Abstract
Microbial communities in the gut have been hypothesized to play key roles in the health of the host organism. Exploring the relationship between these populations and disease states has been a focus of the human microbiome project. However, the biological roles of the compounds produced by the gut bacteria are largely unknown. We hypothesize that these compounds act as metabolic exchange factors—mediating inter- and intra- species interactions in the microbiome. This view is supported through this review of known bacterial metabolic exchange factors and evidence for uncharacterized metabolic exchange factors in the gut. The impact of model systems and technological developments in exploring this hypothesis are also discussed. Together, these investigations are revolutionizing our understanding of the gut microbiome—presenting the possibility of identifying new strategies for treating disease in the host.
Exploring the chemistry and biology of microbes within the gut microbiome
Microbes represent the principle reservoir of biomass, genetic, and chemical diversity in the world [1,2]. Microbial communities can exist within host organisms as commensalists, mutualists, and parasites impacting agriculture, the environment, and human health. These complex relationships between host and symbiont are played out in part through metabolic exchange—the transfer of information with diffusible molecules. This exchange may be especially relevant in the gut microbiome, where bacterial cells outnumber the host by a factor of ten or more. A primary goal of the human microbiome project is cataloguing the organisms that constitute specific microbiomes (e.g. the human gut), and characterizing the role they play in diverse human health concerns such as allergy, inflammation, and obesity [3**]. While these approaches are extremely powerful, they raise further questions such as the identity and biological mechanisms of the molecules that control these interactions. This opinion explores recent progress towards elucidating the factors that mediate these interactions within gut microbiomes (See Figure 1).
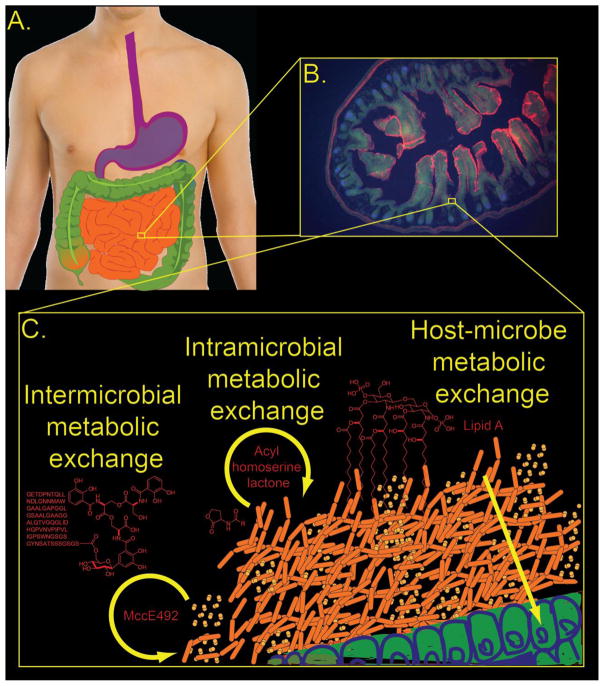
Figure 1. The bacterial chemical repertoire as metabolic exchange factors within the gut microbiome.
A. The human digestive tract is populated by diverse bacteria, forming the microbiome, from esophagus to anus. B. A fluorescence confocal microscopy image of the small intestine. C. A schematic illustrating hypothetical metabolic exchange between host, Escherichia coli and Klebsiella pneumonia. Metabolic exchange can be intraspecies (e.g. quorum-sensing), interspecies, or host-symbiont communication.
We collectively frame the study of the chemistry and biology of these inter- and intra- species interactions as metabolic exchange—the study of the particular molecules that mediate the key biological interactions in a system. Metabolic exchange describes the key molecules in the system which effect multi-cellular behavior in both the microbes and host. We highlight representative examples of known gut bacterial metabolic exchange factors and their biological roles (See Figure 2). Further we illuminate recent research suggesting a biological role for metabolic exchange factors within the microbiome, gut microbiome model systems, and technological advancements towards characterizing metabolic exchange (See Figure 3). These diverse explorations illustrate that the chemical exchange driven by the bacterial chemical repertoire mediates key aspects of biology within the gut microbiome.
