-1 siRNAs. Taken together, the combination of PS2 and 2’-OMe modification triggers an unusual stereoelectronic effect in the backbone of the siRNA, which leads to increased RISC loading. This, along with an increase in resistance to nuclease degradation as well as a decrease in Tm, provides a likely explanation for enhanced RNAi activity by MePS2-1 modification.
In vivo application of MePS2-1 siRNA
Having identified a promising chemical modification that could significantly enhance the stability and silencing activity of siRNAs, we next characterized its potential off-target immunotoxicity. C57BL/6-derived dendritic cells were treated with UM control siRNA (siCon), UM siEphA2, MePS2-1 siCon, and MePS2-1 siEphA2. Interferon alpha (IFN-α) levels, a surrogate marker for non-specific induction of innate immunity, were similar for all four sequences examined (Fig. 3e). No difference in Toll like receptor 7 (TLR7)-induced IFN-α or tumor necrosis factor alpha (TNF-α) levels was observed between UM siEphA2 and MePS2-1 siEphA2 at all time-points examined (Supplementary Fig. 9). Of note, serum IFN-α and TNF-α levels were both undetectable at 2, 6, and 24 h following intraperitoneal (i.p.) administration of 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) nanoliposomes-delivered UM-siEphA2 (UM-siEphA2-DOPC) or MePS2-1 siEphA2 (MePS2-1-siEphA2-DOPC) in mice.
The efficacy of the MePS2-1 siRNA in vivo was next assessed using an orthotopic mouse model of ovarian cancer (SKOV3ip1). The effect of EphA2 knockdown in tumors was first examined following a single injection. While some degree of knockdown was achieved with MePS2-1-siEphA2-DOPC at the dose of 1.25 μg, (Fig. 3f), >2.5 μg per dose was determined to be necessary to achieve consistent and significant gene silencing in tumors. At the dose of 2.5 μg, measurement of EphA2 by Western blot of tumor lysate revealed superior gene silencing effect of MePS2-1-siEphA2-DOPC compared to UM-siEphA2-DOPC (80% vs. 30% reduction, respectively). We next treated mice with 2.5 μg of UM-siCon-DOPC, UM-siEphA2-DOPC, MePS2-1-siCon-DOPC, and MePS2-1-siEphA2-DOPC twice weekly (n=10). After four weeks of therapy, the animals were sacrificed and necropsies were performed. UM-siEphA2-DOPC reduced the tumor burden compared to UM-siCon-DOPC group (50% reduction), however, the difference is not statistical significant. In contrast, MePS2-1-siEphA2-DOPC exhibited significant reduction in tumor weight (Fig. 3g, 70% reduction) compared to the MePS2-1-siCon-DOPC treatment group. No group showed decreased body weight, one major indicator of toxicity (Fig. 3h). Overall, these results demonstrate superior gene silencing and therapeutic effects with MePS2-1-siEphA2-DOPC compared to UM-siEphA2-DOPC.
We next evaluated the pharmacokinetic profile and biodistribution pattern of MePS2-1-siEphA2-DOPC following a single i.p. injection in tumor-bearing mice. The level of siRNAs accumulated in tumors was found to be significantly higher than that observed in other major organs, including lung, heart, liver, spleen, kidneys, and brain (Fig. 3i). The localization of DOPC nanoliposomes to tumors was further confirmed using Cy5.5-labeled siRNAs (Supplementary Fig. 10). This ability of DOPC nanoliposomes to preferentially deliver siRNAs to tumors while avoiding significant uptake in other organs is similar to that observed with several other systems, including pRNA nanoparticles31. The pharmacokinetic analysis of MePS2-1-siEphA2-DOPC showed an Area Under the Curve (AUC)0-72 of 477 pg*h mL-1, Clearance (CL) of 5.23 L h-1, and Volume of distribution (Vd) of 26.3 L. Distribution half-life of MePS2-1-siEphA2-DOPC was 3.7 min and the elimination half-life was 9.3 h.
Targeting GRAMD1B for ovarian cancer treatment
Having demonstrated that MePS2-1 siRNA has superior gene silencing activity compared to its UM counterpart for an established target, we next assessed its silencing, and in turn therapeutic capability for a novel ovarian cancer target gene. Since acquired chemoresistance is a major contributor to patient mortality from ovarian cancer (OvCa), we sought to identify targets playing substantial roles in this process. SiRNAs against approximately 22,000 individual genes were evaluated for their lethality and ability to induce taxane sensitivity in taxane-resistant epithelial ovarian cancer cells (SKOV3-TR) (Supplementary Data 1). High correlation between cell viability score was observed across triplicate plates (median coefficient of variation = 8.6%), indicating high data precision in the screen.
A total of 178 genes were identified to have reduced cell viability upon siRNA treatment and showed further cell death when combined with paclitaxel (Supplementary Data 2). Importantly, the dose of paclitaxel used in the screen was IC30 of that achieved in parental taxane-sensitive SKOV3ip1 cells to permit identification of genes that, when silenced, can dramatically sensitize cells to taxane treatment. Out of 178 hits, eight genes were identified to be up-regulated in ovarian cancer epithelial cells compared to normal ovarian surface epithelial cells32 (Fig. 4a). Within this group, we focused on three genes associated with the highest lethality upon knockdown when combined with paclitaxel: GRAMD1B, RBBP6, and SLC23A1. We found that taxane-resistant ovarian cancer cell lines (HeyA8-MDR, SKOV3-TR) showed elevated levels of GRAMD1B mRNA and protein relative to their sensitive counterparts (HeyA8, SKOV3ip1) (Fig. 4 b,c). The expression of GRAMD1B remained unchanged for platinum resistant A2780-CP20 cells when compared to the parental line (A2780), indicating the lack of involvement of this gene in platinum resistance. GRAMD1B was also confirmed to be highly elevated in HeyA8 and SKOV3ip1 compared to the non-transformed HIO-180 ovarian epithelial cells. RBBP6 and SLC23A1 mRNA expression levels, though elevated in some ovarian cancer lines, were not found to be increased in resistant cell lines examined (Fig. 4b).
Figure 4. Targeting GRAMD1B for ovarian cancer treatment.
