Abstract
Background
PRDM proteins are evolutionary conserved Zn-Finger transcription factors that share a characteristic protein domain organization. Previous studies have shown that prdm1a is required for the specification and differentiation of neural crest cells in the zebrafish.
Results
Here we examine other members of this family, specifically prdm3, 5, and 16, in the differentiation of the zebrafish craniofacial skeleton. prdm3 and prdm16 are strongly expressed in the pharyngeal arches, while prdm5 is expressed specifically in the area of the forming neurocranium. Knockdown of prdm3 and prdm16 results in a reduction in the neural crest markers dlx2a and barx1 and defects in both the viscerocranium and the neurocranium. The knockdown of prdm3 and prdm16 in combination is additive in the neurocranium, but not in the viscerocranium. Injection of sub-optimal doses of prdm1a with prdm3 or prdm16 Morpholinos together leads to more severe phenotypes in the viscerocranium and neurocranium. prdm5 mutants have defects in the neurocranium and prdm1a and prdm5 double mutants also show more severe phenotypes.
Conclusions
Overall, our data reveal that prdm3, 5, and 16 are involved in the zebrafish craniofacial development and that prdm1a may interact with prdm3, 5, and 16 in the formation of the craniofacial skeleton in zebrafish.
Keywords: prdm, craniofacial development, neural crest cell, zebrafish, Morpholino, dlx2a, barx1
INTRODUCTION
Cranial neural crest cells are a multipotent population of cells that originate from the dorsal neural tube and migrate to form multiple derivatives including the craniofacial skeleton, neurons and glia of the peripheral nervous system and melanocytes (LaBonne and Bronner-Fraser, 1998; Le Douarin, 1982). The population of skeletogenic neural crest cells migrates anteriorly around the eye and ventrally into the pharyngeal arches to give rise to cartilage and bone of the face (Graham, 2003; Le Douarin, 1982; Raible et al., 1992; Schilling and Kimmel, 1994; Trainor and Krumlauf, 2001). The cranial neural crest cells that migrate ventrolaterally from the posterior midbrain and the hindbrain move as three distinct streams into the pharyngeal arches. Once there, the cranial neural crest cells interact with other tissue layers in the arches, including the ectoderm, endoderm, and mesoderm, to form the zebrafish viscero- and neurocranium. The viscerocranium consists of cartilages that contribute to the jaw and gills, including Meckel’s cartilage, derived from the first pharyngeal arch, ceratohyal and hyosymplectic from the second arch, and posterior ceratobranchial cartilages from more posterior arches three through seven (Couly et al., 1993; Fraser, 1990; Horstadius, 1946; Le Douarin, 1982; Le Douarin et al., 1993; Le Lie`vre, 1978; Lumsden et al., 1991; Osumi-Yamashita et al., 1994; Schilling and Kimmel, 1994). The more anterior migrating stream of NCCs from the forebrain and anterior midbrain contributes to the anterior portion of the more dorsal neurocranium, a cartilage that supports the brain and auditory capsule (Kimmel and Eberhart, 2008; Wada et al., 2005). The most anterior part of the neurocranium, the ethmoid plate, also represents the zebrafish palate and is connected to the more posterior neurocranium by the trabecular cartilages. Recent fate mapping studies suggest that the medial ethmoid plate is derived from anteriorly migrating neural crest cells while the trabeculae arise from more posterior migrating cells from the maxillary portion of arch 1 (Swartz et al., 2011; Wada et al., 2005). While several genes have been identified as being involved in patterning the craniofacial skeleton, there is still a deficiency regarding our knowledge of the transcriptional control of this process.
PRDM proteins control several critical aspects of development including differentiation, cell growth, and apoptosis and are thus considered to be important regulators of cell fate (Turner et al., 1994; Deng and Huang et al., 2004; Fog et al., 2012; He et al., 1998; Hohenauer and Moore, 2012; Yan et al., 2007). PRDMs are part of a large evolutionary conserved Zn-Finger transcription factor family that has variable numbers of Zn-Finger repeats that mediate DNA binding. In addition, they contain an N-terminal PR domain that is similar to SET domain proteins found in a class of histone methyltransferases that mediate recruitment of histone-modifying enzymes. Because of this, PRDM function is mostly associated with transcriptional repression. There are a total of five subfamilies, which include 17 different genes that have been identified in humans and 15 in the Fugu rubripes fish. The phylogenetic relationships between PRDM genes have been proposed and suggest that while there are similarities and differences between all the family members, subfamilies can be identified based on similar gene sequence and protein structure and number of Zn-finger repeats: PRDM 2, 3, 5, and 16 belong to the same subfamily and PRDM1 shares a relationship with PRDM 4, 10, 15 (Fumasoni et al., 2007).
The founding member of the PRDM family is Prdm1, also called Blimp1 in mammals. Previous studies from multiple labs have shown that Prdm1 is necessary for B-cell development and is required for the differentiation of B-cells into plasma cells (Shaffer et al., 2002; Shapiro-Shelef et al., 2003; Turner et al., 1994), and is further implicated in craniofacial, limb, and germ cell development in mouse (Ohinata et al., 2005; Vincent et al., 2005). In the zebrafish, prdm1a mutant embryos have reduced specification of neural crest cells and Rohon-Beard sensory neurons (Artinger et al., 1999; Roy and Ng, 2004, Hernandez-Lagunas et al., 2005), an absence of slow twitch muscle cells (Beermann et al., 2010), and defects in prechordal plate and fin development (Mercader et al., 2006; Wilm and Solnica-Krezel, 2005). Specifically in craniofacial development, prdm1a is expressed in a large domain covering the posterior pharyngeal arches, which give rise to the posterior viscerocranium and pharyngeal teeth (Birkholz et al., 2009; Sun et al., 2008). In both mouse and zebrafish, prdm1 is required for the formation of the posterior craniofacial structures and glands, such as the thymus, likely functioning by regulating cell proliferation (Birkholz et al., 2009; Robertson et al., 2007; Wilm and Solnica-Krezel, 2005).
While a great deal is known about PRDM1, much less is known about the function of other PRDM proteins during development, although more interest is emerging (Fog et al., 2012; Hohenauer and Moore, 2012; Swartz et al., 2012). PRDM3, also known as EVI-1, is associated with a role in human cancers including acute myelogenous leukemia (Buonamici et al., 2003; Morishita et al., 1988; Russell et al., 1994; Senyuk et al., 2011) and some solid tumors (Brooks et al., 1996; Sunde et al., 2006). Xenopus prdm3 and Mouse Prdm3/Evi1 are expressed in the head folds and first and second pharyngeal arches at late tailbud stage and from E8.5–10.5, respectively (Hoyt et al., 1997; Mead et al., 2005). Mice with a targeted disruption of Prdm3/Evi1 die at E10.5 from cardiovascular and/or placental defects. These embryos also exhibit changes in neural crest–derived structures, including loss of dorsal root and cranial ganglia and hypoplasia of the pharyngeal arches (Hoyt et al., 1997). PRDM5 is inactivated in different types of tumors, suggesting that it functions as a tumor suppressor (Cheng et al., 2010; Duan et al., 2007; Watanabe et al., 2007). In zebrafish, prdm5 is expressed ubiquitously from early cleavage stages and knockdown leads to a cyclopic phenotype with defects in the formation of the head (Meani et al., 2009). In humans, PRDM16 is associated with various disease states including myeloid leukemias and adult T-cell leukemia (Mochizuki et al., 2000; Nishikata et al., 2003), and controls the cell fate choice between brown fat and skeletal muscle cells (Kajimura et al., 2008, 2009; Seale et al., 2007, 2008). In mouse craniofacial development, Prdm16 is expressed in neural crest– derived tissues, similar to Prdm3, as well as in the pharyngeal arches, and the area of the forming palate and Meckel’s cartilage by E11.5. An ENU-derived Prdm16 mutant in mouse has developmental abnormalities that include a hypoplastic mandible and cleft palate (Bjork et al., 2006, 2010a,b; Horn et al., 2011).
Based on the craniofacial skeleton phenotype in prdm1a mutants and the expression of the above-mentioned prdms, we sought to determine the function of other prdm family members during zebrafish craniofacial skeletal development, focusing on prdm3, 5, and 16. Using expression analysis and Morpholino knockdown, we determined the effect of knockdown of individual and combinations of Prdm factors. Our data suggests that the Prdm proteins are necessary for neurocranium patterning, and to a lesser extent, overall patterning of the viscerocranium.
