Abstract
Active maintenance of genome stability is a prerequisite for the development and function of the nervous system. The high replication index during neurogenesis and the long life of mature neurons highlight the need for efficient cellular programs to safeguard genetic fidelity. Multiple DNA damage response pathways ensure that replication stress and other types of DNA lesions such as oxidative damage do not impact neural homeostasis. Numerous human neurologic syndromes result from defective DNA damage signaling and compromised genome integrity. These syndromes can involve different neuropathology, which highlights the diverse maintenance roles required for genome stability in the nervous system. Understanding how DNA damage signaling pathways promote neural development and preserve homeostasis is essential for understanding fundamental brain function.
Introduction
The genesis of the nervous system requires an enormous expansion of highly proliferative neuroepithelium that generates a diverse array of long-lived cell types. Amongst these are specialized neurons that fulfill functionally diverse roles in information processing and signal integration. Equally important are non-neuronal populations of glial cells that provide metabolic and functional support for the nervous system1, 2. A central aspect of neural homeostasis is the need to maintain genomic integrity after damage to DNA during normal cellular activity or during DNA replication. Indeed, DNA damage in the form of double strand breaks can arise spontaneously in the brain as a result of neuronal activity3. Age-related accumulation of DNA damage in the brain can also impact gene expression, which potentially affects processes involving memory and neuronal survival4. More directly, many inherited human syndromes that arise from mutations affecting genome stability are characterized by neuropathology, revealing critical roles for DNA damage surveillance and repair in safeguarding the nervous system5.
The specific requirements for genome maintenance can change substantially in the transition from neurogenesis to nervous system maturation (Figure 1). During neurogenesis a prime source of DNA damage is associated with replication. The genomes of differentiated neural cells, which populate the nervous system for the life of an organism, must be protected against continual DNA damage. This damage can occur for example from reactive chemical species such as those produced by oxidative metabolism, or from transcription-associated damage. Thus, at multiple levels throughout the development and maintenance of the nervous system, there is a constant need to ensure genome integrity. The following sections detail how the many biochemically distinct DNA repair pathways maintain genome integrity during neurodevelopment and in the mature nervous system. Underscoring this is consideration of a variety of human diseases that illustrate how defective DNA damage signaling impacts the nervous system.
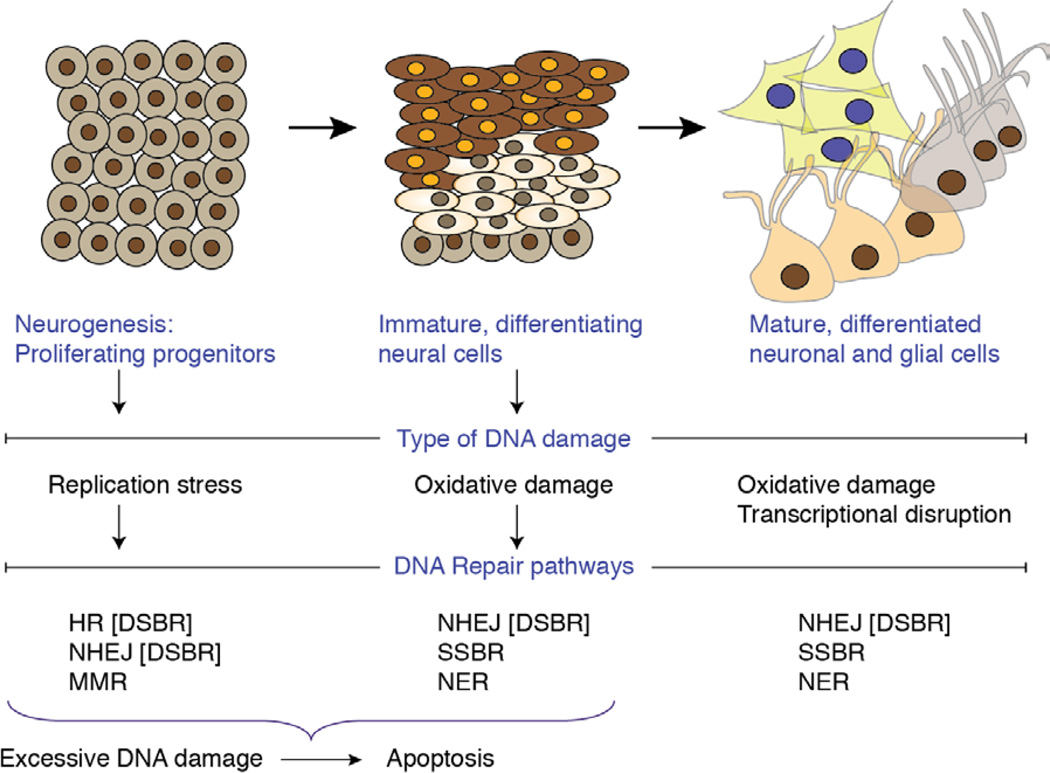
Figure 1. Different DNA repair pathways function during neural development.
Neural development encompasses widespread proliferation, migration and differentiation to generate neurons and glia of the adult nervous system. At different stages of development the nervous system is susceptible to different types of DNA damage. During proliferation, replication associated DNA strand breaks can occur that may require DNA double strand break repair (DSBR), involving homologous recombination (HR) or non-homologous end joining (NHEJ). Mismatch repair (MMR) is also important during replication. HR is dependent on replicated sister chromatids for use as an error-free repair template, and so this pathway is not available in non-replicating or differentiated cells. In non-cycling cells, NHEJ repairs DNA double strand breaks while other types of DNA damage require the single strand break repair (SSBR) pathway or nucleotide excision repair (NER). An alternate outcome to DNA damage in replicating and immature, non-differentiated neural cells is apoptosis, while DNA lesions in differentiated cells do not activate apoptosis but instead can interfere with transcription.