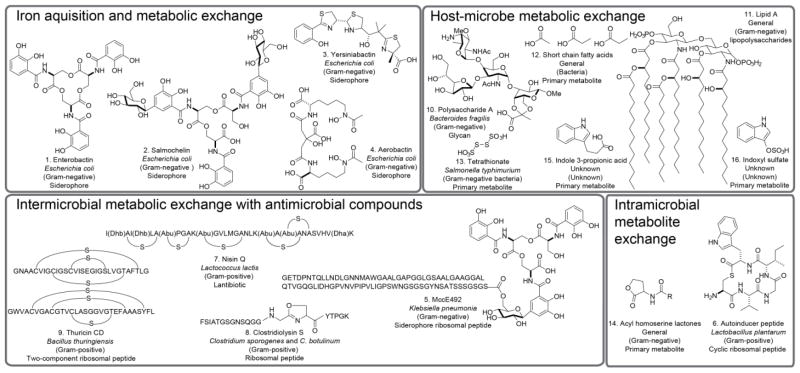
Figure 2. Known gut microbiome metabolic exchange factors.
Compounds are grouped by known biological role including iron acquisition, host-microbe metabolic exchange, intermicrobial metabolic exchange, and intramicrobial metabolic exchange. Compound name, producing bacteria, bacterial group, and biosynthetic origin are noted. -S- is a thioether linkage. Abu, Dha, and Dhb are the non-canonical amino acids aminobutyric acid, dehydroalanine, and dehydrobutyrine.
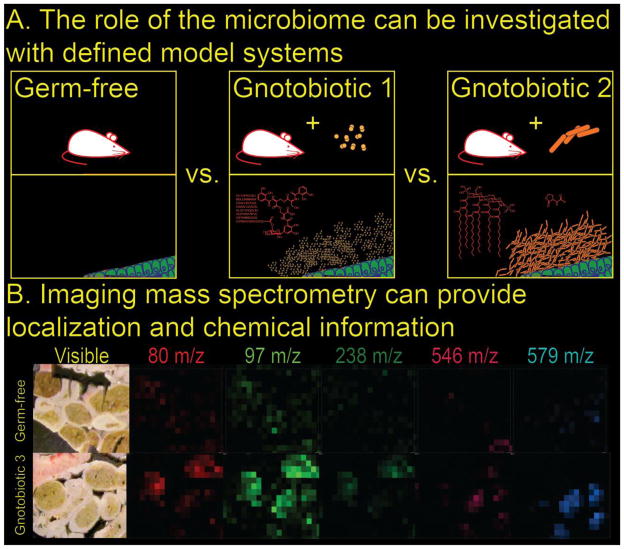
Figure 3. Exploring metabolic exchange factors in the gut microbiome.
A. A hypothetical experiment is comparing germ-free versus gnotobiotic mouse (E. coli and K. pneumonia) gut to determine the role of these strains in host-microbe interactions. B. Imaging mass spectrometry of the mouse gut, comparing germ-free and gnotobiotic animals, allows putative bacterial metabolic exchange factors to be localized and identified chemically without labeling. Each false color image represents a given m/z value (CMR and PCD).
The known bacterial chemical repertoire within the gut microbiome
Bacteria are remarkable synthetic chemists, able to manufacture diverse classes of compounds (e.g. glycans, lipids, nonribosomal peptides, primary metabolites, polyketides, ribosomal peptides, siderophores) stereoselectively in an aqueous environment. This chemical repertoire is a primary tool for bacteria to interact with the environment and the activities of these compounds have been exploited by modern medicine. Clinically relevant examples of bacteria-derived drugs include: the antibiotic erythromycin, the chemotherapeutic ET-743, the cholesterol lowering agent lovastatin, and the immunosuppressant rapamycin—although much less is known about their roles in the natural environment as metabolic exchange factors [4*]. The bacterial chemical repertoire has largely been explored in culturable, free-living organisms from the environment with unculturable microbiome members left under-investigated. Here we review representative gut microbiome derived metabolic exchange factors (See Figure 2).
