Abstract
Background
DDX39B (BAT1) encodes an RNA helicase known to regulate expression of TNF and IL-6. Elevated levels of these two cytokines are associated with increased severity of clinical malaria. The aim of this study was to investigate the relationship between single nucleotide polymorphisms (SNPs) in the DDX39B, TNF and IL6 genes and the clinical outcomes of patients with Plasmodium vivax malaria.
Methods
Cross-sectional investigations were carried out in two regions of the Brazilian Amazon where several studies on the pathogenesis of vivax malaria had been performed. Individuals were categorized according to infection status as well as clinical presentation into the following groups: uninfected, asymptomatic infection, mild infection, or complicated infection. Polymorphisms were identified using PCR restriction fragment-length polymorphism analysis and the restriction enzymes NlaIII or NcoI. The plasma levels of cytokines were determined using ELISA.
Results
The G allele of DDX39B-22C > G was associated with absent or decreased manifestations of malaria and the C allele was a risk factor for disease complications. Study participants heterozygous for TNF-308 (GA) and DDX39B-348 (CT) had higher TNF levels than wild-type participants. Haplotypes that included DDX39B (-22C > G and -348C > T) and TNF polymorphisms were not directly associated with mild or complicated malaria infections; however, haplotypes AGC, ACC, GGT, AGT and ACT were associated with increased TNF levels. Participants with genotype combinations GC/CC/GG/GG and GG/CT/GG/GG (DDX39B-22/DDX39B-348/TNF-308/IL6-176) had decreased and increased risk of mild malaria, respectively, compared with asymptomatic and uninfected participants. GC/CC/GG/GG was linked to decreased TNF and IL-6 levels.
Conclusions
This is the first study to describe patients with DDX39B and IL6 SNPs who had vivax malaria. These findings support the postulation that a set of mutations in immune-related genes is associated with inflammatory mediators and the clinical outcomes of patients with malaria.
Keywords: DDX39B (BAT1), Single nucleotide polymorphisms, Immune response, Plasmodium vivax, Malaria
Background
Plasmodium vivax malaria is a major worldwide threat, with two to three billion people remaining at risk of infection [1]. The clinical outcomes of patients with vivax malaria range from asymptomatic infection to complicated, and potentially lethal disease [2]. The risk for progression of severe Plasmodium falciparum malaria has been thought to be partly accounted for by host genetic factors [3]. These genetic factors are likely to involve several genetic alterations in immune mediators as well as molecules involved in mechanisms of cytoadherence and haemoglobinopathies [4]. Understanding the key host genetic determinants of susceptibility to malaria is critical for developing better therapies and vaccine strategies.
The nuclear protein HLA-B-associated transcript 1 (BAT1) is an RNA helicase encoded by the DDX39B gene (DEAD [Asp-Glu-Ala-Asp] box polypeptide 39B, also known as BAT1). BAT1 has been described as a negative regulator of inflammation by modulating expression of proinflammatory cytokines, such as TNF and IL-6, which suggests that it plays a protective role in several immune-mediated disorders [5]. The -22C > G and -348C > T polymorphisms in the promoter region of DDX39B have been shown to affect its transcriptional activity and the binding of nuclear transcription factors such as YY1 and Oct1 oligonucleotides at the respective positions -348 and -22 relative to the transcription start site of DDX39B[6]. TNF (=TNFalpha or TNFA) and IL-6 are key mediators associated with malaria symptoms, and their levels are increased in proportion to the severity of disease [7-12]. The hypothesis that DDX39B may have an effect on the clinical presentation of malaria by its modulation of the expression of proinflammatory cytokines involved in the pathogenesis of the disease is appealing, but has not yet been tested.