RESULTS
Expression of Prdm Family Members in Zebrafish Craniofacial Skeleton Development
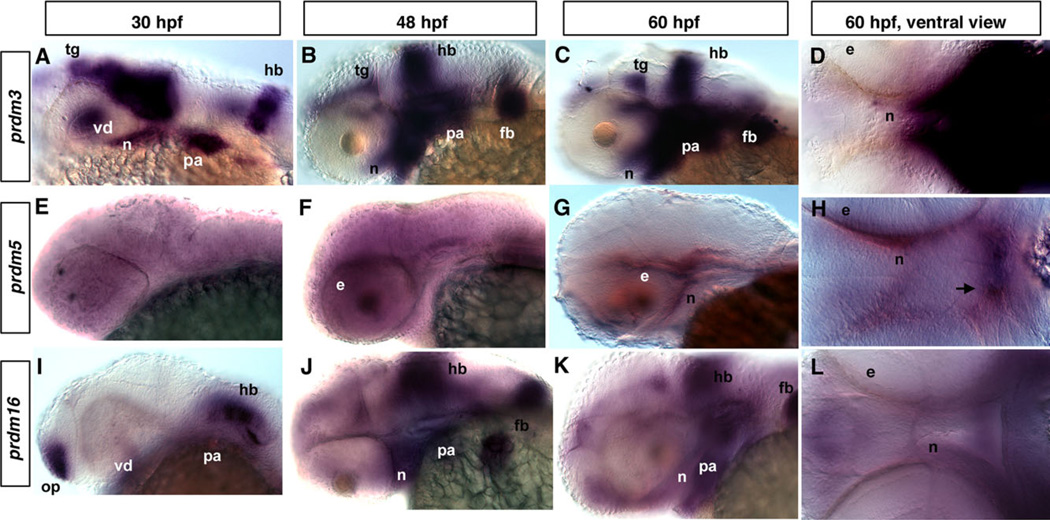
Previous studies have shown that PRDM3, 5, and 16 are associated with different kinds of cancers, but how they function in the development of the zebrafish craniofacial skeleton is not yet known. We performed a series of whole-mount RNA in situ hybridization (ISH) experiments for prdm3, 5, 10, 11, and 16 in zebrafish embryos from 30 hr post-fertilization (hpf) to 60 hpf to determine whether they are expressed at the correct time and place to be important for craniofacial development. A previous study by Sun et al. (2008) reported the expression patterns of many SET domain– containing genes, including prdm1a, b, c, prdm4, prdm15, prdm3, and prdm16 during zebrafish embryogenesis. Consistent with this previous report, we confirmed and extended these findings to examine the expression in the craniofacial region between 30–60 hpf. prdm10 and prdm11 were ubiquitously expressed at low levels during these stages and were not pursued further (data not shown). prdm3 is detected in the tegmentum, ventral diencephalon, hindbrain, pharyngeal arches, and pectoral fin buds in the craniofacial region at 30 hpf (Fig. 1A, and data not shown). At 48 and 60 hpf, prdm3 expression gradually increases and is highly expressed in the pharyngeal arches, as well as in neurocranium and pectoral fin buds (Fig. 1B – D). In a ventral view, regions of the forming neurocranium express prdm3 (Fig. 1D).
Fig. 1.
Expression of different prdms in developing zebrafish embryos. In situ hybridization (ISH) using prdm3 5, and 16 at 30-hpf (A, E, I), 48-hpf (B, F, J), and 60-hpf stages showing lateral (C, G, K) and (D, H, L) ventral views. Note the partially overlapping expression of prdm3 and prdm16. A: Lateral view of whole mount embryo showing the prdm3 expression in the tegmentum, ventral diencephalon, neurocranium, hindbrain, pharyngeal arches at 30 hpf. At 48 hpf (B) and 60 hpf (C, D), prdm3 gradually increases and is highly expressed in pharyngeal arches, and is also expressed in pectoral fin buds, neurocranium, hindbrain, and tegmentum. E–H: prdm5 is not specifically expressed at 30 hpf, but is expressed at a low level in the area behind the eye and neurocranium at 48 and 60 hpf. The arrow in H points to the forming stomodeum in which prdm5 is expressed. I,J: From 30 to 48 hpf, prdm16 expression gradually increases in pharyngeal arches, as well as in neurocranium, pectoral fin buds, hindbrain, and olfactory placode. K,L: At 60 hpf, prdm16 expression decreases in the pharyngeal arches and hindbrain, but is still expressed at a significant level. Anterior is to the left in all panels. e, eye; fb, fin buds; hb, hindbrain; n, neurocranium; op, olfactory placode; pa, pharyngeal arch; tg, tegmentum; vd, ventral diencephalon.
prdm5 is expressed at a low level ubiquitously prior to 48 hpf, but becomes localized and is specifically expressed in the area of the forming neurocranium at 48 and 60 hpf (Fig. 1E–H). From a ventral view, prdm5 is expressed in the area of the forming neurocranium as well as a region anterior to the stomodeum (Fig. 1H, arrow).
prdm16 is expressed in the pharyngeal arches, hindbrain, and olfactory placode at 30 hpf (Fig. 1I). By 48 hpf, prdm16 is expressed at high levels in the pharyngeal arches, neurocranium, hindbrain, and fin bud (Fig. 1J). Similar expression is observed at 60 hpf, albeit at slightly reduced levels, in the area of the pharyngeal arches and neurocranium (Fig. 1K, L). From the expression results presented, it is difficult to determine if there is specific overlap of expression between prdm3, prdm5, and prdm16 as further analysis would be required. However, these results suggest that they are expressed within the same domains within the pharyngeal arches in craniofacial tissues that give rise to the viscerocranium and the neurocranium. Thus, these results indicate that the prdm gene family is expressed and likely functions in both viscerocranium and neurocranium development.
Reduction of prdm3 and prdm16 Results in Craniofacial Skeletal Defects
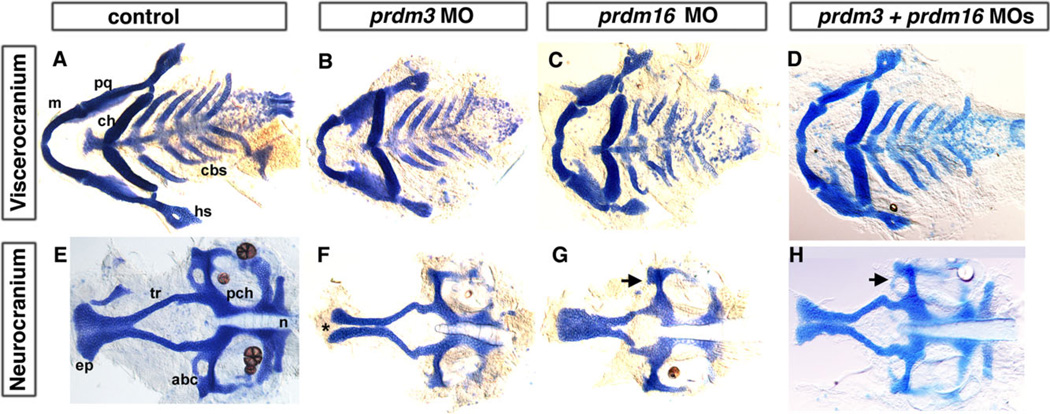
To determine if prdm3 and prdm16 are required for development of the pharyngeal arches and craniofacial skeleton, we knocked down prdm3 and prdm16 in zebrafish embryos by injecting antisense Morpholino oligonucleotides (MO) targeting the splicing site or translation start site of the message. For each, we obtained two differentially targeting MOs, prdm3 e3i3 and i2e3 MO, and for prdm16 ATG and e2i2 MO were injected into tp53M214K −/− or wildtype embryos at the 1- to 4-cell stage. The specificity of the MOs was tested by several methods described in detail in the Experimental Procedures section (see Supp. Figs. S1, S2, which are available online). Both prdm3 and prdm16 MO-injected fish have a normal overall morphology, with slightly smaller heads and a small percentage of embryos had edema (~20%). The embryos with edema were not used for further analysis. We then stained 5-dpf embryos with alcian blue to determine if there are defects in craniofacial skeletal development in prdm3 and prdm16 morphants (for a review of craniofacial skeletal elements see Schilling and Kimmel, 1997). Control injections with p53 MO alone to prevent non-specific cell death or control MO at the 18 ng dose did not result in craniofacial defects (Supp. Fig. S2). Following injection of 6–12 ng of either of the prdm3 splice MOs or 6–8 ng of prdm16 ATG or splice MO, embryos exhibit craniofacial cartilage defects as compared with controls at 5 dpf (n=33/45 for prdm3 e3i3; n=21/24 for prdm3 i2e3; and n=11/16 for prdm16 ATG and n=20/25 prdm16 e2i2; Fig. 2).
Fig. 2.