Siderophores, small molecules involved in microbial iron acquisition, are bacterial metabolic exchange factors present in the gut microbiome. In E. coli differential siderophore production was noted between uropathogenic and commensal gut microbiome strains isolated from patients. Enterobactin (1 in Figure 2) was produced by all strains. Yersiniabactin (2) and salmochelin (3) were predominate among uropathogenic strains whereas aerobactin (4) was predominate among commensal gut strains [5]. The health implications of differential siderophore production between microbiomes are not fully understood.
Microcins are a family of ribosomally-encoded, post-translationally modified antimicrobial peptides, produced by gut associated enterobacteria [6]. The microcin MccE492 (5), derived from a fecal strain of Klebsiella pneumonia, is composed of an 84-amino acid peptide covalently bound to a variant of the siderophore enterobactin—suggesting a Trojan horse mechanism to target bacteria with enterobactin uptake systems [7,8]. Evidence for similar siderophore-peptides has recently been reported in E. coli [9]. The biological role of these metabolic exchange factors in tailoring bacterial populations in the microbiome are not fully characterized.
Numerous ribosomally derived peptides have been identified from the gut microbiome where they act as metabolic exchange factors. Lactobacillus plantarum WCFS1, a cosmopolitan lactic acid bacteria capable of surviving in the gut, has been shown to secrete a cyclic auto-inducing peptide (6)—involved in functions such as gene expression and biofilm adhesion [10]. Nisin Q (7) is an anti-microbial lantibiotic from Lactococcus lactis, a microbe present within the cattle gut which may play a role in tailoring the populations of other bacteria present [11]. Clostridium sporogenes, a common gut bacterium, and C. botulinum, an opportunistic pathogen of the digestive environment of infants, produce a modified peptide clostridiolysin S (8), which has been postulated to play a role in host colonization [12]. Thuricin CD (9) is a two component antimicrobial with an unusual sulfur to α-carbon linkage and was identified from a gut derived strain of Bacillus thuringiensis with activity against C. difficile [13]. The described activities of these metabolic exchange factors are likely only part of the role they play in the gut microbiome.
In addition to the aforementioned activities, the functional roles of some metabolic exchange factors including polysaccharides, lipids, primary metabolites, and inorganic molecules have been well characterized in the gut. The glycan polysaccharide A (10) from Bacteroides fragilis, a human gut symbiont, helps protect against inflammation and infection by opportunistic bacteria in an inflammatory bowel disease model [14*]. The cell membrane component lipopolysaccharides (LPS, 11) can be considered as gut metabolic exchange factors. Mice were treated with antibiotics to alter their gut microbiome populations. LPS production and host absorption increased with antibiotic-induced microbiome population changes. This produced low-level inflammation which is similar to that observed clinically with diabetes and obesity. Gut microbiota can be altered with a high-fat diet often correlated with these disease states [15]. Altered membrane permeability allows LPS to exit the gut and may be influenced by the gut microbiome [16].
Common metabolites, such as short-chain fatty acids (12), are produced by many gut microbiota and act as metabolic exchange factors in host-microbe interactions. Host G-protein coupled receptors recognize these compounds which may play a role resolving inflammation in model organisms for intestinal disease [17*]. The fates of other small metabolic exchange factors in host-microbe interactions have been well characterized. During S. typhimurium invasion inorganic thiosulfates are oxidized to tetrathionate by reactive oxygen species produced as part of the host response. S. typhimurium then metabolizes the resulting tetrathionate (13), promoting its own growth and colonization of the host—illustrating how a pathogenic bacteria can alter the host response [18]. The great diversity of the microbiome chemical repertoire in terms of structure and activity has only begun to be explored—indeed, there is substantial evidence for uncharacterized metabolic exchange factors.