Multiple DNA Repair pathways function in the nervous system
In broad terms, the nervous system can be divided into two different phases that require different cellular strategies to ensure genome integrity. During early development neurogenesis is driven by proliferation, and the high replicative rate of neural progenitors is associated with replication-associated DNA damage5. Like other organs, the nervous system has the full repertoire of DNA repair pathways. Individually, these distinct biochemical pathways respond to specific types of DNA lesions such as DNA single or double strand breaks or DNA cross-links. The biochemical details of each of the main DNA repair pathways have recently been comprehensively reviewed6–13 and in the interest of space, a detailed outline will not be presented.
Importantly, different types of DNA lesions utilize specific biochemical repair pathways. For instance, bulky, helix-distorting lesions such as those induced by ultraviolet radiation rely upon the nucleotide excision pathway (NER)12, while DNA double strand breaks can undergo repair by either homologous recombination (HR) or non-homologous endjoining (NHEJ)7. DNA double strand breaks are particularly hazardous to a cell as they can activate apoptosis or can lead to mutagenic rearrangements. HR requires an available sister chromatid to facilitate error-free repair and so this process occurs during S- or G2-phase of the cell cycle, while NHEJ which involves direct ligation of processed ends of the DNA break can occur at any stage of the cell cycle7, 10. In contrast, the more common lesion of a DNA single stand break is repaired via the XRCC1-mediated base excision repair (BER)/single-strand break repair pathway6. Other pathways that are important include those that repair interstrand crosslinks and the mismatch repair pathway that correct mismatched bases that can form during DNA replication8, 11. The repair pathways listed above are of great relevance in the nervous system as defects in any of these can have a detrimental impact on many facets of neural function5. For instance, perturbation of NHEJ can result in neurodevelopmental defects14, 15, and faulty nucleotide excision repair can cause neurodegeneration or neurodevelopmental issues5, 16–18. In the case of DNA single strand breaks, repair defects can lead to neurodegenerative disease5, 6. An overview of representative neurologic diseases that result from various defects in DNA damage responses is presented in Table 1.
Table 1.
DNA repair pathways disrupted in human syndromes characterized by neuropathology.
| DNA damage | DNA repair pathway | Disease | Neuropathology |
|---|---|---|---|
| Double strand break | Non-homologous end-joining Homologous recombination |
Lig4 syndrome Severe Combined Immunodeficiency (SCID) Fanconi anemia1 |
Neurodevelopmental defects/microcephaly Brain tumors |
| Single strand break | Base excision repair/single strand break repair2 | AOA1 SCAN1 MCSZ |
Neurodegeneration Microcephaly |
| DNA interstrand cross-link3 | Fanconi anemia pathway | Fanconi anemia | Brain tumors Neurodevelopmental defects |
| Helix-distorting lesion/ Bulky adduct |
Nucleotide excision repair Transcription coupled repair |
Xeroderma pigmentosum Trichothiodystrophy Cockayne syndrome |
Neurodegeneration4 Complex |
| Misincorporated nucleotide (during replication) | Mismatch repair | Lynch Syndrome | Brain tumors |
| Defective DNA damage signaling (double strand break signaling) | MRE11/RAD50/NBS1 complex (MRN) Ataxia telangiectasia, mutated (ATM) |
Nijmegen breakage syndrome ATLD Ataxia telangiectasia |
Microcephaly5 Neurodegeneration |
| Replication stress (signaling replication protein A coated DNA) | Ataxia telangiectasia and Rad3-realted (ATR) Pericentrin5 |
ATR-Seckel syndrome Seckel Syndrome6 |
Neurodevelopmental Microcephaly |
Biallelic mutations in the HR factor BRCA2 occurs in a subgroup of FA, highlighting the connections between HR and X-link repair.
The base excision repair pathway is based on Xrcc1-mediated processes, which includes single strand break repair.
This pathway utilizes components of HR (e.g. BRCA2).
Neurodegeneration is only present in some cases of XP and is related to the specific NER component that is mutated.
Microcephaly is characteristic of NBS, although specific mutations in MRE11 result in microcephaly rather than ATLD (REF 34).
Seckel syndrome can rise from defects in the ATR DNA damage-signaling pathway or from defects in the centrosome and shares phenotypic and genetic overlaps with primary microcephaly disorders that arise form centrosome defects.
In addition to key information from inherited human syndromes5, 6, 19, the importance of specific DNA repair pathways during neural development has also been directly shown using gene targeting in mice to disrupt gene function. For example, inactivation of DNA double strand break repair factors involved in HR or NHEJ, or components of the BER pathway, showed a profound effect towards neural development20–25. Neural progenitors at different stages of differentiation and commitment can demonstrate a selective sensitivity towards DNA damage whereby the early born cortical progenitors are hypersensitive to replication-associated DNA damage26. Neural signaling responses to genome insults can also vary depending on cellular differentiation status and tissue type26, 27. These differences in susceptibility to DNA damage may reflect the varied neuropathology characteristic of DNA repair deficiency diseases.
DNA integrity during development is also maintained by the coordinated signaling of pathways that respond to DNA damage by pausing cell proliferation to allow DNA repair, or alternatively, by the activation of apoptosis to eliminate damaged cells and avoid the potential acquisition of mutations. Elimination of progenitors with excessive DNA damage may in some cases be a preferred option, particularly as the nervous system is known to use apoptosis during normal neural development to eliminate over-produced cells28, 29. However, after cessation of neurogenesis DNA repair is still of paramount importance to guard the genome throughout the life of the nervous system.