Reduction in prdm3 and prdm16 results in craniofacial defects. Five-dpf uninjected (A, E), prdm3 (B, F), prdm16 (C, G), and double morphant prdm3 and prdm16 (D, H) larvae stained with alcian blue to detect cartilage following dissection of the viscerocranium and neurocranium. As compared to control (A, E), the flat-mounted 5-dpf prdm3 i2e3 (B, F) and prdm16 e3i3 (C, G) morphant larvae have smaller palatoquadrate, including the pterygoid process of the palatoquadrate, and hyosymplectic of the viscerocranium, shortening of the Meckel’s cartilage (m), widening of the angle between ceratohyals (ch) in the viscerocranium, smaller ethmoid plate (ep), shortened trabeculae of the neurocranium, thinning and smaller of the neurocranium. In prdm3 morphant larvae (20–30%), there is a gap in the anterior edge of the ethmoid plate forming a “cleft” as shown in F. D, H: The combination of sub-optimal doses of prdm3 with prdm16 Morpholino. As compared to control (A, E), combination of 3 ng prdm3 i2e3 with 5 ng prdm16 e3i3 Morpholino (D,H) resulted in a slightly more severe phenotype in the neurocranium (much smaller and thinner neurocranium, smaller ethmoid plate, shortened trabeculae, missing anterior basicapsilar commissure, arrow). However, there is only a small gap in the ethmoid plate. Anterior is to the left. abc, anterior basicapsular commissure; cbs, ceratobranchials; ch, ceratohyal; ep, ethmoid plate; hs, hyosymplectic; m, Meckel’s cartilage; n, notochord; pch, parachordal; pq, palatoquadrate; tr, trabeculae; * indicates cleft in the ethmoid plate.
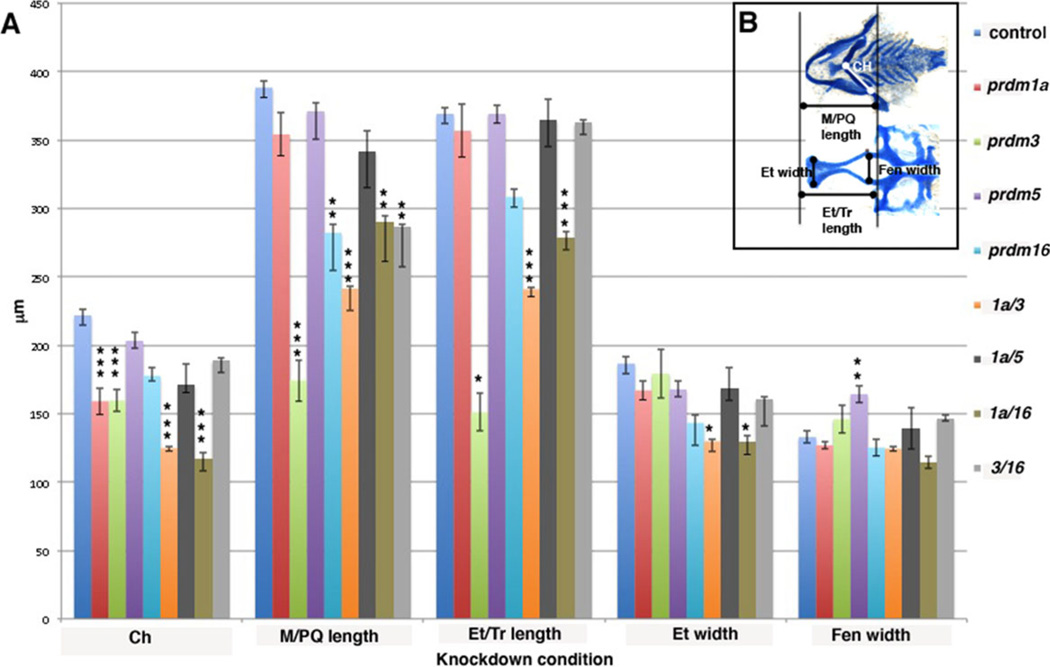
The prdm3 and prdm16 morphant larva have a similar viscerocranium phenotype where defects are observed in the first arch cartilages, including a shortened Meckel’s (m) and palatoquadrate (pq) as compared to unin-jected, tp53M214K −/− or control MO injected larva (Fig. 2A–C, Supp. Fig. S2). Second arch–derived structures also have mild defects; the hyosymplectic (hs) forms normally while the ceratohyals (ch) are often compressed and at a greater angle to one another compared to controls. Thus, the viscerocranium is patterned normally, but has significantly hypoplastic elements. Quantification of the length and width of each craniofacial element was performed and suggests that both prdm3 and prdm16 morphants display a significant reduction in the length of Meckel’s/palatoqua-drate (see Fig. 6, light green bars for prdm3, P < 0.001 and light blue bars for prdm16, P < 0.001). prdm3 also displays a significant shortening of the ceratohyals (see Fig. 6, light green bars, P < 0.001). However, in contrast to prdm1a mutants or morphants, which lack arch 3-7-derived ceratobranchial cartilages (cbs; see Fig. 4B,F), prdm3 and prdm16 morphants exhibit only slight hypoplasia of these elements (Fig. 2A–D, see also Fig. 6).
Fig. 6.
Quantification of cartilage elements in morphant embryos. A: Embryos were dissected and measured in microns (µm; y-axis) for 5 different elements: Ceratohyal (Ch), Meckel’s/palatoquadrate length (M/PQ), Ethmoid plate/trabeculae length (Et/Tr), Ethmoid plate width (Et), and Hypophyseal Fenestra width (Fen) on the x-axis. Each condition is color coded: Control, royal blue, tp53M214K−/− control; red, prdm1a MO; light green, prdm3 MO; purple, prdm5 mutant; aqua, prdm16 MO; orange, prdm1a and prdm3 MOs; dark grey, prdm1a and 5 double mutants; dark green, prdm1a and prdm16 MOs; light grey, prdm3 and prdm16 MOs. Error bars are standard error, and significance was measured by Tukey Kramer one-way Anova posthoc test as compared to control. For Ch, ***P < 0.001. For M/PQ length, **P < 0.01 and ***P < 0.001. For Et/Tr length, *P < 0.01 and ***P < 0.001. For Et width, *P < 0.03, For Fen, **P < 0.002. B: Line diagram on cartilage-stained uninjected embryos illustrating the elements measured: Viscerocranium, top, measuring the CH (white line) and M/PQ length (black line). Neurocranium, bottom, measuring Et/Tr length, Et width, Fen width (black lines, respectively).
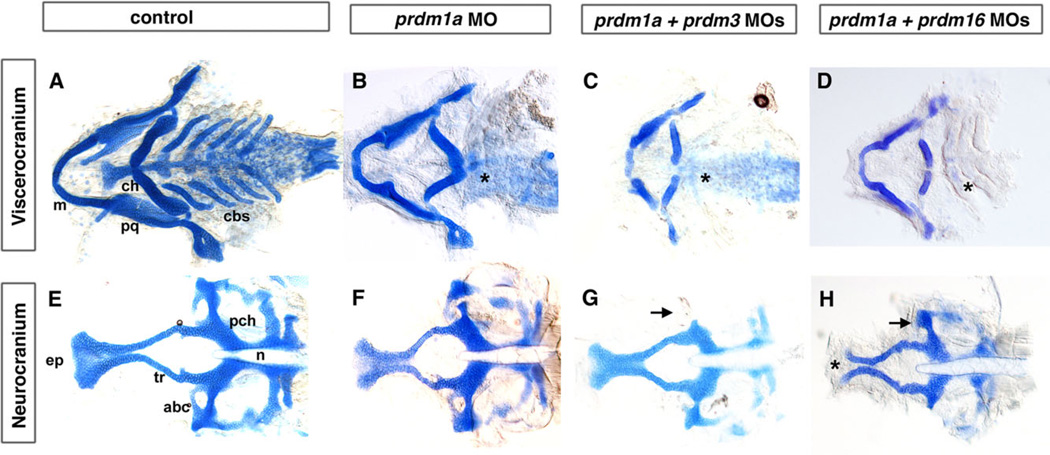
Fig. 4.
Combination of sub-optimal doses of prdm1a with prdm3 or prdm16 Morpholino results in severe craniofacial defects. Alcian blue staining on 5-dpf larvae to detect cartilage formation. Alcian blue–stained uninjected (A, E), prdm1a (B, F), prdm3, prdm1a–prdm3 double (C, G), and prdm1a-prdm16 double (D, H) morphant larvae. As compared to control (A), prdm1a knockdown alone has an inverted ceratohyal (ch) and missing ceratobranchial 2–5 (*) in the viscerocranium (B). In addition, prdm1a morphants have a smaller and narrower neurocranium and slightly shortened trabeculae (F). The combination of sub-optimal doses of prdm1a with prdm3 e3i3 Morpholino resulted in a more severe phenotype in viscerocranium (C, shortening of the Meckel’s cartilage and loss of ceratobranchial 2–5). In the neurocranium, the double morphants have an increasingly smaller and thinner neurocranium, a smaller ethmoid plate, and shorter trabeculae. The combination of suboptimal doses of prdm1a and prdm16 ATG Morpholino led to a more severe phenotype in the neurocranium (H, much smaller and thinner neurocranium with clefting observed in 20–30% of embryos, smaller ethmoid plate, shorter trabeculae) and viscerocranium (D, shortening of the Meckel’s cartilage and absence of ceratobranchial 2–5). Anterior is to the left. abc, anterior basicapsular commissure; cbs, ceratobranchials; ch, ceratohyal; ep, ethmoid plate; m, Meckel’s cartilage; n, notochord; pch, parachordal; tr, trabeculae; * in B–D illustrates missing cbs, in H indicates area of clefting.