“Omics” and other biological investigations suggest a role for uncharacterized metabolic exchange factors
Large “–omic” studies and more targeted investigations indicate the presence of uncharacterized metabolic exchange factors, including secondary metabolites, in the gut. In two separate metagenomic studies numerous (e.g. 65 predicted protein functions[19*] and 217 genes[20]) secondary metabolite biosynthetic genes were annotated. In a metagenomics and metatranscriptomics investigation of the gut microbiomes, secondary metabolite biosynthetic genes were identified at the DNA and cDNA level in 14 cases—indicating the genes are likely active in the microbiome [21], Expression of at least one secondary metabolite biosynthetic protein has been supported in human gut metaproteomics studies [22]. Bioinformatics investigation of published genomes and metagenomes also allows identification of putative metabolic exchange factors. For example, unusual SAM proteins involved in thuricin CD biosynthesis were linked to 15 novel gene clusters—many from gut isolates or opportunistic pathogens of the gut [23].
Gene clusters encoding putative metabolic exchange factors have also been linked to individual members of gut microbiome. C. butyricum, another common gut resident, produces partially characterized phenolic lipids which may possess antitumor activity [24]. Polyketide biosynthetic genes[25] and lantibiotic biosynthetic genes [26], have also been noted in bifidiobacterium—a predominant species in the gut microbiome. The gut microbiome protozoan Blastocystis sp. was also found to contain a putative Type I PKS cluster [27]. These studies illustrate the enormous, underexplored biosynthetic potential for uncharacterized metabolic exchange factors in the gut—each of which may possess novel structural and biological properties.
In many cases, biological activities of uncharacterized metabolic exchange factors were identified prior to biosynthetic potential and/or chemical structure. A gut microbiome derived small molecule inhibits shigatoxin production by pathogenic E. coli—an example of interspecies chemical exchange regulating virulence [28]. Quorum sensing molecules, such as acyl homoserine lactones (14) [29] are well-known for mediating inter- and intra-species bacterial interactions and are necessary for colonization of the cattle rumen by enterohemorrhagic E. coli [30]. Autoinducer 3, is a bacterial chemical exchange factor responsible for activating virulence genes in the microbiome which is still uncharacterized [31]. Clearly such biology should serve as a strong motivation for both characterizing the chemical structure of these factors and identifying further active compounds.
Model systems for exploring the chemistry of gut microbiomes
Developing model systems for studying the human microbiome is critical due to the limitations inherent in studies with human subjects. Non-mammalian systems including D. melanogaster [32], D. rerio [33], and C. elegans [34] may offer advantages in terms of genetics, generational time, and experimental overhead if findings correlate well to investigated pathology. Other insect model systems may offer benefits such low-complexity gut microbiomes [35]. Artificial gut culture systems also hold promise [36]. Mammalian systems such as mice are the standard model of the human microbiome. Specific biological roles for the gut microbiota are often delineated through comparisons between germ-free, gnotobiotic, and specific-pathogen-free animals (See Figure 3A) [37**]. Engineered auxotrophic bacteria can be used with germ-free models to study factors in colonization and decolonization [38]. The function of toll-like receptors which activate the innate immune response by recognizing metabolic exchange factors, differ dramatically in germ-free versus specific-pathogen-free mice [39*]. Differences in brain development between gnotobiotic and specific-pathogen-free animals have been reported [40]. Co-colonization of mice with B. thetaiotaomicron and M. smithii caused an increase in microbial density achieved when compared with colonization by each species alone [41]. In these investigations the metabolic exchange factors that mediate the observed biology are uncharacterized.
When a metabolomics workflow was applied to compare blood between specific pathogen free mice and germ free animals at least 10% of detected blood metabolites were found to vary by at least 50% [42*]. Indole-3-propionic acid (15) and indoxyl sulfate (16) were identified in the blood of specific-pathogen-free mice but not in the germ-free animals [42]. The indole may be produced by the bacteria from tryptophan, transformed to indole-3-propionic acid, and transported to the blood. Liver detoxification enzymes may then transform it to indoxyl sulfate [42]. It is not yet clear how these metabolic exchange factors may mediate host microbiome interactions.