Neural homeostasis in mammals relies on a post-mitotic nervous system that requires constant DNA repair activity during the life of this tissue. The types of DNA damage in non-cycling cells can be different to those encountered during development, as are the pathways available for DNA repair (Figure 1). For example, in non-cycling cells HR is unavailable to repair DNA double strand breaks. In post-mitotic neurons, NHEJ is the sole pathway available to prevent accumulation of DNA double strand breaks30. Accordingly, in the absence of a key NHEJ component, DNA ligase IV, neurons progressively accumulate endogenous double strand breaks, indicating the essential nature of this pathway for preventing DNA damage in the mature nervous system30.
DNA Damage Signaling is required for Neural Development
Coincident with DNA processing and repair are distinct signaling pathways that become activated upon DNA damage. These genome surveillance pathways activate cell cycle arrest to pause proliferation and allow for DNA repair, or ensure genome stability by initiating apoptosis to eliminate damaged cells. The critical role of DNA damage signaling in the nervous system is fulfilled by the DNA damage-activated kinases ATM (ataxia telangiectasia, mutated) and ATR (ATM and Rad3 related) (Figure 2). ATR and ATM play largely separate roles in maintaining genome stability in the nervous system31. These distinct kinases are activated by different types of DNA damage: DNA double strand breaks are detected by the MRN (MRE11-RAD50-NBS1) complex, which promotes autophosphorylation and activation of ATM leading to the subsequent phosphorylation of specific ATM substrates to initiate cell cycle arrest or apoptosis of immature neural cells23, 32, 33. DNA damage-induced apoptosis may be a central function of this kinase in the nervous system27, 34. This pathway is critical in the nervous system as ATM prevents the childhood neurodegenerative syndrome ataxia telangiectasia (A-T)32, 34. Similarly, mutation of components of the MRN complex can lead to an A-T like disease (ATLD) in the case of certain MRE11 mutations, or Nijmegen breakage syndrome (NBS) after NBS1 mutation, which is characterized by microcephaly35–37. The MRN complex is also essential for HR38, and so the disease-causing mutations leading to ATLD or NBS are hypomorphic, which still allow cell replication to proceed. In contrast to ATM, the related ATR kinase is central for signaling DNA replication stress and responds to replication protein A-coated single-stranded DNA, a common lesion formed during replication fork collapse39. ATR is critical to prevent ATR-Seckel syndrome, a neurodevelopmental disease that occurs from hypomorphic mutations of this gene40, 41. In this way, these two kinases function, largely independently, to address common types of DNA damage that occurs during neural development (Figure 2).
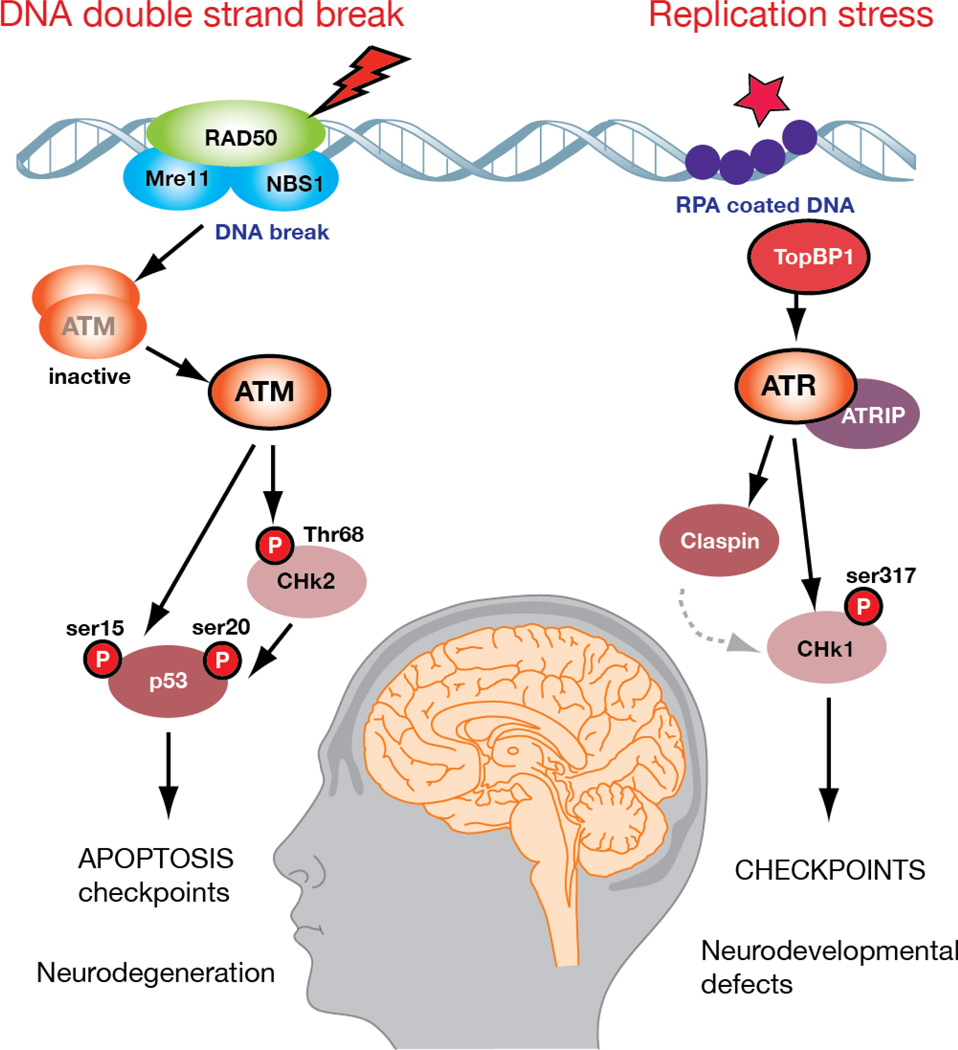
Figure 2. DNA damage signaling in the nervous sytem involves ATM and ATR.