Following prdm3 or prdm16 knockdown, there were specific patterning defects in the neurocranium. In prdm3 morphants, the ethmoid plate was hypoplastic and thinner at the midline, often resulting in a “cleft,” in combination with shortened trabeculae (Fig. 2F, asterisk; see Fig. 6, light green bars, P < 0.01; Eberhart et al., 2008; Swartz et al., 2012). The prdm16 morphant neurocranium has a narrowing in the overall shape of the ethmoid plate coupled with a slightly shortened ethmoid plate/trabecular length. While consistent, these differences were more varied and thus not found to be significant in our quantification (Fig. 2G; see Fig 6, light blue). These data suggest that prdm3 and prdm16 are required for proper neurocranium formation, with a milder effect on the viscerocranium. The effects shown in Figure 2 were for prdm3 i2e3 and prdm16 e2i2. Similar defects of the pharyngeal skeleton can be observed with injection of prdm3 e3i3 splice-blocking MO and prdm16 ATG MO (data not shown). These data suggest that both prdm3 and prdm16 play a role in zebrafish craniofacial development.
Combinatorial Effects of prdm3 and prdm16 in Zebrafish Skeletal Development
Next, to determine whether the roles of different prdm transcription factors during craniofacial cartilage development are redundant, we injected combinations of different prdm Morpholinos. In the combination Morpholino injections, we specifically injected sub-threshold doses of each Morpholino that alone did not result in phenotypes (data not shown), such that the overall Morpholino dose is not too high to cause toxicity. This was done to determine the specific effects of the double knockdown of both genes.
Because prdm3 and prdm16 are in the same subfamily and the knockdown of each results in similar phenotypes, as described above, we determined the combinatorial effect of the knockdown of both. Interestingly, when injected in combination, 6 ng prdm3 i2e3 and 3 ng prdm16 e2i2 splicing Morpholino resulted in an additive effect on the neurocranium, with a smaller and narrower ethmoid plate (n=20/25; Fig. 2D,H). However, we did not observe a significant effect on viscerocranium, since the combined injection results in a similar phenotype as the individual injections (Fig. 2D, H). There was a significant difference in the length of the Meckel’s/palatoquadrate as compared to the control, similar to prdm16 MO alone (see Fig. 6, light grey bars, P < 0.01). Together, these data suggest that prdm3 and prdm16 do appear to act redundantly in the viscerocranium but may have different functions in neurocranium development of the zebrafish.
Gene Expression Domains Within Cranial Neural Crest Cells Are Reduced in prdm3 and prdm16 Morphants
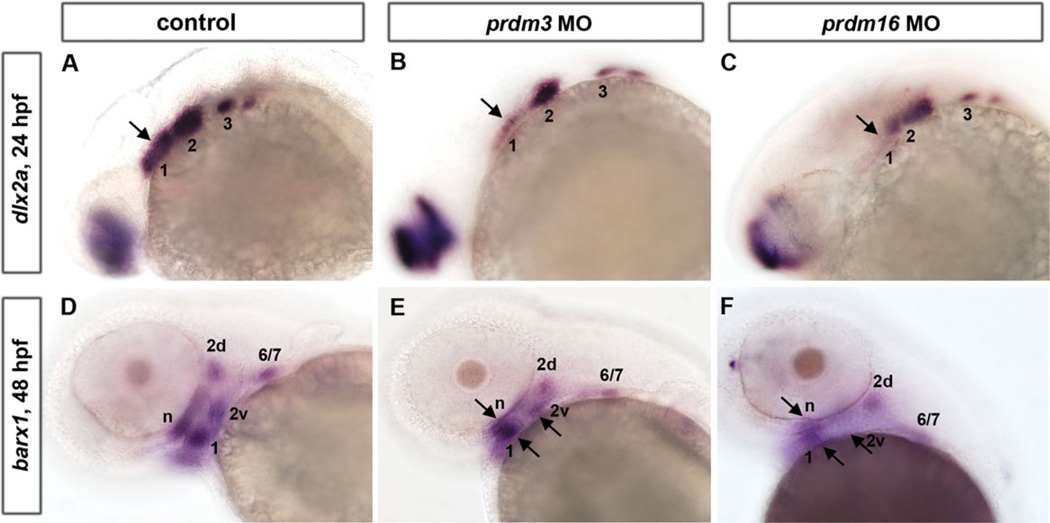
To determine the role of prdm3 and prdm16 on neural crest specific populations, we examined the expression of two genes that are expressed in cranial neural crest cells, dlx2a and barx1. We wanted to examine how early the defects in cranial neural crest cells are and thus contribute to the cartilage defects observed in the prdm3 and prdm16 morphants. We utilized probes specific for the homeodomain-containing transcription factor dlx2a, which is expressed in the post-migratory NCCs of the pharyngeal arches at 24 hpf, and barx1, a marker of cranial NCCs’ condensations as they undergo chondrogenesis, at 48 hpf (Fig. 3). While these two genes were expressed in similar domains in p53-injected Morpholino controls and prdm3 and prdm16 MO-injected embryos, the areas of the expression domains appeared reduced. Expression of dlx2a was substantially reduced in the anterior domain in prdm3 morphants (arrow, Fig. 3A, B), while its expression was reduced throughout the arch region in prdm16 morphants, especially within the first arch (Fig. 3A, C). barx1 is expressed in the neurocranium (n) and arch 1 (1), dorsal and ventral domains within arch 2 (2d and 2v) and a domain representing arches 6 and 7 (6/7). Similar to the expression of dlx2a, barx1 is notably reduced in all the above-mentioned pharyngeal arches in prdm16 morphants and greatly reduced in the anterior pharyngeal arches in prdm3 morphants (Fig. 3D–F, arrows). In prdm16 morphants, the ventral domain within arch 2 (2v) is seemingly absent (Fig. 3F compared to 3D). Interestingly, the overall affect on arch1 is more severe than the other arches, yet all the cartilages are reduced. This may be suggestive of a role for prdm3 and prdm16 in maintaining expression of neural crest genes, and not for the initiation. Consistent with this, we performed live cell imaging of both knockdowns in the tg(sox10:egfp) line and found that there was no significant defects in neural crest migration, in that the same number and pattern of cells were able to migrate to the pharyngeal arches and anteriorly around the eye (data not shown). Together these data suggest that prdm3 and prdm16 are important for cranial neural crest cell development following migration but prior to chondrogenesis.
Fig. 3.
prdm3 and prdm16knockdown causes a reduction in the NCC expression domains of dlx2a at 24 hpf and barx1 at 48 hpf. A–C: Whole mount embryo ISH shows that pharyngeal arch marker dlx2a is expressed in the post-migratory neural crest region in control (A), prdm3 e3i3 (B), and prdm16 ATG (C) morphants at 24 hpf. dlx2a expression in prdm3 and prdm16 morphants shows a significant reduction in the anterior expression domain, and a mild reduction anteriorly relative to controls. D–F: barx1 expression in cranial NCC condensations is significantly reduced in the neurocranium (n) and arch 1 (1), dorsal and ventral domains within arch 2 (2d and 2v), and a domain representing arches 6 and 7 (6/7) in prdm3 (E) and prdm16 (F) morphants at 48 hpf as compared to controls (D). Arrows indicate notably reduced expression domains in prdm3 and prdm16 morphants. Anterior is to the left.
