Abstract
Many efforts have been made in the worldwide quest for a prophylactic HIV vaccine to end the AIDS pandemic, but none has yet succeeded. The lessons learned have repeatedly informed us that the traditional or conventional approaches directly using the pathogens or subunits will not be sufficient for an effective HIV/AIDS vaccine. Recent advances in structure-based technology have shown some promise in the quest for a better immunogen in HIV vaccine development. According to the basic binding structural relationship of an antigen and an antibody, structure-based antigen design could bring some hope for the development of an effective vaccine against HIV.
Keywords: HIV-1, Immunogen design, Vaccine development, Structure-based design, Envelope trimer, Antibody epitope, Germline B-cells
Introduction
Human immunodeficiency virus type 1 (HIV-1) was identified as the causative agent of human acquired immunodeficiency syndrome (AIDS) about thirty years ago [1, 2], but unfortunately we still do not have an effective vaccine to stop this epidemic. Although highly active antiretroviral therapy (HAART) is effective in treating patients by controlling their viral load, it cannot completely eliminate the virus from the body. This is because the virus can reside latently in the resting memory CD4+ T cells, where the latent virus is not sensitive to the drugs [3-8]. When the patients stop HAART, the virus generally rebounds to a high level within a few days. Once an individual is infected by HIV-1, it is almost invariably lifelong. Thus, preventing new infection is critical for ending the HIV/AIDS epidemic.
According to UNAIDS, there are about 34 million people who are currently living with HIV-1 and about 2.6 million people are newly infected each year [9]. Therefore, a vaccine is urgently required to curb the worldwide HIV/AIDS pandemic. Many efforts have been made in searching for an effective vaccine against HIV-1 infection in the past 30 years, but thus far, these efforts have not been successful. Several review articles have been published recently summarizing the basic research and clinical trials which have been conducted [10-13]. Notably, many approaches previously used in successful vaccine development for other pathogens have been applied to HIV/AIDS, but none has been successful [10]. These have included the use of killed or attenuated virus particles (14), HIV-1 subunits of envelope glycoproteins gp160 or gp120 [15-17] and envelope peptides [18-22]. So far the only HIV vaccine trial that has shown some efficacy is the Thai RV144 trial using the subunit gp120 recombinant proteins [23]. However, RV144 only showed 31% protection, which will not be effective enough for actual use as a vaccine, but was merely statistically significant when compared with the control group [23]. Nevertheless, RV144 is considered to be the most successful HIV-1 vaccine trial so far [23]. Therefore, traditional or conventional vaccine approaches by directly using the pathogens (killed or attenuated) or the subunits may not be suitable as an HIV/AIDS vaccine. The fact that broad neutralizing antibodies (bNAbs) have been found in only 10-25% HIV-1-infected people from recent large scale screening [24-28] underscores the difficulty of obtaining antibody-based vaccine-mediated protection using conventional approaches.
Why do the Traditional or Conventional Approaches not Work for Developing an HIV/AIDS Vaccine?
During the past three decades, extensive studies on HIV-1 structure, pathogenicity, and evolution have concluded that the virus has evolved into an immunorecessive form, or so-called “decoy” form, to evade the human immune system surveillance [29-31]. This decoy form of the virus basically has two characteristics: one is that the virus cannot induce a robust response when presented to the human immune system; another is that the viral conserved immunogenic epitopes are not exposed. Since the neutralizing epitopes are hidden or not fully exposed on the surface of the virus, only a minimal level of neutralizing antibodies with no breadth of activity was induced in spite of having a strong antibody response to the HIV-1 envelope glycoproteins in HIV/AIDS patients. This could be one of the reasons why traditional or conventional vaccine immunogens directly using the viruses or their subunits have been ineffective.
HIV-1 is an enveloped retrovirus. The envelope spikes on the viral membrane surface consist of the glycoproteins gp120 and gp41, which are the most prominent viral proteins that directly interact with the human immune system. Studies have shown that Env spikes are sparsely scattered on the viral surface [32-34]. HIV-1 requires two receptors for its entry, i.e. the primary receptor CD4 and a co-receptor CCR5 or CXCR4. The binding of gp120 to the primary receptor CD4 induces a conformational change in gp120 to expose the binding sites in gp120 for coreceptor binding [29, 35, 36]. Receptor binding is a sequential process and the coreceptor binding epitope is not exposed before CD4 binding. Thermodynamic analysis showed that the entropy and enthalpy changes of gp120 upon the binding of CD4 are remarkably large [37]. It was suggested that the gp120 molecule undergoes a major conformational change upon CD4 binding. This large conformational change of gp120 may cause some antibodies to lose their neutralizing activities or reduce their neutralizing capacities, by decreasing their binding affinity or preventing them from binding HIV during the viral entry process.
Extensive studies on the envelope spikes have been carried out to analyze their composition, structure, and immunogenicity. Recent structural studies on the trimeric envelope spikes have opened new avenues for Env trimer-based vaccine design. It has become much clearer that the major loops (V1, V2 and V3) of gp120 are involved in trimer stabilization (Fig. 1) [34, 38, 39]. For a long time, it was thought that the gp120 core structure would form a trimeric body from which the loops might extend. This assumption led to a number of studies in which the loops were eliminated in order to stabilize the structure of the monomeric gp120 or the Env trimer for vaccine design. In fact, the major variable loops (V1, V2, and V3) are packed into the central apex of the trimeric spike body, so changes in the loops can adjust the trimer stability to adapt to the host immune systems. The adjustment could occur through substituting amino acids in the loop region and also through changing the loop length [40-43]. It is apparent that this trimeric packing model also has great advantages for viral evolution and generation of viral diversity. The evolution and adaptation of this virus must have occurred during the passaging of simian immunodeficiency viruses (SIV) to chimpanzees (SIVcpz) and then finally to humans (HIV) [44-50]. It is likely that the virus has gone through an adjustment process to allow it to evade the human immune system.