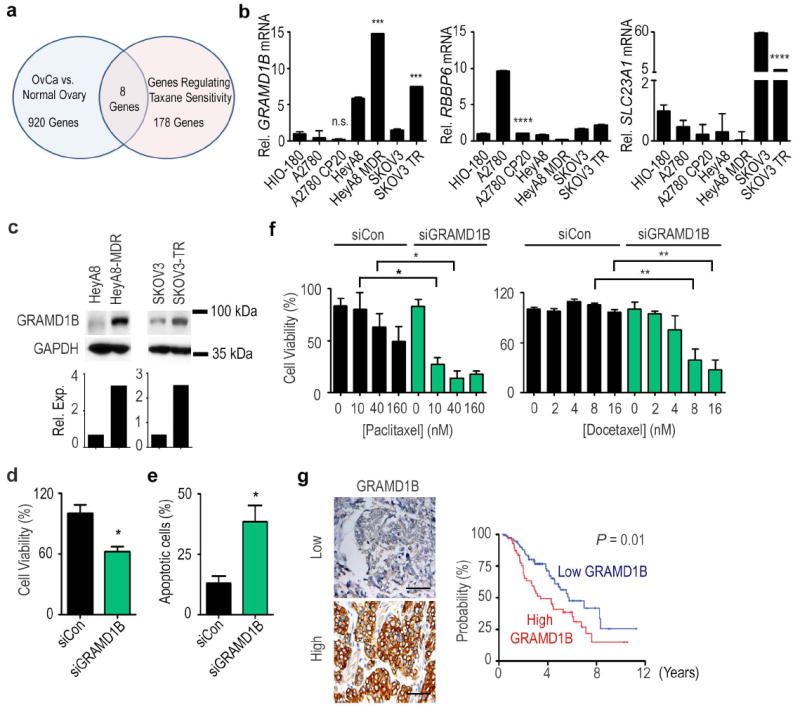
(a) A Venn diagram for identification of novel targets for the treatment of ovarian cancer. (b) Expression of GRAMD1B, RBBP6, and SLC23A1, in a panel of chemosensitive and chemoresistant ovarian cancer cell lines. (c) Protein expression of GRAMD1B in taxane-sensitive (HeyA8 and SKOV3ip1) and taxane-resistant (HeyA8-MDR and SKOV3-TR) ovarian cancer cells (n=1). Effect of GRAMD1B downregulation on (d) cell viablity and (e) apoptosis in HeyA8-MDR cells. (f) Effect of knockdown of GRAMD1B on the sensitivity of HeyA8-MDR cells to paclitaxel or docetaxel treatment. Cell viability is expressed as a percentage of that measured in siRNA monotherapy treated wells. (g) Effect of GRAMD1B protein expression on overall survival in ovarian cancer patients (n=156). Scale bar represents 100 μm. [P-values obtained with (a-f) Student’s t-test or (g) log-rank test; (b) ***, P<0.001 or ****, P<0.0001; compared to the corresponding chemosensitive cell line; (d, e, f) *, P<0.05 or **, P<0.01; compared to cells treated with siCon; bars and error bars represent mean values and the corresponding S.E.M.s (n=3)].
We next examined the effect of silencing GRAMD1B in the taxane-resistant cell line with the highest target expression, HeyA8-MDR (Supplementary Data 3, Supplementary Fig. 11, and Fig. 4 d,e). GRAMD1B silencing via introducing siGRAMD1B into cells using Lipofectamine 2000 exhibited both lethality as monotherapy (40% decrease in cell viability and 3-fold increase in apoptosis, Fig. 4 d,e) and also taxane sensitization at 10 nM paclitaxel (>75% vs. <5% cell death, siGRAMD1B plus paclitaxel vs. siCon plus paclitaxel) (Fig. 4f). Treatment with docetaxel produced a similar pattern. Significant taxane sensitization was also observed when another GRAMD1B-targeting siRNA was transfected into cells using another transfecting reagent, Lipofectamine RNAiMax (Supplementary Fig. 12). These results indicate the potential of GRAMD1B silencing in overcoming taxane resistance in ovarian cancer.
Given the preclinical efficacy after targeting GRAMD1B, we subsequently assessed the impact of tumoral GRAMD1B expression on patient survival. Immunohistochemical GRAMD1B expression was analyzed against the clinical characteristics of a cohort of 156 women with mostly advanced epithelial ovarian cancer (Supplementary Table 2). High tumoral GRAMD1B expression was significantly associated with several indicators of aggressive disease, including suboptimal cytoreductive surgery (50% vs. 33%, P=0.04, Chi-square test), progressive or recurrent disease (92.6% vs. 74.5%, P=0.004, Chi-square test), lower median progression-free survival (PFS, 0.9 vs. 1.72 yrs., P=0.001, log-rank test), and lower overall survival (OS, 3.59 vs. 6.14 yrs., P=0.01, log-rank test, Fig. 4g). GRAMD1B expression remained an independent predictor of survival following multivariate analysis accounting for age, stage, grade, extent of cytoreduction, and GRAMD1B expression (P=0.02, Cox proportional-hazards model).
We further investigated the relationship between tumoral GRAMD1B expression and chemotherapy response in a subset of ovarian cancer patients (n=32) who had suboptimal tumor debulking at the time of surgery (i.e., >1 cm residual disease). Cancer Antigen 125 (CA125) was used as a measure of drug response33. Patients who had CA125 level normalized to 35 at the end of 6 cycles of chemotherapy were considered responsive to treatments. It was found that 89% of patients with high tumoral GRAMD1B protein expression levels responded poorly to chemotherapy (i.e., CA125 > 35 at the end of 6 cycles of chemotherapy). In contrast, patients whose tumor had low GRAMD1B expression responded significantly better to chemotherapy (P<0.001, Chi-square test), with 91.3% of patients having normalized CA125 at the end of the therapy. Median PFS was 0.61 and 2.07 years for patients with high vs. low tumoral GRAMD1B expression, respectively (Supplementary Fig. 13).