Single nucleotide polymorphisms (SNPs) in the TNF (-308G > A) and IL6 (-176G > C) genes may regulate the plasma levels of these cytokines; however, the mechanism of regulation is not yet fully understood [13-15]. Given the major role of the host immune system in P. vivax infection, the aim of this study was to determine whether mutations in the DDX39B, TNF and IL6 genes were associated with the clinical outcomes of patients with vivax malaria. The frequency of SNPs in DDX39B (-22C > G and -348C > T), TNF (-308G > A) and IL6 (-176G > C) were compared between P. vivax-infected study participants who exhibited different clinical outcomes, including asymptomatic infection, mild malaria, complicated malaria, and no infection. Associations between these immune-related mutations and plasma levels of TNF, IL-6, C-X-C motif chemokine 10 (CXCL10), and C-reactive protein (CRP) were also tested. The results reported here revealed that a combination of DDX39B, TNF and IL6 host genotypes were associated with manifestations of malaria, mainly by altering plasma levels of TNF and IL-6.
Methods
Study participants
This report describes series of patients from two distinct studies. The first study performed retrospective analyses of cryopreserved heparinized blood samples from participants living in riverine communities of the state of Rondônia, in the Brazilian Western Amazon, who were recruited between 2006 and 2007, as previously described [7,16-23]. Malaria was diagnosed using two methods: 1) microscopic examination of a thick blood smear performed by professionals at the Brazilian National Foundation of Health (FUNASA); and 2) polymerase chain reaction (PCR) performed at the Oswaldo Cruz Foundation (FIOCRUZ), Salvador, Brazil, as previously described [16-18]. The study included individuals who had been living in the endemic area for more than six months. Exclusion criteria included conditions known to interfere with the parameters evaluated in this report, such as coinfections and chronic diseases: P. falciparum infection confirmed by nested PCR; documented or referred viral hepatitis (hepatitis A, B, C, D virus [HAV, HBV, HCV, HDV]); chronic alcoholism; human immunodeficiency virus (HIV) infection; yellow fever; dengue; leptospirosis; tuberculosis; Hansen disease; visceral leishmaniasis; cancer and/or other chronic degenerative disease; sickle cell trait; and the use of hepatotoxic and immunosuppressant drugs. A total of 257 participants were enrolled in this first part of the study. As reported previously, all asymptomatic participants infected with P. vivax who were identified by active case detection were monitored for 30 days for the evaluation of malaria manifestations [16-18]. Participants who were positive for P. vivax infection but remained without acute febrile signs for 30 days were considered to be asymptomatic cases. Those individuals with positive parasitaemia and with mild symptoms were considered to have mild vivax malaria. Thus, the individuals from this region were divided into three groups as follows: mild malaria (n = 76), asymptomatic malaria (n = 104) and uninfected controls (n = 77). All symptomatic cases were treated following the guidelines of the National Foundation of Health, Brazil, and received chloroquine for three days and primaquine (0.5 mg/kg/day) for seven days. The asymptomatic carriers were treated after the monitoring period, as reported previously [17]. This first part of the study was approved by the Ethics Committee of the São Lucas University, Rondônia, Brazil; and all participants provided written informed consent.
Participants with complicated Plasmodium vivax malaria
Because there was a small number and unavailable blood samples of malaria cases (n = 9) with signs/symptoms of complicated disease among the first series of patients recruited in Rondonia, in order to evaluate the role of DDX39B (BAT1) polymorphism in complicated cases of vivax malaria, a second series of patients (second part of this report) was recruited from the state of Amazonas in the Western Brazilian Amazon between 2009 and 2013. Individuals of all ages who were hospitalized with an unidentified acute febrile syndrome at the reference hospital from the Fundação de Medicina Tropical Dr Heitor Vieira Dourado (FMT-HVD), Amazonas, Brazil, were tested for malaria using microscopic examination of a thick blood thick smear, and those with PCR-confirmed P. vivax were recruited. Patients were excluded for the following conditions: microscopic or molecular diagnosis of P. falciparum or P. vivax and P. falciparum malaria (mixed infection), serologic diagnosis of viral hepatitis (HAV, HBV, HCV, and HDV), HIV, or leptospirosis. Patients with vivax malaria with primaquine-induced haemolysis (patients taking primaquine with decreasing haemoglobin levels to <10 g/dL and reticulocyte counts >1.5%, or increased indirect bilirubin levels after primaquine treatment) were also excluded. The study participants from this second series had either mild signs/symptoms of acute malaria (mild malaria, n = 69) or clinical complications, as listed in Table 1 (complicated vivax malaria, n = 31). Complicated vivax malaria was defined according to the criteria for severe malaria from the World Health Organization (WHO) or based on the presence of hyperbilirubinaemia (serum total bilirubin > 51.3 μmol/L) [24]. Although all the participants in this second study were PCR positive, some of these study patients with complicated malaria (just for this particular group) had already started therapy for malaria before blood samples were taken, which might have affected the results of the laboratory and parasitaemia evaluations shown in Table 1. This second part of the study was approved by the Ethics Committee of the FMT-HVD, and all participants provided written informed consent.