ATM (ataxia telangiectasia, mutated) and ATR (ATM and RAD3 related) are DNA damage-activated kinases that response to specific (and different) types of DNA lesions. ATM responds to DNA double strand breaks while ATR responds to replication protein A (RPA)-coated single strand DNA. In response to DNA damage ATM is activated by the MRN complex and is converted from an inactive dimer to an active kinase that phosphorylates numerous substrates including p53 and Chk2 to activate apoptosis, or to initiate cell cycle arrest. In contrast, inresponse to DNA damage during replication, ATR is activated by TopBP1 in an ATRIP-dependent manner to phosphorylate various substrates including Claspin and Chk1 which promote cell cycle checkpoint activation. Although functional interactions between ATM and ATR have been suggested, these two related kinases function largely independently in response to different types of DNA damage. Inactivation of ATM can lead to neurodegeneration, while hypomorphic mutation of ATR can lead to neurdevelopmental defects and ATR-Seckel syndrome.
Neurogenesis and Susceptibility to DNA Damage
Neural progenitors are particularly sensitive to DNA damage and their relative susceptibility can vary depending on the stage of development. For example, TopBP1 (Topoisomerase II binding protein 1), a protein essential for maintaining DNA integrity during replication42, and a key activator of the DNA damage response kinase ATR (Figure 2) is critical for survival of early-born neural progenitors26. Mice in which TopBP1 was inactivated in early cortical progenitors using Emx1-cre showed substantial neurodevelopmental abnormalities throughout the cortex that resulted from widespread apoptosis of newborn progenitors26. Notably, when TopBP1 was deleted in later stage cortical progenitors using Nes-cre, cortical progenitor loss was markedly reduced; this was despite TopBP1 deletion by either Nes-cre or Emx1-cre resulting in similar amounts of DNA damage in E13.5 neural progenitors31. Thus the striking difference in cortical phenotype after deletion via the different cre-drivers was not due to different amounts of DNA strand breaks, but rather the enhanced susceptibility of earlier-born cortical progenitors to DNA damage after TopBP1 loss. This enhanced sensitivity of earlier progenitors to DNA damage may indicate a lower threshold for apoptosis compared with those progenitors generated at later developmental stages. The propensity for apoptosis after DNA damage in very early progenitors is a preferred way to maintain genome integrity rather than risk progenitor expansion with misrepaired DNA43.
Differential susceptibility of progenitors to DNA damage at different stages of cellular differentiation may be linked to cell cycle dynamics, an important feature that characterizes the changing properties of neural progenitors. Cell-cycle regulation of neural progenitors is a key aspect of cortical development, and involves dynamic changes in G1-phase and S-phase duration44–47. The length of these cell-cycle phases are linked to progenitor proliferation compared to differentiation, as G1-phase lengthening is associated with differentiating progenitors (Figure 3). A key difference between proliferating and neurogenic progenitors is the length of S-phase; an approximately 3-fold longer S-phase is characteristic of neural progenitor expansion compared with progenitors committed to the neurogenic lineage44. This suggests that S-phase duration is an important determinant of proliferative capacity of cortical progenitors. Proliferative progenitors that are not lineage-restricted likely have a greater need for repair of DNA replication-associated damage to avoid transmission of genetic errors during early progenitor expansion.
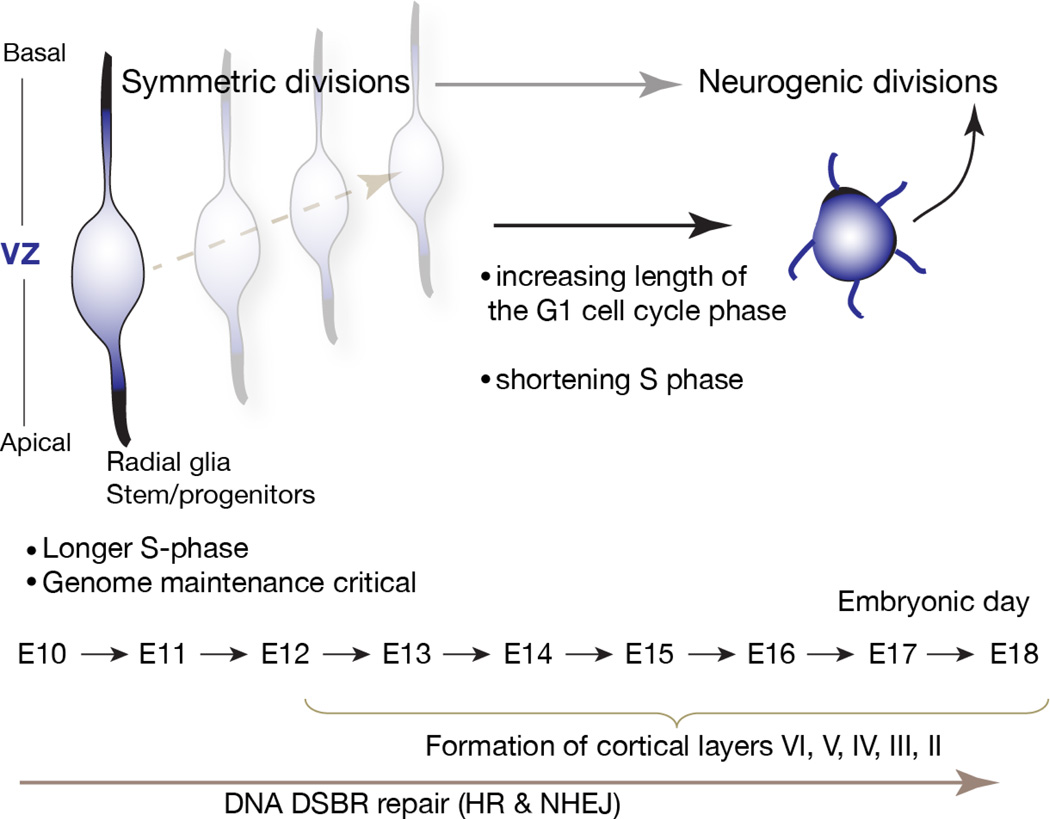
Figure 3. The cell cycle of cortical progenitors change during development.