Fig. (1).
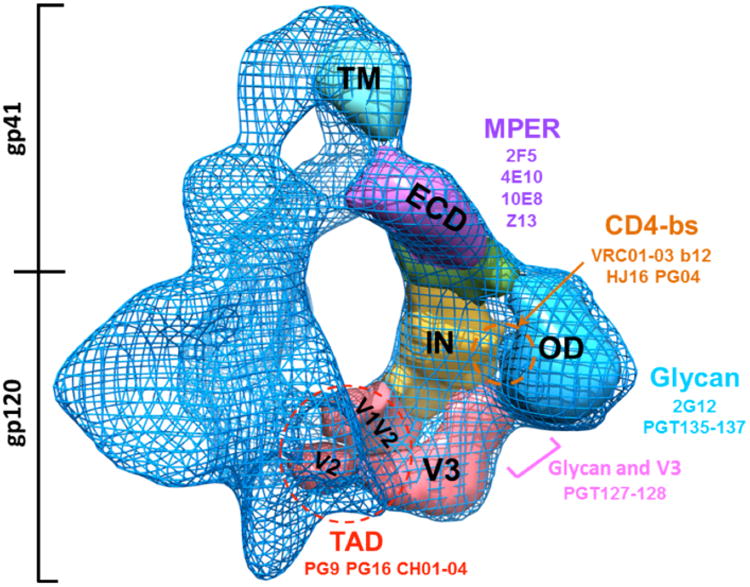
The HIV-1 Env trimer structure and the binding sites of broadly neutralizing antibodies (bNAbs) [13, 34, 140]. The picture is adapted from EMD-5418 at a resolution about 11Å in the Electron Microscopy Data Bank. The domain structures are shown in different colors. TM, transmembrane domain; ECD, Ectodomain of gp41; IN, inner domain of gp120; OD, outer domain of gp120, V1, V1 loop and etc.; TAD, trimer association domain. MPER, membrane-proximal external region; CD4-bs, CD4 binding sites; Glycan, glycosylation sites.
Another characteristic of the HIV-1 immune evasion is the glycosylation of the envelope glycoprotein gp120. The gp120 is heavily glycosylated with about 25-30 glycan molecules, which are mostly of high-mannose forms. Thus, most of the envelope surface is covered by glycans that form a protective shield. This is a major barrier for HIV-1 vaccine development, because the heavy glycan shielding can protect the virus from neutralizing antibodies. Recently, however, some antibodies such as PGT128 were found to be able to penetrate the glycan shield [51-55].
Because traditional or conventional methods used the whole or part of the pathogen as immunogens for vaccination, they cannot overcome the various decoy forms of HIV-1. In order to solve these problems, other approaches must be sought to overcome immune evasion by the viruses. One such approach is the use of modern structural biology to design novel vaccines against HIV-1 infection. The structure-based immunogen design is an innovative and revolutionary method that could help with HIV/AIDS vaccine development.
Structure-Based Immunogen Design for Developing an HIV/AIDS Vaccine
Structural modeling and simulation methods have been broadly used for protein design to study the structure and function of proteins. These could be used to structurally modify and design the immunogens to increase their immunogenicity and to induce specific neutralizing antibodies. The structure-based design approaches may 1) expose the conserved sites, 2) stabilize a conformation of the immunogen, 3) mimic an antibody epitope. The structure-based design of a vaccine could be described as one form of reverse vaccinology [56, 57]. This approach does not involve the use of killed or attenuated viral particles or the subunits for the vaccine, but uses the structural information of the pathogens for the development of an effective vaccine. Structure-based design of HIV-1 vaccines can be classified into four major categories and they are described below, and also summarized in Fig. (2).
Fig. (2).
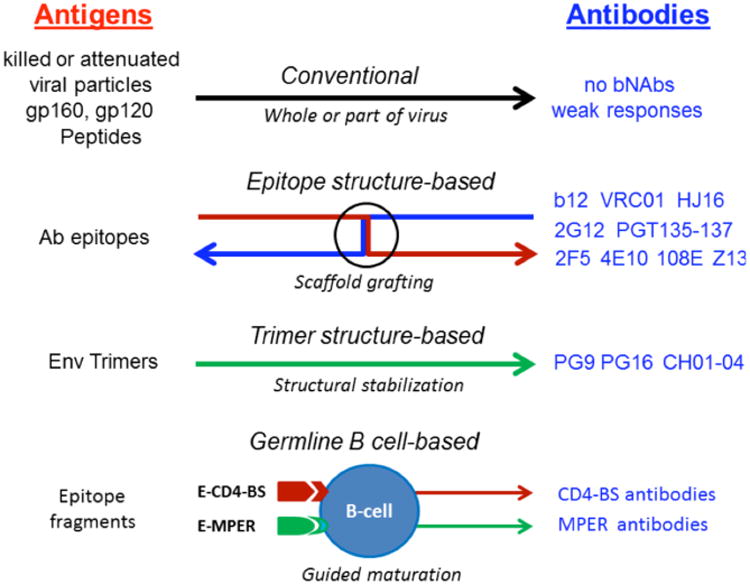
Current vaccine approaches for making preventive vaccines against HIV-1/AIDS [11, 13, 136]. The conventional vaccines directly use the whole pathogens or part of the pathogens as antigens for immunization, such as killed or attenuated viral particles, subunits (gp160 and gp120) of viral surface proteins, or peptides of the surface proteins. The epitope structure-based approach is based on reverse vaccinology [57] for developing vaccines, and it must utilize the known neutralizing epitopes for conducting antigen design. The trimer structure-based approach uses the envelope trimer structure for antigen design [58]. Some trimer-specific antibodies such as PG9 and PG16 could also provide useful information for designing the trimer-based antigens [26, 141]. The germline B cell-based approach is more relevant to the native immune response in inducing specific neutralizing antibodies against HIV-1 infection [132]. bNAbs, broadly neutralizing antibodies; Ab antibody, Env, envelope, E-CD4-bs, epitope of CD4-binding site; E-MPER, epitope of membrane proximal external region.