Table 1.
Characteristics of the second series of participants with complicated vivax malaria: Amazonas, Brazil
| |
|
|
Clinical presentation at admission |
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|---|
| Patient no. | Gender | Age (years) | Major manifestation | Secondary manifestation | Previous treatment | Parasitaemia | Disease duration (days) | Haemoblogin (g/dL) | Total bilirubin (μmol/L) |
| 1 |
M |
59 |
hyperbilirubinaemia |
|
No |
6,222 |
7 |
16.9 |
403.56 |
| 2 |
F |
38 |
hyperlactemia |
|
Yes |
0 |
10 |
10.9 |
30.78 |
| 3 |
M |
31 |
hyperbilirubinaemia |
|
No |
3,298 |
10 |
11.0 |
54.72 |
| 4 |
F |
37 |
hyperbilirubinaemia |
|
No |
67,467 |
7 |
8.9 |
51.30 |
| 5 |
M |
18 |
hyperbilirubinaemia |
|
No |
1,848 |
4 |
13.5 |
165.87 |
| 6 |
M |
45 |
severe anaemia |
hyperbilirubinaemia |
No |
243 |
10 |
6.5 |
138.51 |
| 7 |
F |
40 |
severe anaemia |
|
No |
33,553 |
10 |
7.0 |
27.36 |
| 8 |
M |
66 |
convulsion |
|
Yes |
185 |
14 |
13.0 |
* |
| 9 |
M |
17 |
hyperbilirubinaemia |
|
No |
226 |
6 |
11.9 |
53.01 |
| 10 |
M |
31 |
hyperbilirubinaemia |
|
No |
5,937 |
15 |
13.5 |
165.87 |
| 11 |
M |
1 |
severe anaemia |
|
No |
38,712 |
9 |
5.0 |
8.55 |
| 12 |
M |
26 |
hyperbilirubinaemia |
|
No |
128 |
2 |
10.6 |
61.56 |
| 13 |
F |
4 months |
convulsion |
prostration |
No |
108,033 |
7 |
11.7 |
* |
| 14 |
M |
19 |
severe anaemia |
|
No |
0 |
17 |
6.5 |
15.39 |
| 15 |
M |
50 |
respiratory failure |
|
No |
310 |
5 |
10.1 |
8.55 |
| 16 |
F |
35 |
severe anaemia |
hyperbilirubinaemia |
No |
34,673 |
1 |
4.8 |
138.51 |
| 17 |
F |
41 |
respiratory failure |
|
No |
24,815 |
8 |
9.9 |
39.33 |
| 18 |
F |
18 |
hyperbilirubinaemia |
|
No |
670 |
5 |
11.7 |
61.56 |
| 19 |
M |
44 |
severe anaemia |
|
Yes |
0 |
7 |
6.9 |
13.68 |
| 20 |
F |
23 |
severe anaemia |
|
Yes |
264 |
8 |
6.4 |
6.84 |
| 21 |
M |
5 |
prostration |
|
No |
291 |
15 |
8.2 |
6.84 |
| 22 |
M |
21 |
hyperbilirubinaemia |
|
Yes |
832 |
7 |
9.7 |
102.6 |
| 23 |
M |
12 |
respiratory failure |
|
Yes |
0 |
6 |
7.8 |
15.39 |
| 24 |
F |
74 |
respiratory failure |
|
No |
325 |
5 |
11.7 |
29.07 |
| 25 |
M |
37 |
respiratory failure |
|
Yes |
0 |
10 |
7.1 |
17.10 |
| 26 |
F |
26 |
hyperbilirubinaemia |
|
No |
254 |
9 |
11.3 |
140.22 |
| 27 |
M |
53 |
hyperbilirubinaemia |
|
No |
45,833 |
6 |
11.8 |
59.85 |
| 28 |
F |
36 |
hyperbilirubinaemia |
|
No |
0 |
3 |
7.9 |
85.50 |
| 29 |
F |
15 |
severe anaemia |
|
Yes |
8,894 |
8 |
6.9 |
11.97 |
| 30 |
M |
22 |
hyperbilirubinaemia |
|
No |
3,054 |
7 |
15.1 |
73.53 |
| 31 | M | 25 | hyperbilirubinaemia | Yes | 27,954 | 3 | 15.0 | 138.51 | |
Severe anaemia was defined as haemoglobin levels below 7 g/dL for adults and below 5 g/dL for children and hyperbilirubinaemia by serum total bilirubins >51.3 μmol/L. Respiratory failure was defined as tachypnea, shortness of breath, mental confusion clinical signs of hypoxaemia (central and/or peripheral cyanosis). Previous treatment indicates participants who already started malaria therapy before the blood sample collection. *Total bilirubin was not measured in these individuals.