Combinatorial Effects of prdm1a With prdm3 or prdm16 in Zebrafish Skeletal Development
Because of our interest in prdm1a, we wanted to determine the effect on craniofacial cartilage when combinations of other prdm factors are knocked down in combination with prdm1a itself. Alcian blue staining of 5-dpf prdm1a morphant larvae reveals as previously shown, an inverted ceratohyal (ch), missing ceratobranchial cartilages 2–5 (cb2–5) (n=27/39; Fig. 4B, F) (Birkholz et al., 2009). The ceratohyal was significantly shortened, while there is a slight shortening of the trabeculae in prdm1a morphants compared to controls (see Fig. 6, red bars, P < 0.001). Injection of 0.25 ng of prdm1a and 3 ng of prdm3 e3i3 MO together led to a more severe phenotype than the higher doses of either Morpholino alone. The viscerocranium was further shortened, particularly Meckel’s cartilage and the palatoquadrate, and the ceratohyals was significantly shortened (see Fig. 6, orange bars, P < 0.001 for both). Similarly to prdm1a knockdown alone, ceratobranchials 2–5 were absent. The neurocranium was much smaller and narrower, the ethmoid plate appeared truncated, and the trabeculae were shortened (n=11/15; Fig. 4C, G; P < 0.001). The width of the ethmoid plate was also significantly shorter as compared to control embryos (see Fig. 6, P < 0.03). These data suggest that knockdown of prdm1a and prdm3 has an additive effect on the development of the cartilages of the viscerocranium and a synergistic effect on the development of neurocranium.
Next we asked if there was a similar interaction between prdm1a and prdm16. Injection of two different doses of prdm1a (0.25 and 0.35 ng) and prdm16 ATG (4 and 6 ng) MOs show a dose-responsive effect on craniofacial cartilage development. Injection of the lower dose combination results in a viscerocranium phenotype similar to the prdm16 morphant alone (not shown), along with a reduction of Meckel’s cartilage. With the higher dose combination of 0.35 ng prdm1a and 6 ng prdm16 MO, we observe a more severe phenotype (Fig. 4D, H). In the viscerocranium, there is a greater shortening of the Meckel’s cartilage with a significant change in the length of the Meckel’s/ palatoquadrate and in the angle and length of the ceratohyal cartilages (see Fig. 6, dark green bars; P < 0.01 and P < 0.001, respectively), and an absence of ceratobranchials 1–5 as compared to control larvae (n=28/60; Fig. 4A, E). In the neurocranium, we observe a much smaller and narrower neurocranium with a truncated and clefted ethmoid plate and significantly shorter ethmoid plate/trabeculae (see Fig. 6, dark green bars, P < 0.001). These data suggest that prdm1a and prdm16 have a similar effect on the viscerocranium, while the neurocranium phenotype appears to be stronger in combination than the single morphants alone. Interestingly, the anterior basicapsular commissure that forms at the lateral edge of the parachordal cartilage is missing in both double morphants while slightly reduced in the singly injected morphants (Fig. 4G,H, arrow). Together these data suggest that while prdm1a may interact with prdm3 and prdm16, these prdms do not appear to act redundantly in viscerocranium in zebrafish.
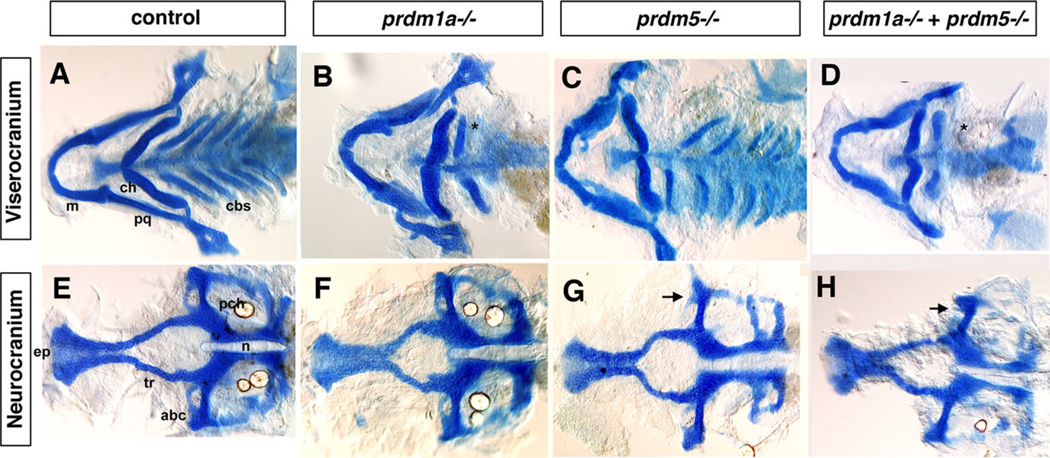
Combinatorial Effects of prdm1a and prdm5 on Craniofacial Development
prdm5 is in the same subfamily as prdm3 and prdm16 and thus may also be important in craniofacial development. In zebrafish, prdm5 has been shown to play a role in early development downstream of Wnt signaling (Meani et al., 2009), but its function in craniofacial development has not been elucidated. We obtained a viral insertional mutant of prdm5, prdm5hi61Tg, which is predicted to be a null mutation (see Experimental Procedures section below for details) and examined its craniofacial phenotype by staining with alcian blue at 5 dpf (Amsterdam et al., 1999, 2004; Amsterdam and Hopkins, 1999). prdm1a mutants have a similar phenotype as the prdm1a Morphants described above, with a significant shortening of the ceratohyal (Figs. 5B,F, 6, red bars). We observe a slight shortening of the viscerocranium in prdm5 mutants, while the neurocranium is shortened and narrower in these larvae (Fig. 5C, D). This results in a significant increase in the width of the hypophyseal fenestra, the space between the trabeculae (Fig. 6, purple bars). To determine the double mutant phenotype, we crossed the prdm5hi61Tg and prdm1am805 fish lines together resulting in ~6.25% embryos displaying a prdm5hi61Tg-prdm1am805 double homozygous mutant phenotype, confirmed by genotyping. In the double mutant, we do observe a more severe phenotype in the overall size of the viscerocranium, and there is an additive phenotype in the neurocranium, which appears smaller and with a shortened ethmoid plate and trabeculae (Fig. 5D, H). None of the elements were significantly changed from control larvae (Fig. 6, dark grey bars). In summary, we conclude that prdm3, 5, and 16 are involved in the zebrafish craniofacial skeleton development and prdm1a may interact with all three to properly pattern the head skeleton.
Fig. 5.
The viscerocranium and neurocranium of prdm5hi61Tg-prdm1am805 double mutant exhibit more severe phenotypes than single mutants. Double prdm1am805 prdm5hi61Tg heterozygotes were crossed to obtain the prdm5hi61Tg-prdm1am805 double mutant embryos. A–D: Flat-mounted Alcian blue staining of the viscerocranium and neurocranium in control, prdm1am805, prdm5hi61Tg mutant, and prdm5hi61Tg-prdm1am805 double mutant. As compared to control (A), the prdm1am805 (B) mutant larva is missing the ceratobranchial 2–5 cartilages (cb2–5) as was also observed in the prdm1a morphant. The prdm5hi61Tg (C) mutant larva has a slightly shortened Meckel’s cartilage (m) in the viscerocranium. The prdm5hi61Tg-prdm1am805 double mutants (D) have a more severe phenotype in the viscerocranium (shortening of the Meckel’s cartilage, greatly widening of the angle between ceratohyals [ch], and absence of ceratobranchials 2–5). E–H: Flat mount of the neurocranium in the control (E), prdm1am805 mutant (F), prdm5hi61Tg mutant (G), and the prdm5hi61Tg-narrowmindedm805 double mutant (H). The neurocranium and ethmoid plate (ep) are slightly smaller and trabeculae are shortened in prdm1am805 mutant and prdm5hi61Tg mutant (F,G). In prdm5−/−, the hypophyseal fenestra is significantly wider and the anterior basicapsilar cartilage is missing (G, arrow). The double mutant shows further reduction of the neurocranium, smaller ethmoid plate, shorter trabeculae, and missing basicapsilar cartilage (H, arrow). Anterior is to the left. abc, anterior basicapsular commissure; cbs, ceratobranchials; ch, ceratohyal; ep, ethmoid plate; m, Meckel’s cartilage; n, notochord; pch, parachordal; tr, trabeculae; * in B,D illustrates missing cbs.
DISCUSSION
In this study, we have determined that prdm3, prdm5, and prdm16 play a role in zebrafish craniofacial development. We have shown that the prdms are expressed at specific developmental time points in multiple tissues and that targeted knockdown both singly, and in combination, leads to cartilage defects in the neurocranium and the anterior viscerocranium. While several other studies have shown a role for PRDM proteins in the immune system and cancer, few studies focus on their role in development. This is the first systematic study focusing on the roles of different members of the prdm gene family in zebrafish craniofacial development.
