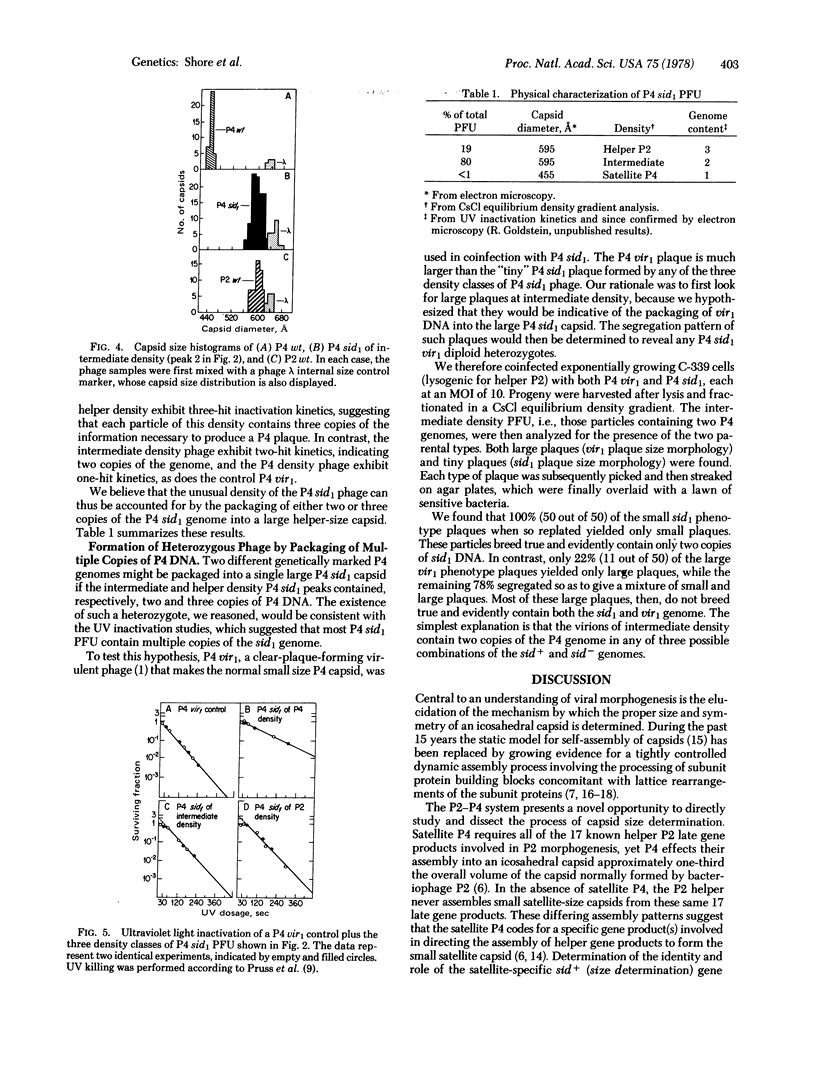
Abstract
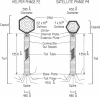
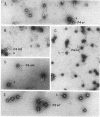
Satellite bacteriophage P4 requires all morphogenic gene products provided by a helper phage, such as coliphage P2, to assemble its own capsid, which is one-third the volume of the larger helper capsid. We have isolated a satellite phage P4 sid (size determination) mutant that is unable to direct the assembly of the small wild-type-size P4 capsid. Instead, this mutant produces P4 plaque-forming units with large P2-size capsids which contain two or three copies of the P4 sid1 genome. P4 sid1 is evidently mutated in a protein that is specifically responsible for determining the precise size and symmetry of the structure into which the helper P2 gene products will assemble. In addition, we have found that the physical size of the genome does not appear to play an essential role in the proper assembly of the icosahedral capsid, since the majority of the P4 sid1 plaque-forming units do not contain a complete capsidful of DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett K. J., Marsh M. L., Calendar R. Interactions between a satellite bacteriophage and its helper. J Mol Biol. 1976 Sep 25;106(3):683–707. doi: 10.1016/0022-2836(76)90259-x. [DOI] [PubMed] [Google Scholar]
- CASPAR D. L., KLUG A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Casjens S., King J. Virus assembly. Annu Rev Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- Gibbs W., Goldstein R. N., Wiener R., Lindqvist B., Calendar R. Satellite bacteriophage P4: characterization of mutants in two essential genes. Virology. 1973 May;53(1):24–39. doi: 10.1016/0042-6822(73)90462-5. [DOI] [PubMed] [Google Scholar]
- Goldstein R., Lengyel J., Pruss G., Barrett K., Calendar R., Six E. Head size determination and the morphogenesis of satellite phage P4. Curr Top Microbiol Immunol. 1974;(68):59–75. doi: 10.1007/978-3-642-66044-3_3. [DOI] [PubMed] [Google Scholar]
- Hendrix R. W., Casjens S. R. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J Mol Biol. 1975 Jan 15;91(2):187–199. doi: 10.1016/0022-2836(75)90159-x. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M., Simon L. D., Six E. W., Walker D. H., Jr Some morphological properties of P4 bacteriophage and P4 DNA. Virology. 1971 Apr;44(1):67–72. doi: 10.1016/0042-6822(71)90153-x. [DOI] [PubMed] [Google Scholar]
- King J., Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974 Sep 13;251(5471):112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Amos L. A., Klug A. Correlation between structural transformation and cleavage of the major head protein of T4 bacteriophage. Cell. 1976 Feb;7(2):191–203. doi: 10.1016/0092-8674(76)90018-0. [DOI] [PubMed] [Google Scholar]
- Lengyel J. A., Goldstein R. N., Marsh M., Calendar R. Structure of the bacteriophage P2 tail. Virology. 1974 Nov;62(1):161–174. doi: 10.1016/0042-6822(74)90312-2. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Six E. W. Replication of bacteriophage P4 DNA in a nonlysogenic host. Virology. 1971 Jan;43(1):1–7. doi: 10.1016/0042-6822(71)90218-2. [DOI] [PubMed] [Google Scholar]
- Pruss G., Goldstein R. N., Calendar R. In vitro packaging of satellite phage P4 DNA. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2367–2371. doi: 10.1073/pnas.71.6.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Six E. W., Klug C. A. Bacteriophage P4: a satellite virus depending on a helper such as prophage P2. Virology. 1973 Feb;51(2):327–344. doi: 10.1016/0042-6822(73)90432-7. [DOI] [PubMed] [Google Scholar]
- Six E. W. The helper dependence of satellite bacteriophage P4: which gene functions of bacteriophage P2 are needed by P4? Virology. 1975 Sep;67(1):249–263. doi: 10.1016/0042-6822(75)90422-5. [DOI] [PubMed] [Google Scholar]
- Sunshine M. G., Thorn M., Gibbs W., Calendar R., Kelly B. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology. 1971 Dec;46(3):691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Martin K. V., Calendar R. On the sequence similarity of the cohesive ends of coliphage P4, P2, and 186 deoxyribonucleic acid. Biochemistry. 1973 May 22;12(11):2119–2123. doi: 10.1021/bi00735a016. [DOI] [PubMed] [Google Scholar]