Cortical progenitors undergo symmetric divisions during E10-12 and at this stage have a long S-phase of around 8 hours. The transition to neurogenic divisions results in a shortened S-phase and as progenitors undergo differentiation the G1 phase lengthens. Chromatin states involving high mobility group A proteins are also important for maintaining an open chromatin conformation in early cortical progenitors. Genome maintenance is paramount in early-born apical neural progenitors, and an increased S-phase length likely allows enhanced genome surveillance to ensure a pristine genome. Cortical layers form progressively from E12; VZ is the ventricular zone. DNA repair via homologous recombination (HR) and non-homologous end-joining (NHEJ) is important to ensure genome integrity during progenitor proliferation and differentiation. Figure adapted from reference 25.
Additionally, recent findings indicate that chromatin state is important for the neurogenic potential of cortical progenitors48. Chromatin accessibility is likely an important feature of early progenitors prior to neurogenic division and may be critical for rapid proliferation and the undifferentiated state. An important mediator of chromatin condensation in cortical progenitors is the high mobility group A proteins, that promote chromatin opening by competitive interaction with histone H149, 50. Correspondingly, Hmga1 levels are increased in cortical progenitors during neurogenesis and the presence of this protein can regulate neurogenic potential in these cells48. Chromatin state is also an important determinant of the DNA damage response and DNA repair26, 51–56. More generally, regulation of chromatin structure is a key feature of neural development and later function of the nervous system57–60. How chromatin state directly affects DNA damage signaling in neural during development and in the mature brain is not yet clear, but will undoubtedly impact aspects of neural genome stability.
Genome Stability in the Mature Nervous System
Genome stability mechanisms in the mature nervous system differ to those during neurogenesis because of the lack of cell division and the absence of replicative DNA damage and repair via HR (Figure 1). For instance, DNA damage in the mature nervous system can involve DNA breaks arising from oxidative stress, which may have as a main consequence perturbation of cellular homeostasis or an impact upon transcription61–64. Furthermore, as distinct to proliferating and immature neural cells that are proficient for DNA damage-induced apoptosis43, differentiated cells do not typically engage apoptosis after DNA damage65, 66. Additionally, within the mature nervous system there are many non-neuronal cells and the response to DNA damage in these are likely similar to neurons. Studies using cortical astrocytes clearly show that these cells mount a normal DNA damage response26, 31. DNA damage is also strongly linked to aging and cognitive decline, processes that highlight the impact in the mature brain of accumulating DNA lesions that may arise from attenuated DNA repair processes17.
Oxidative DNA damage is a primary consideration as the brain metabolizes 20% of all consumed oxygen67, a substantial portion of which is required for basic cellular house-keeping functions in addition to synaptic and cognitive functions68. A key lesion that impact the mature brain are breaks in one strand of DNA, termed DNA single strand breaks. This type of damage is repaired by either the XRCC1-dependent single strand repair pathway/BER or by transcription-coupled repair, a component of the nucleotide excision pathway12, 18, 62. The relevance of single strand break repair in the brain is highlighted by multiple neurologic syndromes arising from inherited defects in components of this pathway (Figure 4). This lesion is considered the most common insult to DNA in the cell and in the order of a thousand breaks may occur per cell per day6. The close relationship between oxidative stress and DNA strand breaks in the nervous system was shown by suppression of DNA damage in mice with disabled NHEJ repair after reduction of the cellular oxygen tension, or the increase in breaks by overexpressing the antioxidant enzyme superoxide dismutase69. An additional source of endogenous DNA double strand breaks may arise via normal topoisomerase-II-beta function during transcription70. These site-specific transcription-associated DNA breaks localize to gene promoters and involve components of the DNA repair machinery, and may contribute to accumulated DNA damage found in the aged brain4.
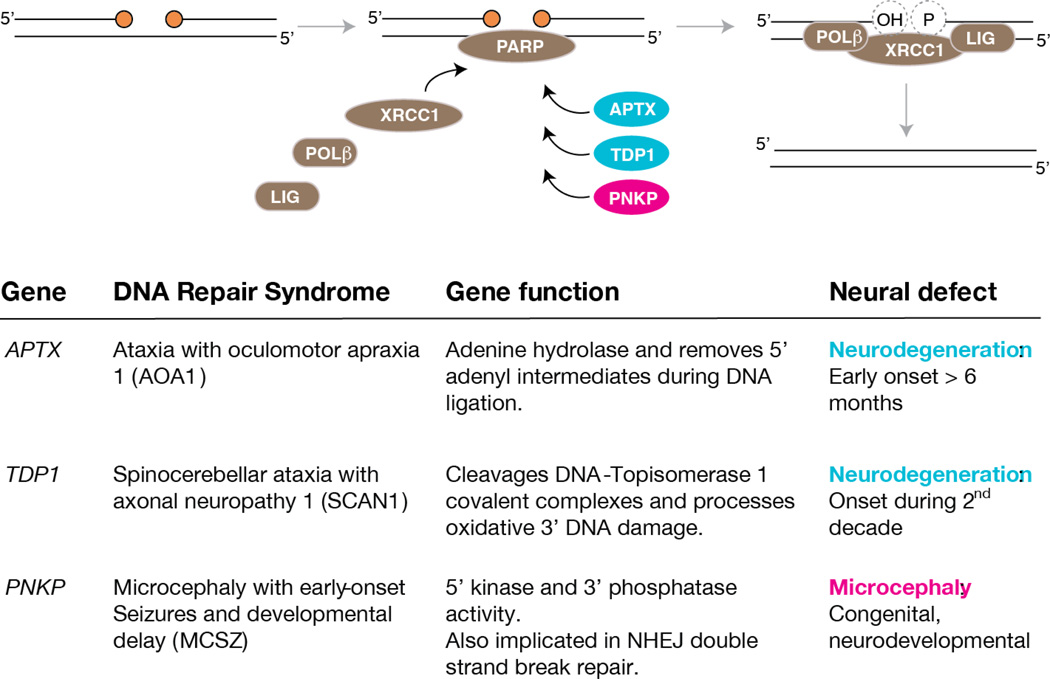
Figure 4. Defective DNA single strand break repair can result in syndrome with varied neuropathology.