Genotyping
DNA was extracted from 200 μL of peripheral blood using a standard Qiagen DNA blood mini kit (Valencia, CA, USA) according to the manufacturer’s protocol. The SNPs at positions -22 (C > G) (rs2239527; G ancestral allele) and -348 (C > T) (rs2239528 ;C ancestral allele) in the promoter region of DDX39B and at position -176 of IL6 (G > C) (rs2234683; G ancestral allele) were typed using PCR restriction fragment-length polymorphism analysis with the restriction enzyme NlaIII (New England Biolabs), according to protocols previously published by Ramasawmy et al.[25] and Yalcin et al.[26], respectively. TNF-308 (G > A) (rs1800629; G ancestral allele) polymorphism was also evaluated using PCR restriction fragment-length polymorphism analysis with the restriction enzyme NcoI (New England Biolabs), as published previously [27]. DDX39B and TNF PCR products were electrophoresed on 10% polyacrylamide gels and IL6 products were separated by electrophoresis on 1% agarose gels under nondenaturing conditions. The PCR products were then detected by staining with ethidium bromide and visualized under ultraviolet illumination.
Plasma measurements
The plasma levels of IL-6, CXCL10, and TNF were measured using a cytometric bead array system (BD Biosciences Pharmingen, Franklin Lakes, NJ, USA) according to the manufacturer’s protocol. CRP levels in plasma were measured using the turbidimetric immunoassay method, performed at the Federal University of Bahia and Faculdade São Lucas, Brazil.
Statistical analysis
Categorized variables (genotypes, alleles, haplotypes, and genotype combinations) were compared using the Chi-square test or Fisher exact test in 2x2 contingency tables along with the relevant odds ratio (OR) and 95% confidence interval (CI). Univariate linear regression analysis was performed to assess the associations between combinations of genotypes and malaria symptomatology. Ordinal variables were evaluated using the Mann–Whitney (between two groups) or Kruskal-Wallis test followed by the Dunn multiple comparison test or trend analysis (when more than two groups were compared). Hardy-Weinberg equilibrium (HWE) was assessed for the different groups by comparing the observed number of different genotypes with those expected under HWE for the estimated allele frequency. The power of this study was calculated based on a medium effect size, a significance level of 0.05, and four degrees of freedom (Chi-square test). A power of 98.3% was found for the first series of participants (n = 257) and a power of 71.10% for the second series of participants (n = 110). Statistical analyses were performed using GraphPad Prism (version 5.0b) software (GraphPad Software, San Diego, CA, USA) or R version 2.15.1 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Baseline characteristics of first series of participants
All different groups of vivax malaria infection had a slightly majority of women. Participants with asymptomatic malaria were older than the uninfected participants and those with mild malaria (P = 0.0264; Table 2). Participants with asymptomatic malaria reported a higher number of previous infections (median 15.0, interquartile range (IQR) 12.00-18.75) than the uninfected participants or those with mild infection (P < 0.0001; Table 2). In addition, participants with asymptomatic P. vivax infection reported living for a longer time in the endemic area (71.15% more than ten years) than those in the other groups (P = 0.0030; Table 2). These results were expected, since this set of patients was a subsample of a larger cohort of individuals where similar results were found [7,16-23].