Therapeutic effect of MePS2-1 siGRAMD1B in vitro and in vivo
Having established the importance of targeting GRAMD1B in ovarian cancer treatment, we next examined if MePS2-1 modification can significantly enhance the silencing efficiency of UM siGRAMD1B. We found that MePS2-1 siGRAMD1B efficiently silenced GRAMD1B both at the mRNA (70%, data not shown) and protein (85%) level in HeyA8-MDR cell line in the presence of serum (40 nM, Fig. 5a). This level of GRAMD1B knockdown was significantly better than that achieved by UM siGRAMD1B (85% vs. 50%). Consistent with our findings using siEphA2 and siPTMS (Fig. 2d,e, Fig. 3b,c, Supplementary Fig. 5), MePS2-1 modification also significantly enhances the serum and intracellular stabilities of siGRAMD1B (Supplementary Fig. 14) and increases its binding to Ago2 protein when compared to UM siGRAMD1B (Fig. 5b). Let-7a was used as a normalizing control for the Ago2 binding study as its expression is not altered by GRAMD1B silencing (Supplementary Fig. 15).
Figure 5. Therapeutic effect of MePS2-1 siGRAMD1B in vitro and in vivo.
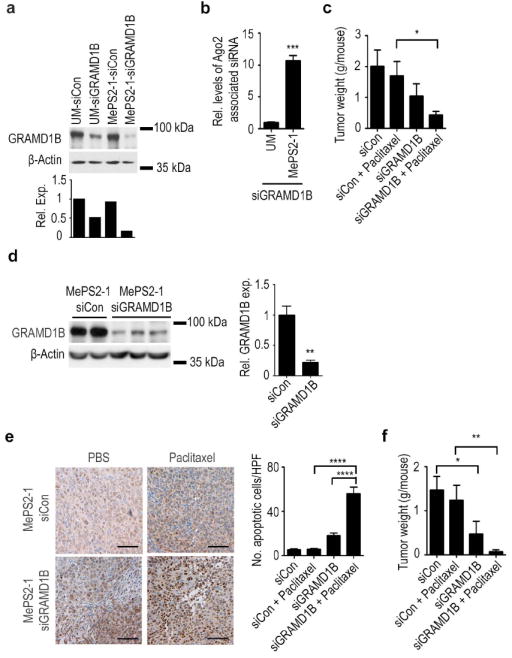
(a) Knockdown of GRAMD1B using UM and MePS2-1 modified siRNAs (40 nM, 10% FBS-containing media) (n=1). (b) Intracellular binding of UM and MePS2-1 siGRAMD1B to Ago2 in HeyA8-MDR cells (50 nM, serum-free condition). (c) Effect of MePS2-1 siGRAMD1B on tumoral response to paclitaxel treatment in HeyA8-MDR orthotopic ovarian cancer mouse model. (d) Knockdown of GRMAD1B by MePS2-1-siGRAMD1B-DOPC in HeyA8-MDR tumors. (e) Caspase 3 activity in tumors treated with MePS2-1-siCon-DOPC or MePS2-1-siGRAMD1B-DOPC +/- paclitaxel. Scale bar represents 100 μm. (f) Effect of MePS2-1 siGRAMD1B on tumoral response to paclitaxel treatment in SKOV3-TR orthotopic ovarian cancer mouse model. [P-values obtained with Student’s t-test; (b) ***, P<0.001, compared to UM si GRAMD1B (n=3); (c, f) *, P<0.05 or **, P<0.01, compared to the corresponding control groups (n=10); (d) **, P<0.01, compared to MePS2-1-siCon-DOPC group (n=2-3); (e), ****, P<0.0001, compared to paclitaxel or MePS2-1-siGRAMD1B-DOPC monotherapy groups (n=10); bars and error bars represent mean values and the corresponding S.E.M.s].
Moving into an orthotopic mouse model, we injected HeyA8-MDR cells into the peritoneal cavity and, after one week, mice were divided into 4 treatment groups: (a) MePS2-1-siCon-DOPC, (b) MePS2-1-siGRAMD1B-DOPC, (c) MePS2-1-siCon-DOPC plus paclitaxel, or (d) MePS2-1-siGRAMD1B-DOPC plus paclitaxel. Dosing was twice weekly for siRNA and weekly for paclitaxel (at a sub-therapeutic 100 μg dose), followed by tumor excision and quantification after five weeks of treatment. No significant reduction in tumor burden was observed for sub-therapeutic paclitaxel with MePS2-1-siCon-DOPC treatment group, as expected (Fig. 5c). However, MePS2-1-siGRAMD1B-DOPC alone resulted in 50% reduction in tumor burden, consistent with our in vitro findings (Fig. 4d). Combining it with paclitaxel induced a 55% further reduction in total tumor weight compared to MePS2-1-siGRAMD1B-DOPC monotherapy, indicating the potential for taxane sensitization after GRAMD1B silencing. The reduction of GRAMD1B levels in tumors by MePS2-1-siGRAMD1B-DOPC was significant, with the protein level being down-regulated by 80% in tumors from mice treated with MePS2-1-siGRAMD1B-DOPC (Fig. 5d). This down-regulation of GRAMD1B in tumors was also significantly associated with the induction of cleaved caspase 3 (Fig. 5e). This is consistent with the increased level of apoptotic cells observed following GRAMD1B silencing in vitro (Fig. 4e). Replicating the experiment with a different GRAMD1B-targeted siRNA sequence in SKOV3-TR, another taxane-resistant ovarian cancer model, produced a similar therapeutic effect (Fig. 5f). In this model, mice treated with both MePS2-1-siGRAMD1B-DOPC and paclitaxel had an 80% further decrease in tumor burden compared with mice treated with MePS2-1-siGRAMD1B-DOPC alone, which again indicated the role of GRAMD1B in taxane resistance. Importantly, in both of the models tested, there was no alteration in mouse body weights in any of the treatment groups (Supplementary Fig. 16).
Discussion
Recent advances in high-throughput genomic and computational tools have facilitated the identification of new targets with translational potential34. Combining novel MePS2 siRNA chemistry with a whole genome lethality screen aimed at identifying novel genes involved in taxane resistance, we demonstrate, for the first time, the feasibility of using highly potent MePS2 modified siRNAs for effective therapeutic targeting. We show that GRAMD1B, a gene identified through our high throughput screening and analyses, has both high biological and clinical importance. Using it as a model system, we demonstrate that robust gene silencing in tumors can be achieved following systemic administration of MePS2 modified siRNAs. This ultimately led to significant re-sensitization of chemoresistant ovarian tumors to taxane therapy.