Prdm Proteins Play a Redundant Role in Neurocranium Development
The neurocranium is the structure that consists of the anterior ethmoid plate and the posterior trabeculae, as well as the posterior parachordal cartilage (Cubbage and Mabee, 1996). The neurocranium is both neural crest- and mesoderm-derived, consisting of the neural crest–derived anterior structures, including the ethmoid plate and trabeculae, and the posterior parachordal cartilage from the mesoderm. Fate mapping of the anterior migrating stream that migrates around the eye gives rise to the medial ethmoid plate cartilage while the cells that migrate more posteriorly into the dorsal part of arch 1 contribute to the trabeculae (Kimmel and Eberhart, 2008; Wada et al., 2005). These populations require shh signaling: Mutations in pathway members or treatment with cyclopamine, to inhibit shh signaling, cause defects in the ethmoid plate and trabeculae (Wada et al., 2005). Our results with prdm3 and prdm16 double morphants suggest they may act downstream of shh signaling. It is not yet known if neural crest cells and/or mesoderm contribute to the anterior basicapsular commissure. This structure seems to be sensitive to prdm dosage and sits at the base of the brain behind the palate. The anterior neurocranium cartilages make up the base of the brain in zebrafish, and because there is no true nasal cavity in fishes, the anterior neurocranium is considered the zebrafish equivalent to the secondary palate in mammals (Eberhart et al., 2008; Swartz et al., 2011). Thus, understanding the development of this structure may help us understand normal palate formation as well as the etiology of cleft lip with or without palate, which result from defects in palate morphogenesis. The phenotypes we observe in the single prdm knockdowns include an overall hypoplastic neurocranium. However, in the double knockdowns, the ethmoid plate is not as broad at the tip and the trabeculae are shortened. In addition, the anterior basicapsular commissures are absent. Interestingly, we observe either a reduction in the number of cells and/or a lack of chondrogenesis in the medial ethmoid plate often resulting in a cleft, which may suggest an effect specifically on the anterior migrating neural crest cell population. To address this, we tried to let some of the embryos survive to 6–7 days post-fertilization (dpf), however, many are severely defective by this time point. As described above, these neurocranium phenotypes are similar to what is observed following treatment with cyclopamine at later stages, 36–48 hpf, perhaps suggesting that prdm3 and prdm16 may be functioning downstream of shh at this specific developmental time (Wada et al., 2005). Further experiments are required to resolve whether prdms are modulated by Shh, but together these data suggest that the prdm genes have redundant roles in development of the neurocranium.
Prdm’s Proteins Have a Moderate Role in Viscerocranium Development
We have shown that prdm morphant larva appear to have mild phenotypes in the viscerocranium, with a shortening of Meckel’s cartilage and a hypoplastic palatoquadrate. Second arch structures also have moderate defects, with the hyosymplectic forming normally, while the ceratohyals are often compressed and the angle between both ceratohyals is greater than normal. Arches 3–7 are the least affected in prdm morphants, with the exception of prdm1a, which is critical for the proper formation of the posterior cartilages (Birkholz et al., 2009). We do observe a more significant affect on Meckel’s cartilage with increasing doses of prdm1a Morpholino, suggesting that the overall levels of Prdm proteins may be important (Hernandez-Lagunas and Artinger, unpublished observation). In mice, an embryonic mutation in Prdm1 causes hypoplastic posterior arches, which is consistent with a role in craniofacial skeletal development, though this has not been directly tested. While the roles of Prdm3 and 5 have not been specifically addressed in mouse craniofacial development, a Prdm16 ENU allele, causing frame shift and premature termination, exhibits a cleft palate (Bjork et al., 2010b). Prdm16 expression begins at E9.5 in the pharyngeal arch region and is expressed in the palatal shelves at E13.5-E14.5 and may play a role in chondrogenesis and bone formation (Horn et al., 2011). Our findings in zebrafish that knockdown of prdm16 in combination with other prdms results in neurocranium defects is consistent with this previous report in mouse.
Evolutionary Conservation of prdm Expression and Function
In terms of sequence, there is high conservation between all prdm paralogs across vertebrates. What seems to differ is the number and position of the zinc finger DNA-binding domains. prdm paralogs have similar but somewhat different expression patterns and function. For example, prdm1 expression in vertebrates is fairly conserved, in that it is expressed in the somites, posterior pharyngeal arches, limb buds, and retina across species in which it has been analyzed (Chang et al., 2002; Wilm and Solnica-Krezel, 2005). This complex expression pattern suggests that PRDM1 is required in a variety of developmental processes. Indeed, studies in both mouse and zebrafish models indicate it is important in the development of B-cells (Messika et al., 1998; Turner et al., 1994), germ cells (Ohinata et al., 2005; Vincent et al., 2005), neural crest (Bikoff et al., 2009; Hernandez-Lagunas et al., 2005; Olesnicky et al., 2010; Roy, 2004) and muscle cells (Baxendale et al., 2004). A recent study in lamprey showed that prdm1 is expressed at the neural plate border and in the gill arches, suggesting it is expressed similarly to its gnathostome orthologs (Nikitina et al., 2011). However, the two close paralogs of prdm1a in zebrafish, prdm1b and prdm1c, have partially lost some of the prdm1a expression domains. prdm1b, like prdm1a, is highly expressed in somites and in retina, but prdm1c shows a more ubiquitous expression pattern (Sun et al., 2008) and neither is expressed in the developing face. prdm4, 10, and 15 are also members of this family and are, for the most part, expressed ubiquitously in zebrafish (Fumasoni et al., 2007; Sun et al., 2008).
The subfamily that contains prdm3, 5, and 16 shares some similarities in expression but also exhibits some differences. Our results showed that the expression of prdm3 and prdm16 in zebrafish is partially overlapping, with both being expressed in hindbrain, telencephalon, pharyngeal arches, pectoral fin buds, and the neurocranium. prdm16 alone is expressed in the olfactory placode. In the fin bud, expression of prdm3 is faint at 30 hpf while prdm16 is already highly expressed by 30 hpf, suggesting that expression of prdm16 is earlier than that of prdm3 in this domain. Both prdm3 and prdm16 are expressed in the pronephric duct, and prdm16 is expressed in the olfactory placode as well (Fig. 1 and data not shown). The similarities and differences in expression of prdm3 and prdm16 imply that they may be functionally redundant in domains where both are expressed, such as the developing brain, pharyngeal arches, and pectoral fin buds, but have independent functions in other domains. Within the same cluster, the expression of prdm5 is specifically localized to the pharyngeal arches and neurocranium. Meani et al. (2009) demonstrated that prdm5 is ubiquitously expressed during cleavage stages, with higher levels of expression in the central nervous system during somitogenesis and overall lower levels in the rest of the embryo. Our results suggest there is some functional redundancy between the prdm paralogs but that there may also have been some divergence in gene expression and function within the prdm gene family.
EXPERIMENTAL PROCEDURES
Zebrafish Maintenance
Zebrafish were maintained according to Westerfield (2007) and embryos were staged according to Kimmel (Kimmel et al., 1995). Lines included wild type AB and TAB (ZIRC), prdm1a mutant fish line (narrowmindedm805from our lab), prdm5hi6Tg (Amsterdam et al., 2004), a p53 mutant fish line, tp53M214K (Berghmans et al., 2005). The prdm1am805 mutant strain (narrowminded) was isolated from a small-scale in situ hybridization screen described in a previous publication (Artinger et al., 1999), and is currently on a mixed background. Control embryos are wildtype or heterozygous prdm1a+/− unless otherwise noted. tp53M214K−/− fish were obtained by incrossing tp53M214K−/+heterozygotes and genotyped by PCR according to a previous publication and are used to eliminate the non-specific effects of Morpholinos (Berghmans et al., 2005; Johnson et al., 2011).
Genotyping and Mutant Lines
The prdm5hi61Tg fish line was identified in a viral insertion screen carried out in the Hopkins laboratory. The insertion is predicted to be a full null since the insertion is in the first exon shortly after the ATG (A. Amsterdam, personal communication). To genotype single zebrafish, adults are anesthetized in 0.15% Tricaine and tail fins clipped and transferred into 50-µl aliquots of lysis buffer. Genomic DNA was extracted at 98°C for 10 min, incubating with 5 µl proteinase K (10 mg/ml) at 55° C for 1 hr, and denaturing with Proteinase K at 98° C for 10 min. The three primer sequences for genotyping prdm5hi61Tg fish line to distinguish wild type (575 bp only), heterozygotes (455 and 575 bp), and homozygous mutants (455 bp only) are 5′-CAG TGT AAA CCT TTC TTA ACT GTG TTT C-3′, 5′-GCT AGC TTG CCA AAC CTA CAG GT-3′,and 5′-GAC AGT GAC ATG GAT GAT CAG C-3′. The primer sequences for genotyping prdm1am805 fish line are forward 5′-TTC AGT CAA GAC CTA AGC CCG C-3′ and reverse 5’-CAA AAA CAT CTT AAG GAA GAG GGC AG-3′, followed by a 2-hr digest with the restriction enzyme fokI, which cuts only the WT allele product.
Whole Mount In Situ Hybridization (ISH)
Conventional whole-mount ISH was performed as described in Thisse (Thisse and Thisse, 1998; Thisse et al., 1993) and Johnson et al. (2011), using DIG-labeled (Roche, Indianapolis, IN) antisense RNA probes: barx1 (Sperber and Dawid, 2008), dlx2a (Jackman et al., 2004), prdm3, 5, 10, 11, and 16 (Sun et al.J, 2008). BM Purple (Roche) was used as a substrate for the alkaline phosphatase reaction.