Envelope Trimer Structure-Based Immunogen Design
It has been known for a long time that trimer-based immunogens generally produce better neutralizing antibodies than subunit-based immunogens [58]. However, it is much more challenging to generate trimer-based immunogens. The envelope trimer consists of the surface envelope glycoprotein gp120 and the transmembrane glycoprotein gp41, but there are no inter-molecule covalent bonds between gp120 and gp41, therefore, it is more difficult to generate a soluble Env trimer for vaccine design. Several methods have been tested for generating Env trimers, such as using a GCN4 trimeric motif [59-61], a T4 bacteriophage fibritin trimer motif (62), using the disulfide bonds to build the soluble cleaved gp140 trimers (63-65), or the V2-loop deleted modification gp140 trimers [66, 67]. The soluble gp140s were able to induce stronger immune responses in animal models than gp120 monomers, and the immune sera were confirmed to have higher neutralizing activities [64, 68-72] although this is not thought to be adequate for vaccine-induced HIV-1 protection.
With the advent of recent structural findings on the trimer architecture of the HIV-1 envelope trimeric spikes using the cryo-electron microscopy (cryo-EM) approaches [32, 38, 73-75], there is now more detailed information on how to stabilize an env-trimer for a vaccine immunogen. With the envelope trimer structure at 11Å resolution based on the cryo-EM, the locations of most domains and the loops can be distinguished (Fig. 1) [38], and a recently published report at 6Å resolution allows most of the secondary structure elements of the HIV-1 Env trimer to be located [34]. This has now provided more information on the structural basis in the stabilization of the molecule, such as the trimer association domain (TAD), which consists of the V1V2 loops and a central mini-barrel, making direct trimeric contacts from the three subunits of gp120. The TAD presumably includes the twin-cysteine motif of the V2-loop of simian immunodeficiency virus, which may form the disulfide bonds in the subunit (intra-) or between the subunits (inter-)(76). Stabilizing the trimer structure appears to be a good strategy for developing a trimer-based vaccine. There are two main methods for the design of a trimer-based vaccine. One is to stabilize the native trimer conformation without other modifications [62, 64, 68, 77, 78]. The rationale of this design is based on the findings that most of the trimer spikes are nonfunctional and unstable [38, 79]. Because the native trimer structure has an open and flexible architecture (Fig. 1), the gp120 subunits can easily fall apart from the gp41 subunits. It is estimated that there are very few particles with stable or functional envelope trimeric spikes [32, 33, 79]. Therefore, generating more stable trimers should result in better immunogens for HIV-1 vaccines. Another strategy for trimer-based immunogen design is to expose the hidden conserved epitope sites [29-31] and this may include removing some glycans to increase the exposure of the conserved epitopes for antibody targeting and to increase its antigenicity [80-83].
The virus-like particle (VLP) design is mainly for stabilizing the envelope trimer structures on a native particle resembling a virion [84, 85]. VLPs are generated essentially by mimicking the native viral particle structure. VLPs are able to be self-assembled, but are non-replicating and non-pathogenic due to the absence of viral genetic elements. VLP-based vaccines are much safer to use than attenuated HIV-1 forms, and can also be engineered easily by the structure-based approaches. Recently, HIV-1 VLPs have been successfully produced from Drosophila cells [86], and even in plants [87]. The VLP-based immunogen design offers a new approach for developing a safe and effective HIV-1 vaccine [85, 88, 89].
Antibody Epitope Structure-Based Immunogen Design
The epitope structure-based design utilizes the antibody epitope for immunogen design. It is based on the notion that the epitope-based immunogen should elicit antibodies that can bind the same epitope on the viral spike. Recent advances in identifying neutralizing antibodies have provided renewed hope to develop an effective vaccine against HIV/AIDS. High through-put technology for large scale screening of neutralizing antibodies, in combination with the B-cell evolutionary genomics, have led to the isolation of a number of broad neutralizing antibodies (bNAbs) [25], such as VRC01-03 (90), PG04 (91), HJ16 (92), PG9 and PG16 (26), CH01-04 (93), and PGT121-137 [94]. For some of these bNAbs, the structures have also been solved with the binding epitopes of the envelope glycoproteins of HIV-1. The structural information from the co-crystalized structures of the binding complex of antibodies and antigens are especially valuable for epitope structure-based design [11, 95, 96].