Most of the chemical modification strategies, to date, have focused on improving siRNA stability in serum7. While this is critical, improving siRNA potency at target sites following systemic administration still remains the major obstacle to clinical translation of siRNAs5,6. Through a systematic approach, we have identified a unique combination of 2’-OMe and PS2 which significantly improves stability and potency of siRNAs simultaneously (Fig. 6). We first confirmed that the increase in potency could not be explained by the design’s enhanced stability, consistent with previous findings that this mechanism has a limited role within rapidly-dividing cells35, and then investigated other potential causes.
Figure 6. A schematic representation of mechanisms by which MePS2 modification enhances siRNA activity.
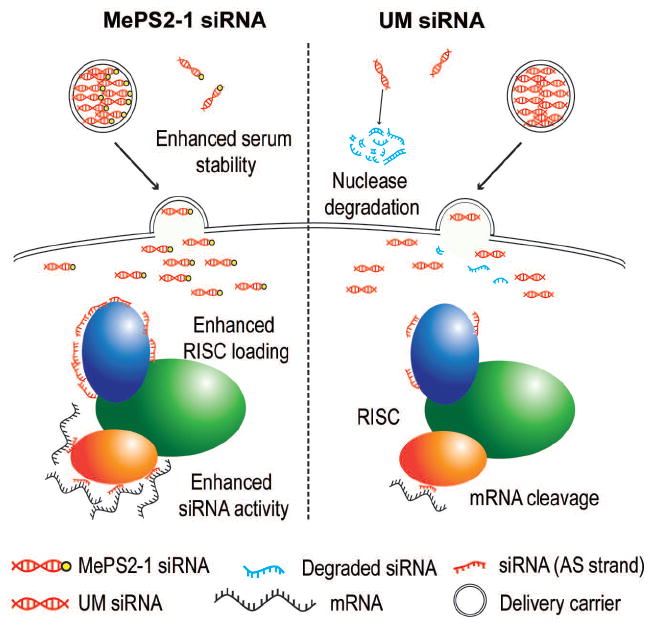
MePS2-1 modification enhances serum stabilty of siRNAs and promotes the loading of the antisense strand into RISC.
We discovered that MePS2 modification further enhances the loading of siRNAs to RISC, compared to PS2 modification, resulting in enhanced silencing. While the addition of methyl groups typically elevates Tm, we show here that the addition of 2’-OMe to PS2-modified siRNAs does not increase the Tm of the siRNA duplex, suggesting a unique property arising from this combination. The computational modeling also indicates an unusual favorable stereoelectronic effect on the siRNA backbone following MePS2 modification. These results indicate that MePS2 modification can improve the interaction of siRNA with RISC via several mechanisms, namely the thiol-protein interaction26,27,36, modulation of Tm, and altered stereoelectronics. This is in contrast to currently existing modifications, where RISC loading efficiency is dependent on alteration in Tm, thus leading to variable silencing effects depending on the Tm of the sequences to be modified20,37. Hence, MePS2 modification has a unique potential to provide more consistent, sequence-independent, silencing effects compared to existing chemistries.
In summary, we show that 2’-OMe and PS2 combination not only significantly increases the serum stability of siRNAs but also triggers a more potent intrinsic RNAi response than the individual modifications. In addition to its achiral nature which favors clinical translation, we have demonstrated the ability of MePS2 modification to overcome major obstacles of bringing siRNA therapeutics into the clinic. It is anticipated that further understanding the unique and advantageous interaction between 2’-OMe and PS2 moieties will facilitate the rational design of highly potent and stable siRNAs in the near future.
Methods
Cell lines
The ovarian cancer cell lines, SKOV3ip1 and HeyA8, were obtained from ATCC and were maintained in RPMI 1640 supplemented with 15% fetal bovine serum and 0.1% gentamicin sulfate (Gemini Bioproducts; Calabasas, CA)38. SKOV3-TR and HeyA8-MDR were kind gifts from Dr. Isaiah Fidler, Department of Cancer Biology, University of Texas M. D. Anderson Cancer Center and Dr. Michael Seiden, Department of Medicine, Massachusetts General Hospital, respectively.
SiRNA synthesis
SiRNAs were synthesized at AM Biotechnologies. All sequences are listed in Supplementary Table 3. Modified and unmodified 21-nt RNAs (sense and antisense strands) were synthesized on the 1 μmole scale on an Expedite 8909 DNA/RNA Synthesizer using commercial available 5’-DMT-2’-O-TBDMS nucleoside (ABz, CAc, GAc, and U) phosphoramidite monomers and 5’-DMT-2’-O-Methyl nucleoside (ABz, CAc, GAc, and U) phosphoramidite monomers as well as in house produced 5’-DMT-2’-O-Methyl nucleoside (ABz, CAc, GAc, and U) thiophosphoramidite monomers15, 16,28. All oligonucleotides were synthesized in DMT-off mode. After completion of the synthesis, the solid support was suspended in ammonium hydroxide/methylamine (AMA) solution (prepared by mixing 1 volume of ammonium hydroxide (28%) with 1 volume of 40% aqueous methylamine) and heated at 65 °C for 15 min to release the product from the support and to complete the removal of all protecting groups except the TBDMS group at the 2’-position. The solid support was filtered, and the filtrate was concentrated to dryness. The obtained residue was resuspended in 115 μL of anhydrous DMF and then heated for 5 min at 65 °C to dissolve the crude product. Triethyal amine (TEA, 60 μL) was added to each solution, and the solutions were mixed gently. TEA·3HF (75 μL) was added to each solution, and the tubes were then sealed tightly and incubated at 65 °C for 2.5 h. The reaction was quenched with 1.75 mL of DEPC-treated water. Purification was performed on an Amersham Biosciences P920 FPLC instrument fitted with a Mono Q 10/100 GL column39. The structures of the modified RNAs were confirmed by ESI-MS and 31P-NMR. The assembly of the resulting duplexes was confirmed on a 4% agarose gel electrophoresis. For in vivo experiments, ammonium counter cation was replaced by sodium cation.