Whole Mount Skeletal Staining
Cartilage was stained according to Walker and Kimmel, 2007, with some modifications. Embryos were incubated in egg water containing 0.003% (w/v) 1-phenyl-2-thiourea (PTU, Sigma-Aldrich, St. Louis, MO) to prevent pigment formation from 24 until 5 dpf. Five-dpf larvae were fixed in 2% PFA/PBS for 1 hr at room temperature, washed in 100 mM Tris pH 7.5/10 mM MgCl2, and stained overnight in 0.04% alcian blue (Anatech Ltd., Battle Creek, MI) in 80% EtOH/ 100 mM Tris pH 7.5/10 mM MgCl2. Larvae were then rehydrated in 80% EtOH/100 mM Tris pH 7.5/10 mM MgCl2, followed by 50 and 25% EtOH/ 100 mM Tris pH 7.5, cleared in 25% glycerol/0.1% KOH, and stored in 50% glycerol/0.1% KOH.
Morpholino Injections
Morpholinos knockdown was accomplished by injecting embryos at the 1-to 4-cell-stage with a mixture of 2.5% fluorescein dextran (10,000 MW, lysine fixable, Invitrogen). Morpholinos to prdm1a (0.5-ng splice blocking Morpholino to intron 2 splice donor, Baxendale et al., 2004), prdm3 (6 ng splicing e3i3 MO, 5′-TAG AAG TAA ATG AGT GTT ACC TGC A-3′ and 12 ng splicing i2e3 MO, 5′-TCA ACC CTG CTG ATG TTA AAC TTC T-3′) and prdm16 (8 ng ATG blocking Morpholino, 5′-CCA GAC AGA ACT TCA CAT TGC CCA T-3′ and 6 ng splice blocking e2i2 MO 5′-ACT CAC ACT ATC ACC CAC CTT ATC A-3′). All MOs were ordered from and designed by Gene Tools, LLC (Philomath, OR). All Morpholinos were injected first into wildtype embryos at several doses to determine a dose that was appropriate for these experiments, and without toxicity. A prdm3 ATG MO could not be designed because in the current annotation of the genome, the ATG start site is ambiguous and thus we designed two splice MOs. Because both prdm3 and prdm16 MOs have not been tested before, we verified that the phenotypes observed in this morphant were the result of reduced expression rather than an off-target by performing several controls: (1) Two independently targeting MOs were injected for each (sequences above) in at least three separate experiments and have similar phenotypes; (2) We determined that splice junction targeted Morpholinos indeed interfered with splicing. mRNA from 8–10 pooled, 48-hpf embryos injected with 12 ng prdm3 i2e3 MO or 6 ng prdm16 e2i2 MO or 10 uninjected or tp53M214K injected embryos was isolated using the RNAeasy Micro Kit (Qiagen, Chatsworth, CA) and reverse transcribed into cDNA using the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). RT-PCR of the prdm3 was performed using this cDNA with a forward primer 5′- GAC CTA AGT CTT GGC AGA C −3′ and reverse primer 5′- GAG CTG AAG GAT TCC AGC −3′, and prdm16 forward primer 5′- GTG CAC GAG TGC AAA GAC TG −3′ and reverse primer 5′- GCT TGA CAC TGC TGT GTA TG 3′, while (β-actin was amplified with forward primer 5′-CAT CAG GGT GTC ATG GTT GGT −3′, and reverse primer 5′-TCT CTT GCT CTG AGC CTC ATC A −3′. Both splice-blocking MO interfered with splicing (Supp. Fig. S1). prdm3 MO caused exon skipping of exon 3, creating a band of 211 bp compared to uninjected/tp53M214KI embryos that maintain exon 3 and a size of 302 bp. In addition, a ~450-bp band was also observed, possibly creating a fragment containing some of intron 2. prdm16 MO also caused exon skipping of exon 2, resulting in a 192-bp band compared to 340 bp in uninjected/tp53M214KI embryos. In both cases, some of wildtype RNA remains, but we believe that we have significant knockdown of both prdm3 and prdm16 in these assays. (3) A standard control MO sequence 5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′ was injected at a relatively high dose of 18 ng without a craniofacial phenotype (Supp. Fig. S2); (4) Rescue experiments were attempted for prdm3 and prdm16. For prdm3, because of the problem in the genome annotation, we were unable to clone the full-length coding region for mRNA synthesis. Thus, a rescue was not attempted. For prdm16, 100 pg of prdm16 mRNA was injected along with prdm16 i2e2 MO. In 60% of embryos (n=12/19), we did observe a rescue in the viscerocranium, size of the ethmoid plate, and the anterior basicapsular commissure, suggesting a partial rescue (Supp. Fig. S2). (5) To suppress p53-mediated, MO-induced cell death, all MO injections were carried out in both tp53M214K−/−embryos or TAB fish line co-injected with 2 ng p53 MO. p53 MO sequence 5′-GCG CCA TTG CTT TGC AAG AAT TG-3′ (Gene Tools, LLC) (Robu et al., 2007). The control embryos presented in all figures are tp53M214K−/− or p53 MO-injected embryos and were identical to that of wildtype embryos. Together, these results provide strong support for the specificity of our Morpholinos.
Imaging Analysis and Quantification
Embryos processed for in situ hybridization were mounted in 80% glycerol or 3% methylcellulose and imaged using an Olympus BX51WI compound microscope. Embryos stained for cartilage with Alcian Blue were sometimes dissected and flat-mounted as described (Javidan and Schilling, 2004) and imaged with the above compound microscope. For quantification of cartilage elements, embryos were flat mounted and images acquired at 10× magnification. Elements were measured in Adobe Photoshop, and Tukey Kramer two-way Anova posthoc test for pairwise comparison calculated as compared to uninjected wildtype or p53 MO injected control using Prism 5 software (GraphPad, San Diego, CA). This correction is used to normalize for an unequal variance shown in Figure 6 in error bars illustrating the standard error. We compared the wildtype length/width of the elements to each morphant condition, and indicated statistical significance by an asterisk (*). The significance of each morphant condition was calculated to each other’s Morpholino injected condition, but is not indicated in Figure 6. Wildtype or tp53M214K−/− fish were measured (n=11) and each Morphant singly injected or in combination (n=3–8 for each condition).
Supplementary Material
Key findings.
prdm3, 5, 16 are expressed in the zebrafish pharyngeal arches.
Knockdown of prdm3 and prdm16 results in neurocranium and viscerocranium defects and, in combination, is additive in the neurocranium.
prdm1a with prdm3 or prdm16 Morpholinos together leads to more severe phenotypes.
prdm5 mutants have defects in the neurocranium and prdm1a and prdm5 double mutants show more severe phenotypes.
ACKNOWLEDGMENTS
We thank Dr. Ting-Xi Liu (Shanghai Institute of Hematology) for generously providing plasmids for the prdms ISH probes and Letitia Kwok for piloting some of the initial experiments. We appreciate the excellent fish care provided by Morgan Singleton, the sharing of the tp53M214K mutant fish line by Dr. Bruce Appel, and technical assistance from Laura Hernandez-Lagunas and Christopher Johnson. We also thank Christopher Johnson and Kristi LaMonica for critically reading the manuscript. This work was supported by Zebrafish Core grant NIH P30-NS048154 and NIH R01-DE17699 to K.B.A.
Grant sponsor: NIH; Grant numbers: NS048154, R01DE17699.
Footnotes
Additional supporting information may be found in the online version of this article.