Single strand breaks are a frequent type of DNA lesion in the nervous system. These lesions are repaired by an XRCC1-mediated pathway that includes polymerase β and ligases to complete repair after end-processing factors process damaged DNA termini to faciliate ligation. Factors such as TDP1 processes 3’ termini and APTX processes 5’ lesions involving adenylation of DNA, while PNKP can process both 5’ and 3’ termini. Defects in APTX, TDP1 lead to human neurodegeneratiove syndromes while disruption of PNKP leads to microcephaly, not neurdegeneration. Notably, despite these factors participating in the same DNA repair pathway the resultant neuropathology is distinct.
One notable feature of the mature nervous system is ongoing neurogenesis in certain brain regions including the lateral ventricle and the hippocampus71, 72. Recent data indicate adult hippocampal neurogenesis occurs at the rate of seven hundred new neurons per day (around a 1.75% turnover) suggesting that this ongoing process is an important feature of hippocampal function73. These new hippocampal neurons will also require genome maintenance during proliferation, and therefore will be competent for HR and other DNA damage response pathways typical of neurogenesis.
Neurologic disease results from loss of genome stability
It is clear from the varied neuropathology present in multiple DNA repair deficient human syndromes that the nervous system is highly susceptible to different types of DNA damage (Table 1). These syndromes can affect different brain regions and impact the nervous system at different developmental times resulting in a spectrum of neuropathology spanning neurodegeneration, neurodevelopmental disorders or brain tumors5.
As discussed, a prime source of DNA lesions during neurogenesis is replication-associated damage and this contributes to defects leading to neurodevelopmental syndromes. In the mature brain the high oxygen consumption and free radicals produced by cellular metabolism can lead to abundant single-strand breaks, which can compromise genetic integrity and perturb cellular homeostasis leading to interference with transcription6, 18. Defects in any of the steps involved in responding to these types of damage can result in syndromes marked by neurologic disease5, 6.
While identification of the gene mutation responsible for a disease will indicate the affected DNA repair pathway, this alone may not necessarily be sufficient to explain the resultant neuropathology. For example, defects in individual components of DNA single strand break repair might be expected to result in similar phenotypes. Three distinct diseases that result from defective single strand break repair are ataxia with oculomotor apraxia 1 (AOA1) in which the nucleotide hydrolase Aprataxin (APTX) is defective, spinocerebellar ataxia with axonal neuropathy 1 (SCAN1) in which tyrosyl-DNA phosphodiesterase 1 is mutated and microcephaly with seizures (MCSZ) resulting from mutations in polynucleotide kinase/phosphatase (PNKP)74–77. APTX processes 5’ adenylation intermediates78, while TDP1 cleaves topoisomerase-1-DNA complexes and can also resolve oxidative DNA lesions6 and the dual 5’kinase/3’phosphatse activity of PNKP restores the ends of a DNA strand break for ligation (Figure 4)79.
The phenotype of AOA1 and SCAN1 are characterized by spinocerebellar ataxia and neurodegeneration, although the onset of AOA1 is early, around 2–4 years while in SCAN1 a later onset neurodegenerative phenotype occurs in the teenage years75–77. Additionally, both syndromes show cerebellar atrophy and progressive degenerative changes including peripheral neuropathy, although AOA1 manifests oculomotor apraxia while this defect is not present in SCAN1. A complicating aspect in directly comparing these syndromes is the limited number of SCAN1 individuals identified, as this disease has currently been linked to multiple individuals from a single family, while AOA1 is a common recessive ataxia in certain geographical areas75–77. In contrast to AOA1 and SCAN1, MCSZ is neurodevelopmental rather than neurodegenerative74. In this disease the function of PNK is severely compromised because of hypomorphic PNKP mutations and an associated reduction in protein levels, which results in a DNA repair defect80. Thus, despite faulty end-processing of DNA damage underlying these diseases, the phenotypes are quite distinct, raising the question of why there are such differences between these syndromes.
Given that the disease phenotypes above correlate with the relative requirements for each factor in processing DNA damage, and that all three factors interact with XRCC1, a central scaffolding protein that enhances the relative repair rate of SSBs6, then it may be that the lesions that require APTX occur more frequently than those dependent upon TDP1 for repair. In general, the differences are unlikely to reflect tissue-specificity because cerebellar dysfunction underpins the age-related cerebellar ataxia in both syndromes75–77. However, other differences between AOA1 and SCAN1, such as oculomotor apraxia, may involve tissue-specific utilization of each DNA repair factor. With regard to PNKP, the 3’-phosphatase activity of this enzyme is a critical for global rates of single strand break repair, and a high percentage of this type of break after endogenous oxidative stress will utilize PNKP79, 81. This predicts that loss of PNKP would show heightened DNA repair deficiency than either APTX or TDP1 loss and a worsened neurodegenerative phenotype than AOA1 or SCAN1. Because MCSZ results in pronounced microcephaly at birth, this may indicate a higher relative importance for PNKP repair functions associated with increased DNA damage during neurogenesis. Additionally, because PNKP has also been linked to DNA double strand break repair79, and defects in this pathway can also lead to microcephaly82, perhaps the main developmental function for PNKP is repair of this lesion? Thus, the small brain size in MCSZ may result from DNA double strand break-induced apoptosis during development5. However, this explanation is not easy to reconcile because deficiency in the repair of DNA double strand breaks impacts tissues outside of the nervous system, and this is not the situation with MCSZ74. The ongoing development of mouse models for these and other diseases will provide important reagents to determine the molecular basis for the different neurologic phenotypes resulting from various DNA lesions.