In vitro siRNA transfection and luciferase assays
Cells were transfected with 10-100 nmol L-1 of specified siRNAs using Lipofectamine 2000 or Lipofectamine RNAiMax reagent (Invitrogen) at 3 μL reagent : 1 μg siRNA ratio21. Cells were treated with siRNAs for 4 h in serum-free or 10% FBS-containing media before incubation in fresh complete media for the specified timeframe.
GoClone pLightSwitch luciferase reporter constructs containing AS and S siRNA target sequences were obtained from SwitchGear genomics (Menlo Park, CA). AS construct contains the following sequence: 5’-CAAAGGGTGGGACCTGATGCAGAACATCA TGAATGACATGCCGATCTACATGTACTCCGTGTGCAACGTGATGTCTGGCGAC-3’. S construct contains the following sequence: 5’-ACCTGATGCAGAACATCATGAACAT GTAGATCGGCATGTCATACTCCGTGTGCAACGTGATGTCT-3’. SKOV3ip1 cells were transfected with FuGENE HD reagent in a 96-well plate with specified siRNAs (100 nM) along with S or AS reporter construct, and Cypridina TK control construct (pTK-Cluc)40. After 24 h of transfection, luciferase activity was obtained using LightSwitch Dual Luciferase assay kits. Luciferase activity was normalized with the Cypridina TK control construct. All assays were performed in triplicate.
Quantitative reverse transcription–polymerase chain reaction
RNA was isolated from cells, Ago2 pull down products, blood samples, and tissues using Trizol (Invitrogen) according to the manufacturer’s protocol. Using 1 μg of RNA, cDNA was synthesized by using a Verso cDNA kit (Thermo Scientific) as per the manufacturer’s instructions. Analysis of mRNA levels was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) with SYBR Green-based qRT-PCR. PCR was performed with reverse-transcribed RNA and 100 ng μL-1 of forward and reverse primers in a total volume of 20 μL38. All primer sequences for mRNA detection are listed in Supplementary Table 4. Each cycle consisted of 15 s of denaturation at 95 °C and 1 min of annealing and extension at 60 °C (40 cycles). Stem loop PCRs for siRNA detection were performed using TaqMan miRNA assays followed by SYBR Green-based qRT-PCR41. Stemloop primers used to synthesize cDNA are listed in Supplementary Table 5. Respective forward primers as well as a common reverse primer used for qRT-PCR are listed in Supplementary Table 4. For miRNA quantification, total RNA was isolated using Trizol (Invitrogen) extraction. TaqMan miRNA assays (Applied Biosystems) were used for reverse transcription and qRT-PCR was performed according to the manufacturer’s instructions. For microRNA detection, Ambion assay probe sets were used according to the manufacturer’s protocol. The relative amount of mRNA/siRNA in each sample was normalized to β-actin mRNA, RNU44, RNU6B, or respective microRNAs.
Melting profiles of siRNAs
Complementary RNA strands were mixed in 10 mM Tris-HCl buffer (pH 7.4) with 100 mM NaCl, and 0.1 mM Ethylenediaminetetraacetic acid (EDTA) at a final concentration of 2 μM. Annealing was performed at a temperature gradient of 1.5 °C min-1, from 85 to 15 °C. Melting profiles were measured using a temperature gradient of 1 °C min-1, from 15 to 85 °C, with the detector set at 260 nm (n=3-6).
Circular Dichroism measurements
Circular Dichroism (CD) spectra were recorded on a CD6 dichrograph (Jabin-Yvon) using cells with 0.5 cm path length, 2 nm bandwidth, and 1-2 s integration time. Each spectrum was smoothed with a 25-point algorithm. The spectra from 200-340 nm were recorded at 25 °C in the same buffer as in the melting experiments (n=3). The concentration of the two complementary RNA oligonucleotides was ca. 2 μM.
Western blot
Protein lysates were prepared from cultured cells or tumors using modified RIPA buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton, 0.5% deoxycholate) plus 25 μg mL-1 leupeptin, 10 μg mL-1 aprotinin, 2 mM EDTA, and 1 mM sodium orthovanadate38. Membranes were probed with primary antibodies against EphA2 (1:1000, Millipore, 05-480), β-actin (1:5000, Sigma, A5316), vinculin (1:2500, Sigma, V9131), GAPDH (1:5000, Sigma, G8795), Ago2 (1:100, Wako Chemicals, 015-22031), and GRAMD1B (1:500, Sigma, HPA008557) in 5% skim milk in TBS-T. Quantification was performed using ImageJ. Full scans of important Western blots are provided in Supplementary Fig. 17.
Intracellular Ago2 binding study
Cells were lysed at 7 h post-transfection using ice-cold lysis buffer (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 0.5% Triton, 2 mM EDTA, 1 mg mL-1 heparin, and protease inhibitor cocktail)42,43. Ago2 antibody (Wako Chemicals, 015-22031, 20 μL) was absorbed onto magnetic protein G Dynabeads (Invitrogen, 40 μL) by incubation at 4 °C for 2 h on a rotator. The beads were then washed twice with lysis buffer to remove unbound antibodies. Following washing, beads were incubated with protein lysate (360 μg) overnight at 4 °C on a rotator. Following removal of unbound proteins, bound material was eluted from beads using 50 μL of 0.1 M glycine (pH 2.3) for 15 min at RT. Eluted fractions were neutralized immediately with an equal volume of 1 M Tris-HCl (pH 8) and then treated with 20 U of proteinase K for 10 min at 65 °C44. The amount of Ago2-bound siRNA was then quantified using stem loop PCR technique. For the biotin-siRNA pull down experiment, strepavidin beads were blocked and incubated with cell lysates at 4 °C for 2 h42. The amount of siRNA-associated Ago2 was quantified via Western blotting.
Serum stability assay
SiRNAs (1.33 μg) were incubated in 10 μL of 10% FBS in PBS at 37 °C for up to 24 h. All samples were separated in 20% polyacrylamide gels. The percentage of intact siRNA present was quantified using ImageJ.