The membrane-proximal external region (MPER) of gp41 is a well-known vulnerable region as target of neutralizing antibodies [97], such as 2F5 [98], 4E10 [99], 10E8 [100] and Z13e1 [99]. The MPER neutralizing antibodies 2F5 and 4E10 appear to be autoreactive [101, 102], but it is not clear whether the autoreactivity or polyreactivity is detrimental for vaccine purposes. It is interesting that another MPER monoclonal antibody 10E8 was not found to bind phospholipids and is not autoreactive [100]. Because the MPER-binding antibodies have broad neutralizing capacity, and their binding epitopes are linear, they represent a very attractive solution for structure-based immunogen design. Since the structures of some of the MPER antibodies and their epitopes have been solved [103-105], structure-based design for better immunogens can now be easily conducted. One of the first studies on epitope design was to constrain and stabilize the 2F5 epitope-based fragment as an immunogen [103, 106]. More recently, the protein scaffold grafting approach has been introduced to this design, and the 2F5 linear epitope was grafted onto a protein scaffold [107, 108]. This engineered 2F5 epitopescaffold was used for immunization and elicited antibodies that could recognize the 2F5 epitope and had significantly higher binding affinity. Unfortunately, the antibodies did not result in neutralization of the virus [95, 107, 109]. Other similar MPER neutralizing antibodies 4E10 [110] and Z13e1 [111], were also tested for epitope-based grafting design; the outcomes were similar as 2F5, and the antibodies generated did not possess any strong neutralizing activity. One explanation perhaps is that these antibodies bind to their epitopes in a different conformation from 2F5 and resulted in less potent neutralization activities.
Another target for epitope structure-based vaccine design is the primary receptor CD4-binding site (CD4-bs). A number of bNAbs target this site, such as the well-known monoclonal antibody b12 [112], and the recently discovered monoclonal antibodies VRC01-03 [90], PG04 [91] and HJ16 [92]. From the actual structural binding information of the CD4-bs antibody b12 [112] and VRC01 [113], it was found that their epitopes are mostly located on the outer domain of gp120. However the CD4-binding site is not a linear epitope, and it is discontinuous- or conformation-dependent. Therefore, the CD4-bs antibodies generated are also against discontinuous epitopes which have presented a challenge for structure-based immunogen design. The protein grafting approach was also tested with the CD4-bs antibody b12 epitopes. The designed b12 discontinuous epitope-scaffold from the backbone grafting was able to bind b12 antibody with high affinity. It was also shown that the designed epitope scaffold is structurally similar to the b12 discontinuous epitope [114]. The structure-based design using germ-line genes to target the CD4-binding site has also been tested, and such an approach is described in the following section on B cell-based immunogen design.
Glycan Structure-Based Immunogen Design
The glycans on the surface of HIV-1 virions play an important role in shielding and protecting the virus from the immune response because they function as a steric barrier for the binding of neutralizing antibodies. However, the glycans of HIV-1 have been demonstrated to be immunogenic and immunogenicity [40, 118], and can also be a target for vaccine design. For example, it has been shown that bNAb 2G12 [115] can directly bind to the glycans on the outer domain of gp120. Furthermore, a number of other glycan-associated bNAbs have recently been identified, such as PGT121-137 [94]. These antibodies not only directly interact with HIV-1 glycans on the gp120, but can also penetrate the glycan shield to reach the short beta-strand segment of the gp120 V3-loop [51]. The antibodies of PGT141-145 [116] and PG9 [116] and PG16 (26) have also been found to share a specificity for an N-linked glycan at residue 160 in V1V2 of gp120 [116]. The glycans consist of mostly N-linked oligo-mannoses, and the Manalphal →2Man motif is the primary neutralization determinant (epitope) for antibody 2G12 [117]. The PG9 and PG16 antibodies have an unusually wide and long CDR H3 loop, and can potentially bind to both the glycan and protein backbone of the virus to mediate neutralization. Similarly, the epitopes of PGT121-137 consist of both the glycan and protein backbone. Thus, it is well established that the glycans can also be targeted by neutralizing antibodies. The loss of glycans in HIV-1 gp120 vaccine candidates may result in a loss of their antigenicity [119-125].
The glycan structure and its distribution pattern on the virion are critical for glycan structure-based immunogen design. Therefore, in order to target HIV-1 glycans by vaccine-induced antibodies, one must first determine the types of glycans which are specific on the virion surface and whether they are distinguishable from other glycans present on other proteins. A glyco-conjugate antigen based on the antibody 2G12 recognition motif was used to immunize rabbits, but the elicited antibodies failed to bind the glycans of gp120 [126]. However, using polysaccharide mimicry of the epitope of 2G12 from yeast, it was shown that they were able to elicit 2G12-like antibodies [117]. In addition, high-mannose oligosaccharides from yeast have also been used for immunization; they elicited antibodies that could recognize the glycan structures. These antibodies generated unfortunately did not have neutralizing activities [127, 128]. An engineered triple mutant yeast strain elicited antibodies could bind to gp120 glycans but were not able to bind the cell surface-expressed trimeric envelope [129]. However, when the envelope was expressed in the presence of mannosidase inhibitor, the elicited antibodies gained the ability to bind the trimeric Env and neutralize the HIV-1 viruses. This suggests that the high-mannose glycans may be required for eliciting neutralizing antibodies against HIV-1 [129].