Table 2.
Baseline characteristics of the first series of participants enrolled in the first part of the study: Rondonia, Brazil
| Uninfected (n = 77) | Asymptomatic vivax malaria (n = 104) | Mild vivax malaria (n = 76) | P value | |
|---|---|---|---|---|
|
Male - no. (%) |
36 (46.75) |
47 (45.19) |
36 (47.37) |
0.9548** |
|
Median (IQR*) age (yr) |
35.00 (25.50-45.00) |
42.00 (32.00-49.00) |
36.00 (27.25-50.00) |
0.0264*** |
|
Median (IQR*) of previous malaria episodes |
12.00 (6.00-17.00) |
15.00 (12.00-18.75) |
6.00 (1.00-12.75) |
<0.0001*** |
|
Time residing in the area (yr) |
|
|
|
|
| ≤2 |
17 (22.08) |
24 (23.08) |
24 (31.58) |
0.0011** |
| 3 to 10 |
19 (24.67) |
6 (5.77) |
16 (21.05) |
|
| >10 | 41 (53.25) | 74 (71.15) | 36 (47.37) |
* IQR, interquantile range; **Categorized variables were compared using Chi-square test; ***Ordinal variables were compared using the Kruskal-Wallis test with the Dunn multiple comparison test
Single nucleotide polymorphisms
Genotype and allele distributions of IL6-176G > C and TNF-308G > A polymorphisms were compared in the main clinical groups and in participants stratified according to mild vs asymptomatic vivax malaria infection, infected vs uninfected and asymptomatic vs uninfected (Table 3). No association was found between alleles or genotypes of IL6 and TNF SNPs and the different clinical outcomes of vivax malaria (Table 3). The distributions of IL6-176G > C and TNF-308G > A genotypes in all categories were under HWE. Table 4 shows the results for DDX39B polymorphisms (-22C > G and -348C > T) according to the same categories of participants (Table 4). No differences were observed for the SNPs DDX39B-22C > G and DDX39B-348C > T with regard to mild malaria, asymptomatic malaria, or infection status. The distribution of DDX39B-22C > G in the asymptomatic participants was not under HWE (x2 = 6.10, P = 0.0134), suggesting those participants reflect evolutionary selective pressure. The frequencies of DDX39B genotypes were under HWE in all the other categories of vivax malaria infection.
Table 3.
TNF (-308G > A) and IL-6 (-176G > C) polymorphisms and outcome of vivax malaria infection: first series of participants from Rondonia, Brazil
| |
Genotype |
χ2 |
Allele frequency |
χ2 | GG vs GA + AA χ2 | |||
|---|---|---|---|---|---|---|---|---|
| GG n (%) | GA n (%) | AA n (%) | G (%) | A (%) | ||||
|
TNF-308
polymorphism |
|
|
|
|
|
|
|
|
| Uninfected (n = 77) |
60 (77.92) |
15 (19.48) |
2 (2.60) |
2.12 p = 0.7134# |
135 (87.66) |
19 (12.34) |
0.57 p = 0.7509 |
0.96 p = 0.6178 |
| Asymptomatic malaria (n = 104) |
78 (75.00) |
25 (24.04) |
1 (0.96) |
181 (87.02) |
27 (12.98) |
|||
| Mild malaria (n = 76) |
54 (71.05) |
21 (27.63) |
1 (1.32) |
129 (84.87) |
23 (15.13) |
|||
| Infected vs uninfected |
|
|
|
p = 0.3401*# |
|
|
p = 0.6745* |
P = 0.5316* |
| Mild vs asymptomatic |
|
|
|
p = 0.6059*# |
|
|
p = 4798* |
P = 0.4323* |