Technological developments towards understanding the bacterial inventory in the gut microbiome
Even with a tractable model system, exploring the gut is an incredible technological challenge due to diverse species present, heterogeneous composition, and dynamic population across both cross-section and length. In any given investigation, it will likely be necessary to apply multiple, complementary techniques to begin to characterize the system. For example, a census of a particular microbiome in terms of species and genetic composition can be accomplished through metagenomic sequencing of DNA. This total pool of information can be complimented by Fluorescent In Situ Hybridization (FISH) which allows bacteria to be spatially localized. Putative metabolic exchange factors and/or associated biosynthetic genes can be identified through genome mining [43]. Metabolic exchange factor-related genes identified from metagenomics investigations can be rapidly expressed and screened for activity in vitro to verify predicted activity [44].
Chemically characterizing individual metabolic exchange factors can be accomplished with “omics” techniques [45]. Metaproteomics approaches have the capability to identify polypeptide factors and expression of biosynthetic proteins [22,46]. Metabolomics approaches can be implemented using MS[42,47] or NMR[48*,49] platforms to identify numerous metabolic exchange factors simultaneously. MS based metabolomics have been used to explore changes in human gut microbiome metabolites after small bowel/microbiome transplantation [47]. NMR of mixtures[50] is an emerging technique and has been employed with a “systems-biology” outlook to monitor mice microbiomes [51]. Targeted NMR techniques allow structural characterization of unknown molecules at the nanomole scale using technologies such as cryo-microprobes [52]. While the authors believe “omic” tools and information they provide are invaluable, we also want to provide one word of caution. The large datasets generated are interpreted with complex statistical models, often concealed by software packages. Data must be analyzed with appropriate consideration of errors (e.g. false discovery rates) and the implication of such errors and relevancy of the data when dealing with extremely large data sets, followed by orthogonal, downstream validation of key findings [53]. Nonetheless, recent investigations into the chemistry of the gut microbiome have chemically identified numerous putative metabolic exchange factors. Assigning a biological role for these compounds, however, is an on-going challenge.
Powerful emerging technologies allow metabolic exchange factors to be characterized, however, an unmet challenge in terms of assigning biological activity to these compounds is localizing them within the microbiome. We propose that imaging mass spectrometry (IMS) is one tool that has the potential to solve this challenge (See Figure 3B) [54]. MALDI-IMS utilizes a planar sample (e.g. tissue sections) after matrix application, mass spectra are taken at a series of raster points across the sample. A false color can then be assigned to each m/z value of interest, resulting in a massively multi-channel analysis of unlabeled samples. This technique offers the benefits of spatial localization within a 2D sample, as with confocal microscopy or FISH, superimposed with the specific chemical information supplied by many “-omics” techniques. Combined with germ-free and gnotobiotic animals, a powerful platform for exploring the chemistry of metabolic exchange factors in the microbiome can be envisioned (see Figure 3B). An additional benefit is the ability to study the metabolic output of unculturable organisms. MALDI-IMS has already proven to be a powerful technique for monitoring inter-[54] and intra-[55*] microbial metabolic exchange and various tissues [56]. Another variant of IMS, nanoSIMS, has been used to monitor deposited FISH probes in microbiome samples below the optical diffraction limit [57].
Conclusion
We hypothesize that bacteria produce metabolic exchange factors which mediate intra- and inter- species biological interactions in the gut microbiome and that these molecules are involved in the establishment of the gut microbial community (See Figure 1). This hypothesis is supported by a sampling of the known bacterial chemical repertoire, evidence of biosynthetic capabilities, and described biological activities in the microbiome (See Figure 2). Further studies utilizing model systems such as germ-free and gnotobiotic models (See Figure 3A) strongly suggest a role for the bacterial chemical repertoire in mediating observed biology. Developments in analytical technologies, such as imaging mass spectrometry (See Figure 3B), will help us identify novel metabolic exchange factors and the biology behind these factors within the microbiome. These findings will have substantial impact on the understanding and treatment of diverse human health concerns.