Centrosomes, genome stability and neurologic disease
Multiple microcephaly syndromes have been identified in which the targets of the developmental defects are cortical progenitors, and functional analysis of the gene product in these syndromes strongly implicates the centrosome83, 84. Intriguingly, related syndromes also characterized by microcephaly and neurodevelopmental abnormalities arise directly from mutation of DNA damage response factors such as ATR40, 41. Further, mutations in the centrosomal factor pericentrin can result in Seckel syndrome associated with defective DNA damage signaling85. Collectively, these findings implicate an important connection between the DNA damage response and the centrosome.
The centrosome is a key cellular organelle in cortical progenitors as it can regulate migration and also cellular division symmetry, which is important determinant in symmetric versus neurogenic division86–88. Recent data has shown that centrosomes can also integrate cell cycle checkpoints and DNA repair after genotoxic stress83, 85, 89–91. Additionally, key DNA damage response factors such as ATR and NBS1 and cell cycle checkpoint factors Chk1 and Chk2 can localize to centrosomes after DNA damage to effect cell cycle arrest and modulate mitosis83, 86, 90, 92. In Seckel syndrome arising from specific mutations of ATR (ATR-Seckel syndrome) or centrosomal protein 152 (CEP152) that result in Seckel syndrome (SCKL5), supernumery centrosomes were found after ATR dysfunction and defective DNA damage signaling was observed when CEP152 function was compromised40, 41, 92. Thus a connection is apparent between the DNA damage response, centrosome function and microcephaly. Understanding how the DNA damage response interfaces with centrosome function and neural development will be important for understanding the genesis of human microcephaly and other related neurodevelopmental disorders.
Mitochondrial genome stability in the nervous system
In addition to the nuclear genome, there is also the ~16 kb circular mitochondrial genome, which is also susceptible to DNA damage. While DNA damage and repair occurs in mitochondria, the mitochondrial genome may be most susceptible to replication defects. Faulty mtDNA replication such as that resulting from germline mutations of DNA polymerase γ (POLG) mutations can lead to a wide spectrum of clinically distinct syndromes, many of which affect the nervous system93. In particular, the diversity of different phenotypes including multiple neurologic diseases, which result from specific point mutations in POLG, illustrate the striking outcomes of defective mtDNA replication94. In large part, these replication defects impact components of the mtDNA-encoded electron transport chain and oxidative phosphorylation, and the different phenotypes resulting from POLG mutations may reflect tissue-specific mitochondrial distribution and energy requirements.
Active mtDNA repair processes have been extensively demonstrated by cellular studies95, as has the mitochondrial localization of key BER/single strand break repair factors including those linked to neurodegenerative disease such as Aprataxin96. Thus, defective mtDNA repair may contribute to the phenotype of neurodegenerative diseases95. Moreover, there is a mitochondrial specific version of DNA ligase III that can participate in mtDNA replication and repair. Inactivation of this DNA ligase in the nervous system leads to a profound postnatal loss of cerebellar granule neurons and neuraxis-wide defects leading to pronounced ataxia, underscoring the paramount importance of mtDNA integrity for neural function97.
However, given that there can be hundreds of individual mitochondria in a cell, then mtDNA repair deficiency (if damage is stochastic) will sporadically affect individual mitochondria and the subsequent cellular impact is not as obvious as general mtDNA replication defects. Additionally, the presence of mitochondrial fusion as a means of safeguarding mtDNA integrity98 suggests additional strategies to prevent adverse effects from mitochondrial genome disruption. Nonetheless, the existence of mtDNA repair capacity suggests that this process is important for mitochondrial homeostasis, and associated defects in mtDNA repair could exacerbate the neurodegenerative phenotypes in human DNA repair deficient human syndromes.
Perspectives and conclusions
The occurrence of DNA damage is a feature of nervous system development and maintenance and an armamentarium of DNA repair factors exist to prevent this damage from persisting and having pathogenic consequences. Human DNA repair deficiency syndromes provide direct examples of the need for vigilant DNA repair processes, which are needed to resolve multiple types of DNA damage. Furthermore, the impact upon the nervous system in these syndromes provides critical insight as to how specific DNA repair factors are utilized throughout this tissue. However, many key questions remain unanswered regarding the outcomes of defective DNA damage signaling in the nervous system. For example, disruption of DNA double strand break repair generally results in neuropathology but also includes more general systemic defects, while disruption of DNA single stand break repair leads to pathology that mostly, if not exclusively, involves the nervous system. Furthermore, how dysfunction of these DNA repair pathways lead to different pathology such as neurodegeneration compared with microcephaly remains unclear.
The prevalence of endogenous DNA damage throughout the neuraxis also raises the specter of DNA damage contributing to pathology in other neurologic syndromes. DNA damage has been considered a likely contributing factor in Parkinson’s and Alzheimer’s disease99, 100, and recently DNA double strand break repair deficiency was found to exacerbate the effects of amyloid precursor protein in the context of Alzheimer’s disease, with a potential impact upon synaptic function3. It will be important to determine if DNA lesions impact other neurologic syndromes and if these lesions occur as collateral damage during disease processes. Understanding how DNA damage impacts the nervous system will be essential for developing appropriate therapeutics that can prevent damage or enhance functionality of the DNA damage response pathways.