| Asymptomatic vs uninfected |
|
|
|
p = 0.5866*# |
|
|
p = 8749* |
P = 0.7252* |
| |
Genotype |
χ2 |
Allele frequency |
χ2 |
GG
vs
GC + CC χ2 |
|||
| |
GG n (%) |
GC n (%) |
CC n (%) |
G (%) |
C (%) |
|||
|
IL6-176
polymorphism |
|
|
|
|
|
|
|
|
| Uninfected (n = 77) |
49 (63.64) |
23 (29.87) |
5 (6.49) |
2.16 p = 0.7058 |
121 (78.57) |
33 (21.43) |
1.61 p = 0.4462 |
1.91 p = 0.3854 |
| Asymptomatic malaria (n = 104) |
60 (57.69) |
38 (36.54) |
6 (5.77) |
158 (75.96) |
50 (24.04) |
|||
| Mild malaria (n = 76) |
40 (52.63) |
30 (39.47) |
6 (7.90) |
110 (72.37) |
42 (27.63) |
|||
| Infected vs uninfected |
|
|
|
1.56 p = 0.4579 |
|
|
p = 0.3695* |
p = 0.2702* |
| Mild vs asymptomatic |
|
|
|
1.30 p = 0.5214 |
|
|
p = 0.2618* |
p = 0.2711* |
| Asymptomatic vs uninfected | 0.88 p = 0.6435 | p = 0.0819* | p = 0.4458* | |||||
*In these cases, Fisher exact test was used. #Analysis excluded genotype AA. χ2: coefficient and P value measured using Chi-square test. Infected individuals represent symptomatic plus asymptomatic cases.
Table 4.
DDX39B polymorphisms (-22C > G and -348C > T) and outcome of vivax malaria infections: first series of participants from Rondonia, Brazil
| |
Genotype |
χ2 |
Allele frequency |
χ2 | CC vs CG + GG χ2 | |||
|---|---|---|---|---|---|---|---|---|
| CC n (%) | CG n (%) | GG n (%) | C (%) | G (%) | ||||
|
DDX39B-22
polymorphism |
|
|
|
|
|
|
|
|
| Uninfected (n = 77) |
10 (12.99) |
38 (49.35) |
29 (37.66) |
6.75 p = 0.1496 |
58 (37.66) |
96 (62.34) |
2.35 p = 0.3088 |
0.53 p = 0.7660 |
| Asymptomatic malaria (n = 104) |
15 (14.42) |
64 (61.54) |
25 (24.04) |
94 (45.19) |
114 (54.81) |
|||
| Mild malaria (n =7 6) |
13 (17.10) |
34 (44.74) |
29 (38.16) |
60 (39.47) |
92 (60.53) |
|||
| Infected vs uninfected |
|
|
|
1.48 p = 0.4758 |
|
|
p = 0.3281* |
p = 0.2460* |
| Mild vs asymptomatic |
|
|
|
2.90 p = 0.2339 |
|
|
p = 0.6244* |
p = 0.7028* |
| Asymptomatic vs uninfected |
|
|
|
3.98 p = 0.1364 |
|
|
p = 0.1625* |
p = 0.8307* |
| |
Genotype |
|
Allele frequency |
|
CC
vs
TC + TT χ2 |
|||
| |
CC n (%) |
TC n (%) |
TT n (%) |
|
C (%) |
T (%) |
|
|
|
DDX39B-348
polymorphism |
|
|
|
|
|
|
|
|
| Uninfected (n = 77) |
64 (83.12) |
12 (15.58) |
1 (1.30) |
1.99 p = 0.3691# |
140 (90.91) |
14 (9.09) |
1.00 p = 0.6041 |
1.56 p = 0.4586 |
| Asymptomatic malaria (n = 104) |
83 (79.81) |
21 (20.19) |
0 (0.00) |
187 (89.90) |
21 (10.10) |
|||
| Mild malaria (n = 76) |
57 (75.00) |
19 (25.00) |
0 (0.00) |
133 (87.50) |
19 (12.50) |
|||
| Infected vs uninfected |
|
|
|
p = 0.2397*# |
|
|
p = 0.5342* |
p = 0.4012* |
| Symptomatic vs asymptomatic |
|
|
|
p = 0.2372*# |
|
|
p = 0.3469* |
p = 0.3108* |
| Asymptomatic vs uninfected | p = 0.5594*# | p = 0.8578* | p = 0.7008* | |||||
*In these cases, Fisher exact test was used. #Analysis excluded genotype TT. χ2: coefficient and P value measured using Chi-square test. Infected individuals represent symptomatic plus asymptomatic cases.