Highlights.
Gut microbiome derived metabolic exchange factors have been shown to mediate biology in the gut.
Evidence exists for the biosynthetic capability of the gut microbiome to generate active molecules.
Key described biology may be the result of diffusible molecules produced by the gut microbiota.
Advances in analytical technologies will develop our understanding of microbiome chemistry.
Acknowledgments
C.M.R is supported by the Keck Foundation and the P.C.D. laboratory is supported by US National Institutes of Health (NIH) grants GM094802, GM086283 and AI095125. Portions of Figures 1A–B and 3A incorporated materials from the Wikimedia Project released for use under the Creative Commons License. We would like to thank Frédéric Michel, Kelvinh88, Mariana Ruiz Villarreal, and Seans Potato Business for their contributions to this valuable resource.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Nat Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 3**.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. A description of the human microbiome project project-a key driver for much of the research reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Yim G, Huimi Wang H, Davies Frs J. Antibiotics as signaling molecules. Philos T R Soc B. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. An excellent review of how little is known about the role of metabolic exchange factors in their native context. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson JP, Crowley JR, Pinkner JS, Walker JN, Tsukayama P, Stamm WE, Hooton TM, Hultgren SJ. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5:1–11. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 7.Thomas X, Destoumieux-Garzan D, Peduzzi J, Afonso C, Blond A, Birlirakis N, Goulard C, Dubost L, Thai R, Tabet J-C, et al. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem. 2004;279:28233–28242. doi: 10.1074/jbc.M400228200. [DOI] [PubMed] [Google Scholar]
- 8.Nolan EM, Fischbach MA, Koglin A, Walsh CT. Biosynthetic tailoring of microcin mccE492 post-translational modification affords an antibacterial siderophore-peptide conjugate. J Am Chem Soc. 2007;129:14336–14347. doi: 10.1021/ja074650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassiliadis G, Destoumieux-Garzon D, Lombard C, Rebuffat S, Peduzzi J. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother. 2010;54:288–297. doi: 10.1128/AAC.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturme MHJ, Nakayama J, Molenaar D, Murakami Y, Kunugi R, Fujii T, Vaughan EE, Kleerebezem M, de Vos WM. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J Bacteriol. 2005;187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukao M, Obita T, Yoneyama F, Kohda D, Zendo T, Nakayama J, Sonomoto K. Complete covalent structure of Nisin Q, new natural nisin variant, containing post-translationally modified amino acids. Biosci Biotechnol Biochem. 2008;72:1750–1755. doi: 10.1271/bbb.80066. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez DJ, Lee SW, Hensler ME, Markley AL, Dahesh S, Mitchell DA, Bandeira N, Nizet V, Dixon JE, Dorrestein PC. Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J Biological Chem. 2010;285:28220–28228. doi: 10.1074/jbc.M110.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a post-translationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Nat Acad Sci. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. A well characterized example of the biological role of a microbiome metabolic exchange factor. [DOI] [PubMed] [Google Scholar]
- 15.Cani PD, Bibiloni R, Knauf C, Waget Al, Neyrinck AM, Delzenne NM, Burcelin RM. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 16.Muccioli GG, Naslain D, Backhed F, Reigstad CS, Lambert DM, Delzenne NM, Cani PD. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:1–15. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Di Y, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. An excellent example of host recognition of key metabolic exchange factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. An example of the depth of information available for hypothesis based investigations into the roles of gut metabolic exchange factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Nat Acad Sci. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2008;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 23.Haft DH, Basu MK. Biological systems discovery in silico: Radical SAM protein families and their target peptides for post-translational modification. J Bacteriol. 2011;193:2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaenko G, Moiseeva E, Savelaev O, Molotkovskii Y, Vodovozova E. Antitumor activity of the lipid fraction of the spores of an anaerobic bacterium Clostridium butyricum. Microbiol. 2009;78:580–584. [Google Scholar]