Immunogenicity of siRNAs
Bone marrow progenitor cells were isolated from C57BL/6 mice and were differentiated into dendritic cells in vitro for 7 days using FLT3 ligand (100 ng mL-1; PeproTech). Cells were treated with 75 nM of siRNAs for 6 h in serum-free media using RNAiMax™. A high interferon-inducing siRNA sequence (BP1Mod2; Supplementary Table 3) and CpG2216 (1 μg mL-1, Sigma) were used as positive controls. The level of IFN-α and TNF-α in the culturing media was assessed using murine IFN-α and TNF-α ELISA kits at 2, 6, and 24 h following transfection (R&D Systems). For TLR7 experiments, peripheral blood mononuclear cells (PBMCs) were obtained from wild-type and TLR7-KO C57BL/6 mice and were treated with 75 nM of siRNAs for 6 h. IFN-α and TNF-α levels in the culturing media was measured 24 h post-transfection. For in vivo studies, serum was collected from mice at 2, 6, and 24 h post siRNA treatment and IFN-α and TNF-α levels were measured as described above.
Liposomal nanoparticle preparation
SiRNAs for in vivo delivery were incorporated into DOPC liposomes21. DOPC and siRNA were mixed in the presence of excess tertiary butanol at a ratio of 1:10 (w/w) siRNA/DOPC. Tween 20 was added to the mixture in a ratio of 1:19 Tween 20:siRNA/DOPC. The mixture was vortexed, frozen in an acetone/dry ice bath, and lyophilized. Before in vivo administration, this preparation was hydrated with PBS.
Orthotopic in vivo models of ovarian cancer
Female athymic nude mice (8-12 weeks old) were obtained from the National Cancer Institute, Frederick Cancer Research and Development Center (Frederick, MD). All mouse studies were approved and supervised by the M.D. Anderson Cancer Center Institutional Animal Care and Use Committee. For therapeutic experiments, ten mice were assigned per treatment group. This sample size gave 80% power to detect a 50% reduction in tumor weight with 95% confidence. To establish intraperitoneal tumors, cells were injected into the peritoneal cavity (i.p.) at a concentration of 5×106 cells mL (200 μL per injection). All mice were treated with siRNAs (2.5 or 5 μg for EphA2 and GRAMD1B experiments, respectively) twice weekly beginning at one week following tumor cell injection. Paclitaxel (100 μg) was injected i.p. once weekly. Mice (n=10 per group) were monitored for adverse effects and tumors were harvested after 5 weeks of therapy or when any of the mice began to appear moribund. Mouse weight, tumor weight, and the number/location of tumor nodules were recorded. Tumor tissues was fixed in formalin for paraffin embedding or was snap frozen for lysate preparation21. For gene silencing (1.25 and 2.5 μg), immune toxicity (5 μg), pharmacokinetic (2.5 μg), and biodistribution studies (2.5 μg), single dose of siRNA was administered i.p. once tumors became palpable. Blood and/or tissues samples were collected at specified time points for RNA/protein extraction or ELISA assay. For the fluorescence-based biodistribution study, Cy5.5-labeled siRNAs were administered i.p. and 48 h later, fluorescence imaging of excised tumor and organs was performed using the Xenogen IVIS 200 system40. Cy5.5 fluorophore excitation (678 nm) and emission (703 nm) filters were used. Using Living image 2.5 software, regions of interest were drawn for each organ and the total radiant efficiency ps-1 μW-1cm2 was measured.
Pharmacokinetic analysis
Blood concentrations of siRNA at the indicated times, C(t), were expressed as pg siRNA per mL of blood. Pharmacokinetic parameters were determined by fitting the C(t) with a two-compartment model using the MULTI program45. The area under the blood concentration-time curve (AUC) was calculated by integration of C(t) up to 72 h following administration. Vd was calculated by dividing the injected dose by C(0). CLtotal was calculated by dividing injected dose by AUC.
Microarray and pathway enrichment analysis
HeyA8-MDR cells were treated with siGRAMD1B and total RNA was extracted 48 h following transfection using mirVana RNA isolation labeling kit (Ambion). Five hundred nanograms of total RNA were used for labeling and hybridization on a Human HT-12 v4 Beadchip (Illumina) according to the manufacturer’s protocols46. After the bead chips were scanned with an Illumina BeadArray Reader (Illumina), the microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data (LIMMA) package in the R language environment. The expression level of each gene was transformed into a log2 base before further analysis.
GEOID for the microarray data is GSE54459. The top ten pathways perturbed by siGRAMD1B treatment were identified using NetWalker software47. Each interaction in the global network of biological relationships was scored based on combined assessment of the network connectivity and the input data. Biological pathways were ranked based on geometric P-values.
SiRNA lethality screen
SKOV3-TR cells used for the screen were maintained in antibiotic-free RPMI1640 supplemented with 10% fetal bovine serum48 and were authenticated prior to their utilization in the screen. For the screen, SKOV3-TR (2,000 cells per well) were transfected using siRNAs in individual wells of black 384-well plates for 48 h. Transfection was performed using a pool of 4 individual siRNAs targeting individual genes (siGenome Smart pool, G-004605, Dharmacon) using Dharmafect 1 (Dharmacon)49. SiTOX was used as a control for transfection efficiency. Non-targeting siRNA number 4 (siOTP4, Dharmacon) was used as a transfection control in all experiments. In summary, 0.2 μL of 20 μM siRNA was mixed with 0.2 μL of Dharmafect 1 and 20 μL of Optimem medium (Invitrogen) for 20 min in a well of 384-well plate and overlaid with 80 μL of cell suspension (2,000 cells) in antibiotic-free RPMI1640 supplemented with 10% fetal bovine serum. This gave a final siRNA concentration of 40 nM per well. For each plate, 4 types of control were included: siOTP4 as a negative control (16 wells), siTOX as a control for transfection efficiency (8 wells), cells only (24 wells), and blank wells (16 wells). Following siRNA treatment, cells were treated with paclitaxel (3.5 nM). All experiments were performed at 37 °C in the presence of 5% CO2. Finally, cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, USB Corporation) as per manufacturer’s instruction three days after treatment and signal was detected using a PHERAstar FS plate reader (BMG Labtech). Following data collection, the values in each plate were centered on 100 using median scaling49 and cell viabilities were expressed as percentages of siOTP4 treated samples. Student’s t-test was used to examine chemosensitization following conversion of raw values to log2 scale.