Acknowledgments
P.J.M. is supported by the US National Institutes of Health (NS-37956, CA-96832), a Cancer Center Support grant (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital.
Footnotes
COMPETINGFINANCIALINTERESTS
The author declares no competing financial interests.
References
- 1.Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 5.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 7.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes & development. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiricny J. The multifaceted mismatch-repair system. Nature reviews. Molecular cell biology. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 12.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends in genetics : TIG. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Molecular cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 15.Woodbine L, et al. PRKDC mutations in a SCID patient with profound neurological abnormalities. The Journal of clinical investigation. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anttinen A, et al. Neurological symptoms and natural course of xeroderma pigmentosum. Brain : a journal of neurology. 2008;131:1979–1989. doi: 10.1093/brain/awn126. [DOI] [PubMed] [Google Scholar]
- 17.Borgesius NZ, et al. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci. 2011;31:12543–12553. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nature reviews. Molecular cell biology. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 19.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 21.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. Embo J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 23.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes & development. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frappart PO, et al. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nature medicine. 2005;11:538–544. doi: 10.1038/nm1228. [DOI] [PubMed] [Google Scholar]
- 25.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. Embo J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, et al. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nature neuroscience. 2012;15:819–826. doi: 10.1038/nn.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 28.Nijhawan D, Honarpour N, Wang X. Apoptosis in neural development and disease. Annual review of neuroscience. 2000;23:73–87. doi: 10.1146/annurev.neuro.23.1.73. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, et al. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 30.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, et al. ATR maintains select progenitors during nervous system development. Embo J. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature reviews. Molecular cell biology. 2013;14:197–210. [PubMed] [Google Scholar]
- 33.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews. Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annual review of pathology. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 35.Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA repair. 2004;3:1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 37.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 38.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509, 514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews. Molecular cell biology. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Driscoll M, Dobyns WB, van Hagen JM, Jeggo PA. Cellular and clinical impact of haploinsufficiency for genes involved in ATR signaling. Am J Hum Genet. 2007;81:77–86. doi: 10.1086/518696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 42.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 43.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arai Y, et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nature communications. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caviness VS, Jr, Nowakowski RS, Bhide PG. Neocortical neurogenesis: morphogenetic gradients and beyond. Trends in neurosciences. 2009;32:443–450. doi: 10.1016/j.tins.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 47.Gotz M, Huttner WB. The cell biology of neurogenesis. Nature reviews. Molecular cell biology. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 48.Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nature neuroscience. 2012 doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- 49.Catez F, et al. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. Embo J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 52.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murr R, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 54.Sulli G, Di Micco R, d'Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer. 2012;12:709–720. doi: 10.1038/nrc3344. [DOI] [PubMed] [Google Scholar]
- 55.Sun Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker SA, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Molecular cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature reviews. Molecular cell biology. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 63.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes & development. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 64.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chemical reviews. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 65.Latella L, Lukas J, Simone C, Puri PL, Bartek J. Differentiation-induced radioresistance in muscle cells. Mol Cell Biol. 2004;24:6350–6361. doi: 10.1128/MCB.24.14.6350-6361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fortini P, Ferretti C, Dogliotti E. The response to DNA damage during differentiation: Pathways and consequences. Mutation research. 2013;743–744:160–168. doi: 10.1016/j.mrfmmm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Barzilai A. The contribution of the DNA damage response to neuronal viability. Antioxidants & redox signaling. 2007;9:211–218. doi: 10.1089/ars.2007.9.211. [DOI] [PubMed] [Google Scholar]
- 68.Du F, et al. Tightly coupled brain activity and cerebral ATP metabolic rate. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6409–6414. doi: 10.1073/pnas.0710766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR. Oxygen metabolism causes chromosome breaks and is associated with the neuronal apoptosis observed in DNA double-strand break repair mutants. Curr Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]
- 70.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 71.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 73.Spalding KL, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen J, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nature genetics. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat.Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 76.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat.Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 77.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat.Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 78.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 79.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reynolds JJ, Walker AK, Gilmore EC, Walsh CA, Caldecott KW. Impact of PNKP mutations associated with microcephaly, seizures and developmental delay on enzyme activity and DNA strand break repair. Nucleic Acids Res. 2012;40:6608–6619. doi: 10.1093/nar/gks318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breslin C, Caldecott KW. DNA 3'-phosphatase activity is critical for rapid global rates of single-strand break repair following oxidative stress. Mol Cell Biol. 2009;29:4653–4662. doi: 10.1128/MCB.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O'Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 83.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes & development. 2011;25:2011–2024. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends in genetics : TIG. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffith E, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gruber R, et al. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nature cell biology. 2011 doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 87.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends in neurosciences. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 88.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alderton GK, et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8:725–733. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 90.Shimada M, Komatsu K. Emerging connection between centrosome and DNA repair machinery. J Radiat Res (Tokyo) 2009;50:295–301. doi: 10.1269/jrr.09039. [DOI] [PubMed] [Google Scholar]
- 91.Tibelius A, et al. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalay E, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nature genetics. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 94.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annual review of medicine. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Souza-Pinto NC, Wilson DM, 3rd, Stevnsner TV, Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA repair. 2008;7:1098–1109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sykora P, Croteau DL, Bohr VA, Wilson DM., 3rd Aprataxin localizes to mitochondria and preserves mitochondrial function. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang E, et al. The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death. Nat Cell Biol. 2010;12:563–571. doi: 10.1038/ncb2058. [DOI] [PubMed] [Google Scholar]
- 100.Nouspikel T, Hanawalt PC. When parsimony backfires: neglecting DNA repair may doom neurons in Alzheimer's disease. Bioessays. 2003;25:168–173. doi: 10.1002/bies.10227. [DOI] [PubMed] [Google Scholar]