- 25.Ventura M, Turroni F, Lima-Mendez G, Foroni E, Zomer A, Duranti S, Giubellini V, Bottacini F, Horvath P, Barrangou R, et al. Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl Environ Microbiol. 2009;75:6929–6936. doi: 10.1128/AEM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, Viscogliosi E, Brochier-Armanet C, Couloux A, Poulain J, et al. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011;12:R29. doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–790. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decho AW, Frey RL, Ferry JL. Chemical challenges to bacterial AHL signaling in the environment. Chem Rev. 2011;111:86–99. doi: 10.1021/cr100311q. [DOI] [PubMed] [Google Scholar]
- 30.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, Sperandio V. Chemical sensing in mammalian host-bacterial commensal associations. Proc Nat Acad Sci. 2010;107:9831–9836. doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu JH, Ha EM, Lee WJ. Innate immunity and gut-microbe mutualism in Drosophila. Devel Compar Immunol. 2010;34:369–376. doi: 10.1016/j.dci.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Nat Acad Sci USA. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roeder T, Stanisak M, Gelhaus C, Bruchhaus I, Grötzinger J, Leippe M. Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Devel Compar Immunol. 2010;34:203–209. doi: 10.1016/j.dci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Allen HK, Cloud-Hansen KA, Wolinski JM, Guan C, Greene S, Lu S, Boeyink M, Broderick NA, Raffa KF, Handelsman J. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 2009;28:109–117. doi: 10.1089/dna.2008.0812. [DOI] [PubMed] [Google Scholar]
- 36.Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends in Biotech. 2011 doi: 10.1016/j.tibtech.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 37**.Faith JJ, Rey FE, O'Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. An example of the key role that animals with defined microbiota can play in these investigations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-Like Receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. An exciting investigation into the effects of host recognition of microbiome metabolic exchange factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Nat Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-“archaeal”-bacterial mutualism. Proc Nat Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Nat Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. An example of an approach to identify metabolic exchange factors chemically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nuc Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, et al. Metagenomic discovery of biomass-degrading denes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Li F, Nie L. Integrating multiple -omics analysis for microbial biology: application and methodologies. Microbiol. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 46.Johansson MEV, Thomsson KA, Hansson GC. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp Protein. J Proteome Res. 2009;8:3549–3557. doi: 10.1021/pr9002504. [DOI] [PubMed] [Google Scholar]
- 47.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Nat Acad Sci. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Martin F-PJ, Sprenger N, Montoliu I, Rezzi S, Kochhar S, Nicholson JK. Dietary modulation of gut functional ecology studied by fecal metabonomics. J Proteome Res. 2010;9:5284–5295. doi: 10.1021/pr100554m. An example of an investigation using metabolomics and systems biology to explore the microbiome. [DOI] [PubMed] [Google Scholar]
- 49.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina Jrm, Paris A, Want EJ, de Waziers I, Cloarec O, et al. Colonization-induced host-gut microbial metabolic interaction. mBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. P Nat Acad Sci USA. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin F-PJ, Dumas M-E, Wang Y, Legido-Quigley C, Yap IKS, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molinski TF. NMR of natural products at the 'nanomole-scale'. Nat Prod Rep. 2010;27:321–329. doi: 10.1039/b920545b. [DOI] [PubMed] [Google Scholar]
- 53.Pevzner PA, Kim S, Ng J. Comment on “Protein sequences from Mastodon and Tyrannosaurus rex revealed by mass spectrometry”. Science. 2008;321:1040b. doi: 10.1126/science.1155006. [DOI] [PubMed] [Google Scholar]
- 54.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Micro. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Liu W-T, Yang Y-L, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Nat Acad Sci USA. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. An example of a hypothesis guided investigation illustrating the power of imaging mass spectrometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry-looking beyond classical histology. Nat Rev Cancer. 2010;10:639–46. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 57.Behrens S, Losekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol. 2008;74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]