Germline B-Cell Receptor-Based Immunogen Design
Structure-based vaccine design can be also applied to the germline B-cell receptor-based immunogen design. This is based on the experience gained from in vitro B-cell maturation studies to identify bNAbs. The basis of this approach is to stimulate or activate the germline B-cells using specific immunogens to induce them to enter a specific maturation pathway in order for them to produce more specific antibodies against HIV-1. Germline B-cell receptor-based immunogen design appears to be a more specific approach for inducing or educating the B-cells to make specific antibodies. Recent reports on germline B-cell based immunogen design have focused on modifying the gp120 immunogen for eliciting VRC01-like [130-133] and 2F5-like bNAbs [109, 134]. In addition, the complex-type N-glycan binding antibody PGT121 has also been investigated by inducing differentiation of the germline B-precursor cells [135].
Challenges of Structure-Based Vaccine Design
The use of structural biology has shed some light on making better immunogens for HIV-1 vaccine development (13, 96, 136). The structure-based approach is mainly dependent on the binding structural relationship between an antigen and an antibody. However, one must be aware that it is the natural immune response that is important in generating antibodies specific against incoming antigens or pathogens, and the process of generating an immune response is complex and may not be simply mimicked by biophysical structural relationships [137]. The same antigen or even the same small epitope can induce many different types of antibodies. As mentioned above, with the 2F5 epitope design, there are some 2F5-like antibodies elicited that cannot neutralize HIV-1. This suggests that antibody synthesis or the generation of broadly neutralizing antibodies is a complicated in vivo process that may not be replicated readily by a simple biochemical synthesis process in vitro. That is why one has to carefully evaluate and validate any immunogen based-designed HIV vaccine in the context of an immune system within a host [138].
There are a number of challenges to designing HIV-1 immunogens, and they can be summarized as follows. First, it is important to note that the bNAbs are rare in HIV-1 infected patients, and they often have unusual features. For example, these features include a long CDR H3 loop (b12) or a long and wide CDR H3 loop (PG9, PG16); a sulfated tyrosine on the CDR H3 loop (412d); swapped heavy chains (2G12); or auto-reactive and poly-reactive activities (2F5, 4E10). Most of these structurally uncommon antibodies may not be easily elicited by a normal vaccine or via the normal immune response pathway [139]. Second, for the trimer structure-based design, there is still a need to obtain a higher-resolution structure at an atomic level of a native Env trimer. In addition, how can we stabilize the native or mutant trimers? Since the interaction between gp120 and gp41 is non-covalent, it will always be a challenge in generating stable and cleaved soluble trimers. Third, for the epitope-structure-based design, stabilizing the epitope structure alone or presenting it on a carrier scaffold may be the key to success. However, it will be a challenge to induce the immune system to recognize primarily the neutralizing epitopes and in parallel reduce the induction of other antibodies that are usually non-neutralizing. Fourth, in glycan structure-based design, the binding characteristics of glycan-associated bNAbs are specific for a conformation on the HIV-1 virion. It has been suggested that glycan-targeting antibodies may still need to interact with viral protein epitopes. Some of these antibodies can penetrate the glycan layer and reach the viral protein backbone epitopes. Fifth, in the germline B-cell targeting approach, the engineered antigens should induce or activate B-cells to mature in order to make specific bNAbs against HIV-1. Using such an approach in vitro may not reflect the process in vivo, since B cell maturation in the context of the entire human immune system will be significantly more complex.
In conclusion, HIV-1 has evolved into an immune decoy form which includes glycan shielding, recessing of conserved epitopes and trimer flexibility. These unique Env trimer properties have rendered these proteins poorly immunogenic. This is one of the reasons why natural HIV-1 infection cannot induce a robust immune response by the host to control viral infection. It can also explain why conventional vaccines using the pathogen or subunits as immunogens have not shown any success thus far. The structure-based immunogen design has achieved some successes in eliciting structurally similar antibodies that are able to bind the epitopes, but the elicited antibodies still do not have strong neutralizing activities. The problem we are now facing is how one can use a highly modified antigen to induce broadly neutralizing antibodies against the native and unmodified incoming viral targets. This is currently one of the most challenging questions in the field of structure-based HIV-1 vaccine design.
Acknowledgments
This author would like to thank Dr. Joseph Sodroski at Harvard University and Dr. Charles Wood at the University of Nebraska-Lincoln for reviewing the manuscript; and Dr. Youdong Mao at Harvard Medical School for providing the picture of the HIV-1 trimer structure. This work was supported by a grant (#51783) from the Bill and Melinda Gates Foundation.
Footnotes
Conflict of Interest: The authors confirm that this article content has no conflict of interest.
Patient Consent: Declared none.
Human/Animal Rights: Declared none.