Immunohistochemistry analysis
Staining was performed in paraffin-embedded tumor sections21. For patient samples, tissue microarray (TMA) slides containing 156 ovarian cancer samples were obtained from M.D. Anderson Cancer Center following approval by the Institutional Review Board. Antigen retrieval was performed using citrate buffer (pH 6) or Borg Decloaker solution (Biocare Medical, BD1000 S-250) for GRAMD1B and cleaved caspase 3, respectively. Slides were incubated with primary antibodies against GRAMD1B (1:100 dilution, Sigma, HPA008557) or cleaved caspase 3 (1:200, BioCare Medical, CP229A) overnight, followed by incubation with horseradish peroxidase-conjugated or biotinylated/streptavidin anti-Rabbit secondary antibodies, respectively (Jackson Laboratory, 111-036-047 and BioCare Medical, GR602 H). The stained slides were scored by two investigators blinded to the treatment groups or patient survival information. For in vivo samples, 5-10 random fields at 200x magnification for each tumor were examined. For patient samples, a scoring system, which considered both the intensity of staining and the percentage of cells stained, was used46,50. H scores of >100 and ≤100 were defined as high and low expression, respectively.
Cell viability assays
HeyA8-MDR cells were transfected with siRNAs (100 nM) on day 1 and 3 before paclitaxel or docetaxel treatment. Cell viability was assessed three days following taxane treatment using MTT assay as per manufacturer’s protocol.
Statistical analysis
Kaplan–Meier plots were constructed and a log-rank test was used to determine differences among survival curves according to tumoral GRAMD1B expression level46,50. Contingency tables and Fisher’s exact test or Chi-square test were used to evaluate the relationship between death and categorical variables. Multivariate analyses were performed with the use of a Cox proportional-hazards model to examine the effects of tumoral GRAMD1B expression on death from disease while adjusting for other covariates. Chi-square test was performed to assess the effect of tumoral GRAMD1B expression on drug response. For siRNA lethality screen, student t-test was used to examine chemosensitization following conversion of raw values to log2 scale. For other assays, student’s t-test was performed to examine the difference between control and treatment groups.
Apoptosis assay
Apoptosis was studied using the Annexin V apoptosis detection kit (BD Biosciences) according to the manufacturer’s protocol38. Apoptotic cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences). CellQuest Pro software (BD Biosciences) was used to determine the number of apoptotic cells.
Modeling of siRNA:PAZ domain interactions
Coordinates of the complex between a 9 mer siRNA and the PAZ domain from human Argonaute eIF2c1 were retrieved from the Protein Data Bank (http://www.rcsb.org; PDB ID code 1si229). The program UCSF Chimera51 was used to convert the sequence of the last four nucleotides in the siRNA from the 1si2 crystal structure to match the sequence and chemistry of MePS2-1 (…ACUdCdU-3’ and …AUMeS2GMeS2dTdT-3’, respectively). Changes to reflect the chemistries of the Me-1 and PS2-1 siRNAs were accomplished in a similar manner. Side chain torsion angles of C270 and M273 were adapted in USCF Chimera, and the program was used for measuring distances and producing figures. The complex models generated were not subjected to energy minimization with either molecular mechanics or dynamic approaches. The important role of M273 in stabilizing the siRNA:PAZ interaction is reflected by the change in Kd upon mutation of residue 273 from methionine to alanine29. The side chain of M273 snakes along the terminal base pair of the siRNA duplex, and the mutation to alanine abolishes this stacking interaction and doubles the Kd. The effect of hydrophobic interactions between siRNAs and M273/C270 on Kd was also examined.
Supplementary Material
Acknowledgments
S.Y.W. is supported by Ovarian Cancer Research Fund, Inc., Foundation for Women’s Cancer, and Cancer Prevention Research Institute of Texas training grants (RP101502, RP101489, and RP110595). Portions of this work were supported by the NIH (GM100777-01, GM086937-01, GM084552-04, CA016672, CA109298, P50 CA083639, P50 CA098258, CA128797, U54 CA151668, UH2 TR000943-01, and U24CA143835), NCI-DHHS-NIH T32 Training grant (T32 CA101642), the DOD (OC120547 and OC093416), the Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), the Blanton-Davis Ovarian Cancer Research Program, the RGK Foundation, the Gilder Foundation, the estate of C.G. Johnson Jr, the Betty Anne Asche Murray Distinguished Professorship, and statutory funds of the Centre of Molecular and Macromolecular Studies of the Polish Academy of Sciences. We would like to thank D. Ruder, D. Reynolds, J. Seymour, S. Dorta-Estremera, N. Sadaoui, Drs. M. Ravoori, R. Langley, X.F. Huang, C.H. Lu, J. Filant, W. Cao, and members of the histology, flow cytometry and siRNA screening facilities at the University of Texas M.D. Anderson Cancer Center for technical assistance, provision of study materials, and/or helpful suggestions on the manuscript.
Footnotes
S.Y.W., X.Y., and A.K.S. conceived the project and designed the experiments. X.Y. synthesized the chemically modified siRNAs. S.Y.W, K.G., H.H., M.M., T.M., A.S.N, R.R., L.H., and C.K. participated in in vitro experiments. M.S. and B.N. performed preliminary silencing experiments on MePS2 siRNAs and thermostability study. M.E. performed computational modeling analysis. C.M. and T.H. performed siRNA characterization. S.Y.W. and W.O. designed and performed immunotoxicity assays. S.Y.W., R.S., N.A.J.M, and R.R. designed and performed stem-loop PCR assays. S.Y.W., T.M., S.P., H.D.H., and H.J.C. participated in in vivo experiments. C.R.A. and G.B.L designed and prepared liposomes for in vivo studies. V.S., P.R., C.I., K.B., and S.Y.W. designed and performed computational analyses. A.K.S., S.Y.W., R.A.P., A.S.N, and M.M. analyzed the clinical data. M.T., C.V.P., G.N.A., and K.K. participated in experimental design and/or manuscript preparation. S.Y.W., X.Y., and A.K.S. analyzed all the data and wrote the paper. All authors edited and approved the final manuscript.