REFERENCES
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N. Retrovirus-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 1999;60:87–98. doi: 10.1016/s0091-679x(08)61895-6. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell E, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Beermann ML, Ardelt M, Girgenrath M, Miller JB. Prdm1 (Blimp-1) and the expression of fast and slow myosin heavy chain isoforms during avian myogenesis in vitro. PLoS One. 2010;5:e9951. doi: 10.1371/journal.pone.0009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RP, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, Plasterk R, Zon LI, Look AT. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev. 2009;19:379–385. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Birkholz DA, Olesnicky Killian EC, George KM, Artinger KB. Prdm1a is necessary for posterior pharyngeal arch development in zebrafish. Dev Dyn. 2009;238:2575–2587. doi: 10.1002/dvdy.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork BC, Vieira AR, Faust S, Camper SA, Murray JC, Beier DR. Phenotypic, genetic, and developmental characterization of CPO1, a recessive ENU-induced mouse model of cleft palate. Woodbury, NY: Mouse Molecular Genetics Cold Spring Harbor Press; 2006. p. 27. [Google Scholar]
- Bjork BC, Fujiwara Y, Davis SW, Qiu H, Saunders TL, Sandy P, Orkin S, Camper SA, Beier DR. A transient transgenic RNAi strategy for rapid characterization of gene function during embryonic development. PLoS One. 2010a;5:e14375. doi: 10.1371/journal.pone.0014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010b;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DJ, Woodward S, Thompson FH, Dos Santos B, Russell M, Yang JM, Guan XY, Trent J, Alberts DS, Taetle R. Expression of the zinc finger gene EVI-1 in ovarian and other cancers. Br J Cancer. 1996;74:1518–1525. doi: 10.1038/bjc.1996.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonamici S, Chakraborty S, Senyuk V, Nucifora G. The role of EVI1 in normal and leukemic cells. Blood Cells Mol Dis. 2003;31:206–212. doi: 10.1016/s1079-9796(03)00159-1. [DOI] [PubMed] [Google Scholar]
- Chang DH, Cattoretti G, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Chen XW, Cheng L, Liu YD, Lou G. DNA methylation and carcinogenesis of PRDM5 in cervical cancer. J Cancer Res Clin Oncol. 2010;136:1821–1825. doi: 10.1007/s00432-010-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly GF, Coltey P, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in chickquail chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Cubbage C, Mabee P. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae) J Morphol. 1996;229:121–160. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Deng Q, Huang S. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene. 2004;23:4903–4910. doi: 10.1038/sj.onc.1207615. [DOI] [PubMed] [Google Scholar]
- Duan Z, Person RE, Lee HH, Huang S, Donadieu J, Badolato R, Grimes HL, Papayannopoulou T, Horwitz MS. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol. 2007;27:6889–6902. doi: 10.1128/MCB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog CK, Galli GG, Lund AH. PRDM proteins: important players in differentiation and disease. BioEssays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restriction. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007;7:187. doi: 10.1186/1471-2148-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A. Development of the pharyngeal arches. Am J Med Genet A. 2003;119A:251–256. doi: 10.1002/ajmg.a.10980. [DOI] [PubMed] [Google Scholar]
- He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- Hernandez-Lagunas L, Choi I, Kaji T, Simpson P, Hershey C, Zhou Y, Zon L, Mercola M, Artinger KB. Zebra-fish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev Biol. 2005;278:347–357. doi: 10.1016/j.ydbio.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- Horn KH, Warner M, Pisano MM, Greene RM. PRDM16 expression in the developing mouse embryo. Acta Histochem. 2011;113:150–155. doi: 10.1016/j.acthis.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstadius SaS S. Experimental studies about the determination of the cartilaginous skeleton of the head in Urodeles. Nova Acta R Soc Sci Upsal Ser. 1946;13:1–170. [Google Scholar]
- Hoyt PR, Bartholomew C, Davis AJ, Yutzey K, Gamer LW, Potter SS, Ihle JN, Mucenski ML. The Evi1 protooncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech Dev. 1997;65:55–70. doi: 10.1016/s0925-4773(97)00057-9. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Javidan Y, Schilling TF. Development of cartilage and bone. Methods Cell Biol. 2004;76:415–436. doi: 10.1016/s0091-679x(04)76018-5. [DOI] [PubMed] [Google Scholar]
- Johnson CW, Hernandez-Lagunas L, Feng W, Melvin VS, Williams T, Artinger KB. Vgll2a is required for neural crest cell survival during zebrafish craniofacial development. Dev Biol. 2011;357:269–281. doi: 10.1016/j.ydbio.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Eberhart JK. The midline, oral ectoderm, and the arch-0 problem. Integr Comp Biol. 2008;48:668–680. doi: 10.1093/icb/icn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Induction and patterning of the neural crest, a stem cell-like precursor population. J Neurobiol. 1998;36:175–189. doi: 10.1002/(sici)1097-4695(199808)36:2<175::aid-neu6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Le Douarin N. The neural crest. New York: Cambridge University Press; 1982. [Google Scholar]
- Le Douarin NM, Ziller C, Couly GF. Patterning of neural crest derivatives in the avian embryo: in vivo and in vitro studies. Dev Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- Le Lie`vre C. Participation of neural crest-derived cells in the genesis of the skull in birds. J Embryol Exp Morphol. 1978;47:17–37. [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Mead PE, Parganas E, Ohtsuka S, Morishita K, Gamer L, Kuliyev E, Wright CV, Ihle JN. Evi-1 expression in Xenopus. Gene Expr Patterns. 2005;5:601–608. doi: 10.1016/j.modgep.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Meani N, Pezzimenti F, Deflorian G, Mione M, Alcalay M. The tumor suppressor PRDM5 regulates Wnt signaling at early stages of zebrafish development. PLoS One. 2009;4:e4273. doi: 10.1371/journal.pone.0004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, Fischer S, Neumann CJ. Prdm1 acts downstream of a sequential RA, Wnt and Fgf signaling cascade during zebrafish forelimb induction. Development. 2006;133:2805–2815. doi: 10.1242/dev.02455. [DOI] [PubMed] [Google Scholar]
- Messika EJ, Lu PS, Sung YJ, Yao T, Chi JT, Chien YH, Davis MM. Differential effect of B lymphocyte-induced maturation protein (Blimp-1) expression on cell fate during B cell development. J Exp Med. 1998;188:515–525. doi: 10.1084/jem.188.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Tong L, Bronner ME. Ancestral network module regulating prdm1 expression in the lamprey neural plate border. Dev Dyn. 2011;240:2265–2271. doi: 10.1002/dvdy.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukha-nych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Olesnicky E, Hernandez-Lagunas L, Artinger KB. prdm1a Regulates sox10 and islet1 in the development of neural crest and Rohon-Beard sensory neurons. Genesis. 2010;48:656–666. doi: 10.1002/dvg.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodson W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cell in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, Nutt SL, Bikoff EK. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–1777. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Russell M, List A, Greenberg P, Woodward S, Glinsmann B, Parganas E, Ihle J, Taetle R. Expression of EVI1 in myelodysplastic syndromes and other hematologic malignancies without 3q26 translocations. Blood. 1994;84:1243–1248. [PubMed] [Google Scholar]
- Schilling TF, Kimmel C. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel C. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime` A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyuk V, Premanand K, Xu P, Qian Z, Nucifora G. The oncoprotein EVI1 and the DNA methyltransferase Dnmt3 co-operate in binding and de novo methylation of target DNA. PLoS One. 2011;6:e20793. doi: 10.1371/journal.pone.0020793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and preplasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Sperber SM, Dawid I. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches Dev. Biol. 2008;321:101–110. doi: 10.1016/j.ydbio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Xu PF, Zhou T, Hu M, Fu CT, Zhang Y, Jin Y, Chen Y, Chen SJ, Huang QH, Liu TX, Chen Z. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde JS, Donninger H, Wu K, Johnson ME, Pestell RG, Rose GS, Mok SC, Brady J, Bonome T, Birrer MJ. Expression profiling identifies altered expression of genes that contribute to the inhibition of transforming growth factor-βsignaling in ovarian cancer. Cancer Res. 2006;66:8404–8412. doi: 10.1158/0008-5472.CAN-06-0683. [DOI] [PubMed] [Google Scholar]
- Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK. Examination of a palatogenic gene program in zebrafish. Dev Dyn. 2011;240:2204–2220. doi: 10.1002/dvdy.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz ME, Nguyen V, McCarthy NQ, Eberhart JK. Hh signaling regulates patterning and morphogenesis of the pharyngeal arch-derived skeleton. Dev Biol. 2012;369:65–75. doi: 10.1016/j.ydbio.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Thisse C. High resolution whole-mount in situ hybridization. Zebrafish Sci Monitor. 1998;15:8–9. [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol. 2001;13:698–705. doi: 10.1016/s0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- Turner CA, Mack DH, Davis MM., Jr Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel C. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Toyota M, Kondo Y, Suzuki H, Imai T, Ohe-Toyota M, Maruyama R, Nojima M, Sasaki Y, Sekido Y, Hirat-suka H, Shinomura Y, Imai K, Itoh F, Tokino T. PRDM5 identified as a target of epigenetic silencing in colorectal and gastric cancer. Clin Cancer Res. 2007;13:4786–4794. doi: 10.1158/1078-0432.CCR-07-0305. [DOI] [PubMed] [Google Scholar]
- Westerfield . A guide for the laboratory use of zebrafish (Danio rerio) 5th. Eugene: Univ. of Oregon Press; 2007. The zebrafish book. [Google Scholar]
- Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- Yan J, Jiang J, Lim CA, Wu Q, Ng HH, Chin KC. BLIMP1 regulates cell growth through repression of p53 transcription. Proc Natl Acad Sci USA. 2007;104:1841–1846. doi: 10.1073/pnas.0605562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.