References
- 1.Barre-Sinoussi F, Chermann JC, Rey F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–71. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Salahuddin SZ, Popovic M, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224(4648):500–3. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Dinoso JB, Kim SY, Siliciano RF, Blankson JN. A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy. Clin Infect Dis. 2008;47(1):102–4. doi: 10.1086/588791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinoso JB, Kim SY, Wiegand AM, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2009;106(23):9403–8. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends Mol Med. 2004;10(11):525–31. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Shen A, Zink MC, Mankowski JL, et al. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol. 2003;77(8):4938–49. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Chung C, Hu BS, et al. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest. 2000;106(7):839–45. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UNAIDS. Data and analysis. 2013 http://wwwunaidsorg/en/
- 10.Lifson JD, Haigwood NL. Lessons in nonhuman primate models for AIDS vaccine research: from minefields to milestones. Cold Spring Harb Perspect Med. 2012;2(6):a007310. doi: 10.1101/cshperspect.a007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong PD, Mascola JR, Nabel GJ. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Cold Spring Harb Perspect Med. 2012;1(1):a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong L, Sattentau QJ. Antigenicity and Immunogenicity in HIV-1 Antibody-Based Vaccine Design. J AIDS Clin Res. 2012;S8:3. doi: 10.4172/2155-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton DR, Ahmed R, Barouch DH et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12(4):396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lifson JD, Rossio JL, Piatak M, et al. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res Hum Retroviruses. 2004;20(7):772–87. doi: 10.1089/0889222041524661. [DOI] [PubMed] [Google Scholar]
- 15.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372(9653):1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191(5):654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 17.Pitisuttithum P, Gilbert P, Gurwith M, et al. Randomized, doubleblind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 18.Ghiara JB, Ferguson DC, Satterthwait AC, Dyson HJ, Wilson IA. Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site. J Mol Biol. 1997;266(1):31–9. doi: 10.1006/jmbi.1996.0768. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi E, Joyce JG, Miller MD et al. Vaccination with peptide mimetics of the gp41 prehairpin fusion intermediate yields neutralizing antisera against HIV-1 isolates. Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10655–60. doi: 10.1073/pnas.1004261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cease KB. Peptide component vaccine engineering: targeting the AIDS virus. Int Rev Immunol. 1990;7(1):85–107. doi: 10.3109/08830189009061767. [DOI] [PubMed] [Google Scholar]
- 21.Francis MJ. Peptide vaccines for viral diseases. Sci Prog. 1990;74(293 Pt 1):115–30. [PubMed] [Google Scholar]
- 22.Holley LH, Goudsmit J, Karplus M. Prediction of optimal peptide mixtures to induce broadly neutralizing antibodies to human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1991;88(15):6800–4. doi: 10.1073/pnas.88.15.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 24.Doria-Rose NA, Klein RM, Manion MM, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83(1):188–99. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15(8):866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 26.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simek MD, Rida W, Priddy FH, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83(14):7337–48. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sather DN, Armann J, Ching LK, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83(2):757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyatt R, Kwong PD, Desjardins E, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 31.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 32.Zhu P, Liu J, Bess J, Jr et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–52. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 33.Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6(5):e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y, Wang L, Gu C, et al. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA. 2013;110(30):12438–43. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses. 2002;18(16):1207–17. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 36.Xiang SH, Kwong PD, Gupta R, et al. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76(19):9888–99. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myszka DG, Sweet RW, Hensley P, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97(16):9026–31. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Y, Wang L, Gu C, et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol. 2012;19(9):893–9. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang SH, Finzi A, Pacheco B, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84(7):3147–61. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolk T, Schreiber M. N-Glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med Microbiol Immunol. 2006;195(3):165–72. doi: 10.1007/s00430-006-0016-z. [DOI] [PubMed] [Google Scholar]
- 41.Chaillon A, Braibant M, Moreau T, et al. The V1V2 domain and an N-linked glycosylation site in the V3 loop of the HIV-1 envelope glycoprotein modulate neutralization sensitivity to the human broadly neutralizing antibody 2G12. J Virol. 2011;85(7):3642–8. doi: 10.1128/JVI.02424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Gils MJ, Bunnik EM, Boeser-Nunnink BD, et al. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J Virol. 2011;85(14):6986–95. doi: 10.1128/JVI.00268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curlin ME, Zioni R, Hawes SE, et al. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 2011;6(12):e1001228. doi: 10.1371/journal.ppat.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp PM, Hahn BH. Origins of HIV and the AIDS Pandemic. Cold Spring Harb Perspect Med. 2011;1(1):a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci. 2010;365(1552):2487–94. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–6. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Heuverswyn F, Li Y, Neel C, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444(7116):164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 48.Wain LV, Bailes E, Bibollet-Ruche F, et al. Adaptation of HIV-1 to its human host. Mol Biol Evol. 2007;24(8):1853–60. doi: 10.1093/molbev/msm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F, Bailes E, Robertson DL, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397(6718):436–41. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 50.Starcich BR, Hahn BH, Shaw GM, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45(5):637–48. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 51.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20(7):804–13. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pancera M, Yang Y, Louder MK, et al. N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One. 2013;8(2):e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J, O'Dell S, Ofek G, et al. Somatic Populations of PGT135-137 HIV-1-Neutralizing Antibodies Identified by 454 Pyrosequencing and Bioinformatics. Front Microbiol. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pejchal R, Walker LM, Stanfield RL, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107(25):11483–8. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19(17-19):2688–91. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 57.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–50. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 58.Forsell MN, Schief WR, Wyatt RT. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr Opin HIV AIDS. 2009;4(5):380–7. doi: 10.1097/COH.0b013e32832edc19. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74(12):5716–25. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Florin L, Farzan M, et al. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000;74(10):4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75(3):1165–71. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Binley JM, Sanders RW, Clas B, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beddows S, Schulke N, Kirschner M, et al. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–27. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beddows S, Kirschner M, Campbell-Gardener L, et al. Construction and characterization of soluble, cleaved, and stabilized trimeric Env proteins based on HIV type 1 Env subtype A. AIDS Res Hum Retroviruses. 2006;22(6):569–79. doi: 10.1089/aid.2006.22.569. [DOI] [PubMed] [Google Scholar]
- 66.Bontjer I, Land A, Eggink D, et al. Optimization of human immunodeficiency virus type 1 envelope glycoproteins with V1/V2 deleted, using virus evolution. J Virol. 2009;83(1):368–83. doi: 10.1128/JVI.01404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bontjer I, Melchers M, Eggink D, et al. Stabilized HIV-1 envelope glycoprotein trimers lacking the V1V2 domain, obtained by virus evolution. J Biol Chem. 2010;285(47):36456–70. doi: 10.1074/jbc.M110.156588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grundner C, Li Y, Louder M, et al. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331(1):33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 69.Kang YK, Andjelic S, Binley JM, et al. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine. 2009;27(37):5120–32. doi: 10.1016/j.vaccine.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selvarajah S, Puffer BA, Lee FH, et al. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res Hum Retroviruses. 2008;24(2):301–14. doi: 10.1089/aid.2007.0158. [DOI] [PubMed] [Google Scholar]
- 71.Sundling C, Forsell MN, O'Dell S, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207(9):2003–17. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sundling C, O'Dell S, Douagi I, et al. Immunization with wild-type or CD4-binding-defective HIV-1 Env trimers reduces viremia equivalently following heterologous challenge with simian-human immunodeficiency virus. J Virol. 2010;84(18):9086–95. doi: 10.1128/JVI.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White TA, Bartesaghi A, Borgnia MJ, et al. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol. 2011;85(23):12114–23. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White TA, Bartesaghi A, Borgnia MJ, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6(12):e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohl C, Bowder D, Thompson J, et al. A twin-cysteine motif in the V2 region of gp120 is associated with SIV envelope trimer stabilization. PLos One. 2013;8(7):1–6. doi: 10.1371/journal.pone.0069406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harris A, Borgnia MJ, Shi D, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108(28):11440–5. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pancera M, Lebowitz J, Schon A, et al. Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis. J Virol. 2005;79(15):9954–69. doi: 10.1128/JVI.79.15.9954-9969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol. 2006;80(14):6725–37. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Depetris RS, Julien JP, Khayat R, et al. Partial enzymatic deglycosylation preserves the structure of cleaved recombinant HIV-1 envelope glycoprotein trimers. J Biol Chem. 2012;287(29):24239–54. doi: 10.1074/jbc.M112.371898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu Q, Mahmood N, Shattock RJ. High-mannose-specific deglycosylation of HIV-1 gp120 induced by resistance to cyanovirin-N and the impact on antibody neutralization. Virology. 2007;368(1):145–54. doi: 10.1016/j.virol.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koch M, Pancera M, Kwong PD, et al. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 83.Zhang C, Wan Y, Shi J, et al. Deglycosylation of HIV-1 AE Gp140 enhances the capacity to elicit neutralizing antibodies against the heterologous HIV-1 clade. AIDS Res Hum Retroviruses. 2010;26(5):569–75. doi: 10.1089/aid.2009.0228. [DOI] [PubMed] [Google Scholar]
- 84.Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines. 2010;9(10):1149–76. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 85.Young KR, McBurney SP, Karkhanis LU, Ross TM. Virus-like particles: designing an effective AIDS vaccine. Methods. 2006;40(1):98–117. doi: 10.1016/j.ymeth.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 86.Yang L, Song Y, Li X, et al. HIV-1 virus-like particles produced by stably transfected Drosophila S2 cells: a desirable vaccine component. J Virol. 2012;86(14):7662–76. doi: 10.1128/JVI.07164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scotti N, Rybicki EP. Virus-like particles produced in plants as potential vaccines. Expert Rev Vaccines. 2013;12(2):211–24. doi: 10.1586/erv.12.147. [DOI] [PubMed] [Google Scholar]
- 88.McBurney SP, Young KR, Ross TM. Membrane embedded HIV-1 envelope on the surface of a virus-like particle elicits broader immune responses than soluble envelopes. Virology. 2007;358(2):334–46. doi: 10.1016/j.virol.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 89.Pastori C, Tudor D, Diomede L, et al. Virus like particle based strategy to elicit HIV-protective antibodies to the alpha-helic regions of gp41. Virology. 2012;431(1-2):1–11. doi: 10.1016/j.virol.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Falkowska E, Ramos A, Feng Y, et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J Virol. 2012;86(8):4394–403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corti D, Langedijk JP, Hinz A, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5(1):e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bonsignori M, Hwang KK, Chen X, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walker LM, Sok D, Nishimura Y, et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci USA. 2011;108(50):20125–9. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Azoitei ML, Ban YE, Julien JP, et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J Mol Biol. 2012;415(1):175–92. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burton DR. Scaffolding to build a rational vaccine design strategy. Proc Natl Acad Sci USA. 2010;107(42):17859–60. doi: 10.1073/pnas.1012923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev. 2008;72(1):54–84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muster T, Steindl F, Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–7. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zwick MB, Labrijn AF, Wang M, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haynes BF, Fleming J, St Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308(5730):1906–8. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 102.Yang G, Holl TM, Liu Y, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med. 2013;210(2):241–56. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ofek G, Tang M, Sambor A, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–37. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pejchal R, Gach JS, Brunel FM, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009;83(17):8451–62. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cardoso RM, Zwick MB, Stanfield RL, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22(2):163–73. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 106.Tian Y, Ramesh CV, Ma X, et al. Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints. J Pept Res. 2002;59(6):264–76. doi: 10.1034/j.1399-3011.2002.02988.x. [DOI] [PubMed] [Google Scholar]