Conflict of interest
X.Y. is an employee of AM Biotechnologies LLC. The remaining authors declare no competing financial interests
References
- 1.Tabernero J, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 2.Davis M, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valentino J, et al. Novel small interfering RNA cotargeting strategy as treatment for colorectal cancer. Surgery. 2012;152:277–285. doi: 10.1016/j.surg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, et al. Systemic delivery of E6/7 siRNA using novel lipidic particles and its application with cisplatin in cervical cancer mouse models. Gene Ther. 2011;18:14–22. doi: 10.1038/gt.2010.113. [DOI] [PubMed] [Google Scholar]
- 5.Wu S, McMillan N. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11:639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pecot C, Calin G, Coleman R, Lopez-Berestein G, Sood A. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choung S, Kim Y, Kim S, Park H, Choi Y. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 8.Kenski D, et al. siRNA-optimized modifications for enhanced in vivo activity. Mol Ther Nucleic Acids. 2012;1:e5. doi: 10.1038/mtna.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkov A, et al. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19:191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 10.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein D, Foster E, Huang SB, Weller D, Summerton J. A specificity comparison of four antisense types: morpholino, 2’-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 1997;7:151–157. doi: 10.1089/oli.1.1997.7.151. [DOI] [PubMed] [Google Scholar]
- 12.Stein CA. Exploiting the potential of antisense: beyond phosphorothioate oligodeoxynucleotides. Chem Biol. 1996;3:319–323. doi: 10.1016/s1074-5521(96)90113-1. [DOI] [PubMed] [Google Scholar]
- 13.Raal F, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 14.Prakash T, et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J Med Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, et al. Gene silencing activity of siRNA molecules containing phosphorodithioate substitutions. ACS Chem Biol. 2012;7:1214–1220. doi: 10.1021/cb300078e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesler WT, Caruthers MH. Synthesis of pohosphorodithioate DNA via sulfur-linked, base-labile protecting groups. J Org Chem. 1996;61:4272–4281. doi: 10.1021/jo960274y. [DOI] [PubMed] [Google Scholar]
- 17.Marshall WS, Caruthers MH. Phosphorodithioate DNA as a potential therapeutic drug. Science. 1993;259:1564–1570. doi: 10.1126/science.7681216. [DOI] [PubMed] [Google Scholar]
- 18.Khairuddin N, et al. siRNA-induced immunostimulation through TLR7 promotes antitumoral activity against HPV-driven tumors in vivo. Immunol Cell Biol. 2012;90:187–196. doi: 10.1038/icb.2011.19. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, et al. Asymmetric siRNA targeting the bcl-2 gene inhibits the proliferation of cancer cells in vitro and in vivo. Int J Oncol. 2013;42:253–260. doi: 10.3892/ijo.2012.1691. [DOI] [PubMed] [Google Scholar]
- 20.Elmén J, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landen CJ, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 22.Bramsen J, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sipa K, et al. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rana T. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 25.Chiu Y, Rana T. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckstein F. Phosphorothioate Oligodeoxynucleotides: What Is Their Origin and What Is Unique About Them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 27.Rand T, Ginalski K, Grishin N, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci U S A. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson A, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Ye K, Patel D. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubini P, Zürcher W, Egli M. Crystal structure of an oligodeoxynucleotide duplex containing 2’-O-methylated adenosines. Chem Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 31.Abdelmawla S, et al. Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Mol Ther. 2011;19:1312–1322. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowen N, et al. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genomics. 2009;2:71. doi: 10.1186/1755-8794-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridgewater J, et al. Comparison of standard and CA-125 response criteria in patients with epithelial ovarian cancer treated with platinum or paclitaxel. J Clin Oncol. 1999;17:501–508. doi: 10.1200/JCO.1999.17.2.501. [DOI] [PubMed] [Google Scholar]
- 34.Chin L, Hahn W, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraynack B, Baker B. Small interfering RNAs containing full 2’-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubo T, et al. SiRNAs conjugated with aromatic compounds induce RISC-mediated antisense strand selection and strong gene-silencing activity. Biochem Biophys Res Commun. 2012;426:571–577. doi: 10.1016/j.bbrc.2012.08.128. [DOI] [PubMed] [Google Scholar]
- 38.Vivas-Mejia P, et al. Silencing survivin splice variant 2B leads to antitumor activity in taxane--resistant ovarian cancer. Clin Cancer Res. 2011;17:3716–3726. doi: 10.1158/1078-0432.CCR-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Hodge R, Luxon B, Shope R, Gorenstein D. Separation of synthetic oligonucleotide dithioates from monothiophosphate impurities by anion-exchange chromatography on a mono-q column. Anal Biochem. 2002;306:92–99. doi: 10.1006/abio.2001.5694. [DOI] [PubMed] [Google Scholar]
- 40.Pecot C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng A, et al. Stem-loop RT-PCR quantification of siRNAs in vitro and in vivo. Oligonucleotides. 2009;19:203–208. doi: 10.1089/oli.2008.0176. [DOI] [PubMed] [Google Scholar]
- 42.Le M, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PloS Genet. 2011;7:e1002242. doi: 10.1371/journal.pgen.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei Y, et al. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010;16:2553–2563. doi: 10.1261/rna.2255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goff L, et al. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PloS One. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatakeyama H, Akita H, Maruyama K, Suhara T, Harashima H. Factors governing the in vivo tissue uptake of transferrin-coupled polyethylene glycol liposomes in vivo. Int J Pharm. 2004;281:25–33. doi: 10.1016/j.ijpharm.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 46.Armaiz-Pena G, et al. Src activation by β-adrenoreceptors is a key switch for tumour metastasis. Nat Commun. 2013;4:1403. doi: 10.1038/ncomms2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komurov K, White M, Ram P. Use of data-biased random walks on graphs for the retrieval of context-specific networks from genomic data. PLoS Comput Biol. 2010;6:e1000889. doi: 10.1371/journal.pcbi.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamendola D, Duan Z, Yusuf R, Seiden M. Molecular description of evolving paclitaxel resistance in the SKOV-3 human ovarian carcinoma cell line. Cancer Res. 2003;63:2200–2205. [PubMed] [Google Scholar]
- 49.Ahmed A, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18:109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merritt W, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen E, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