- 107.Ofek G, Guenaga FJ, Schief WR, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107(42):17880–7. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ofek G, McKee K, Yang Y, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84(6):2955–62. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guenaga J, Dosenovic P, Ofek G, et al. Heterologous epitopescaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One. 2011;6(1):e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Correia BE, Ban YE, Friend DJ, et al. Computational protein design using flexible backbone remodeling and resurfacing: case studies in structure-based antigen design. J Mol Biol. 2010;405(1):284–97. doi: 10.1016/j.jmb.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 111.Stanfield RL, Julien JP, Pejchal R, Gach JS, Zwick MB, Wilson IA. Structure-based design of a protein immunogen that displays an HIV-1 gp41 neutralizing epitope. J Mol Biol. 2011;414(3):460–76. doi: 10.1016/j.jmb.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou T, Xu L, Dey B, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou T, Georgiev I, Wu X, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Azoitei ML, Correia BE, Ban YE, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334(6054):373–6. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 115.Trkola A, Purtscher M, Muster T, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McLellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dunlop DC, Bonomelli C, Mansab F, et al. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20(7):812–23. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lavine CL, Lao S, Montefiori DC, Haynes BF, Sodroski JG, Yang X. High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection. J Virol. 2011;86(4):2153–64. doi: 10.1128/JVI.06201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Polzer S, Dittmar MT, Schmitz H, et al. Loss of N-linked glycans in the V3-loop region of gp120 is correlated to an enhanced infectivity of HIV-1. Glycobiology. 2001;11(1):11–9. doi: 10.1093/glycob/11.1.11. [DOI] [PubMed] [Google Scholar]
- 120.Quinones-Kochs MI, Buonocore L, Rose JK. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J Virol. 2002;76(9):4199–211. doi: 10.1128/JVI.76.9.4199-4211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Polzer S, Dittmar MT, Schmitz H, Schreiber M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology. 2002;304(1):70–80. doi: 10.1006/viro.2002.1760. [DOI] [PubMed] [Google Scholar]
- 122.Losman B, Bolmstedt A, Schonning K, et al. Protection of neutralization epitopes in the V3 loop of oligomeric human immunodeficiency virus type 1 glycoprotein 120 by N-linked oligosaccharides in the V1 region. AIDS Res Hum Retroviruses. 2001;17(11):1067–76. doi: 10.1089/088922201300343753. [DOI] [PubMed] [Google Scholar]
- 123.Gray ES, Moore PL, Pantophlet RA, Morris L. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J Virol. 2007;81(19):10769–76. doi: 10.1128/JVI.01106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hemming A, Gram GJ, Bolmstedt A, et al. Conserved N-linked oligosaccharides of the C-terminal portion of human immunodeficiency virus type 1 gp120 and viral susceptibility to neutralizing antibodies. Arch Virol. 1996;141(11):2139–51. doi: 10.1007/BF01718221. [DOI] [PubMed] [Google Scholar]
- 125.Gram GJ, Hemming A, Bolmstedt A, et al. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch Virol. 1994;139(3-4):253–61. doi: 10.1007/BF01310789. [DOI] [PubMed] [Google Scholar]
- 126.Astronomo RD, Lee HK, Scanlan CN, et al. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J Virol. 2008;82(13):6359–68. doi: 10.1128/JVI.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luallen RJ, Fu H, Agrawal-Gamse C, et al. A yeast glycoprotein shows high-affinity binding to the broadly neutralizing human immunodeficiency virus antibody 2G12 and inhibits gp120 interactions with 2G12 and DC-SIGN. J Virol. 2009;83(10):4861–70. doi: 10.1128/JVI.02537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luallen RJ, Lin J, Fu H, et al. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J Virol. 2008;82(13):6447–57. doi: 10.1128/JVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Agrawal-Gamse C, Luallen RJ, Liu B, et al. Yeast-elicited cross-reactive antibodies to HIV Env glycans efficiently neutralize virions expressing exclusively high-mannose N-linked glycans. J Virol. 2010;85(1):470–80. doi: 10.1128/JVI.01349-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hoot S, McGuire AT, Cohen KW, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9(1):e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McGuire AT, Hoot S, Dreyer AM, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210(4):655–63. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jardine J, Julien JP, Menis S, et al. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science. 2013;340(6133):711–6. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109(30):E2083–90. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alam SM, Liao HX, Dennison SM, et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol. 2011;85(22):11725–31. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mouquet H, Scharf L, Euler Z, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109(47):E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schief WR, Ban YE, Stamatatos L. Challenges for structure-based HIV vaccine design. Curr Opin HIV AIDS. 2009;4(5):431–40. doi: 10.1097/COH.0b013e32832e6184. [DOI] [PubMed] [Google Scholar]
- 137.Van Regenmortel MH. Basic research in HIV vaccinology is hampered by reductionist thinking. Front Immunol. 2012;3:194. doi: 10.3389/fimmu.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Van Regenmortel MH. Requirements for empirical immunogenicity trials, rather than structure-based design, for developing an effective HIV vaccine. Arch Virol. 2012;157(1):1–20. doi: 10.1007/s00705-011-1145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pejchal R, Wilson IA. Structure-based vaccine design in HIV: blind men and the elephant? Curr Pharm Des. 2010;16(33):3744–53. doi: 10.2174/138161210794079173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37(3):412–25. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pancera M, McLellan JS, Wu X, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84(16):8098–110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