Association of single nucleotide polymorphisms with inflammatory mediators levels
The SNPs were assessed regarding association with systemic levels of TNF, IL-6, CXCL10, and CRP, which have been associated with vivax malaria manifestations [23]. Participants with the TNF-308 GA genotype or A allele had higher levels of TNF than those with the GG genotype or G allele (P = 0.0347 and P = 0.0296, respectively, Figure 1A,B). Participants with the DDX39B-348 CT genotype or T allele had higher concentrations of TNF than those with the CC genotype or C allele (P = 0.0215 and P = 0.0299, respectively, Figure 1A,B). Polymorphism at DDX39B-22 appeared to correlate with serum concentrations of CRP and CXCL10, but not IL-6. Participants with the DDX39B-22 CC genotype exhibited higher levels of CRP than those with the GC genotype (P = 0.0395, Figure 1C) but this difference was not observed in the allele analysis (P = 0.0622, Figure 1D). No relevant associations were found between genotypes or alleles of DDX39B-22 C > G and plasma IL-6 levels (P = 0.0496 and P = 0.7475, respectively, Figure 1E,F). Furthermore, participants carrying the DDX39B-22 CG genotype had lower CXCL10 levels than those with the GG genotype (P = 0.0294, Figure 1E), although no difference was seen in the allele analysis (P = 0.0898, Figure 1F).
Figure 1.
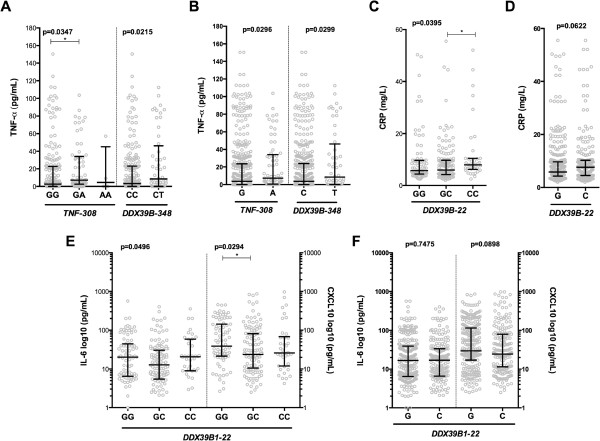
Influence of genetic alterations on levels of inflammatory mediators. Each symbol represents a single patient, and the lines represent medians and interquartile range. (A) Levels of TNF according to genotypes of TNF-308G > A and DDX39B-348C > T polymorphisms. Analysis excluded TT (DDX39B-348C > T), because only 1 uninfected participant had this genotype. (B) Levels of TNF according to alleles of TNF-308G > A and DDX39B-348C > T polymorphisms. (C) CRP plasma levels according to different DDX39B-22C > G genotypes. (D) CRP plasma levels according to different DDX39B-22C > G alleles. (E) IL-6 and CXCL10 levels according to DDX39B-22C > G genotypes. (F) IL-6 and CXCL10 levels according to DDX39B-22C > G alleles. The data were compared using the Mann–Whitney test (comparisons between two groups), the Kruskal-Wallis test with Dunn multiple comparisons or linear trend analysis (comparisons between more than 2 groups). P values are shown in each panel.
Haplotypes and combinations of genotypes
DDX39B and TNF are located in the same major histocompatibility complex (MHC) region on chromosome 6, and therefore assessment of the association of these polymorphisms with the outcomes of malaria can be performed using haplotype analysis. The assessment of haplotypes representing all possible combinations of TNF-308, DDX39B-22 and DDX39B-348 SNPs was unable to identify significant association with any of the clinical categories of vivax malaria infection (Figure 2A). Nevertheless, haplotypes were associated with differential expression of inflammatory mediators in plasma, which could potentially influence the degree of immunopathology and malaria manifestations. TNF-308, DDX39B-22, and DDX39B-348 haplotypes GGC and GCC were linked with lower concentrations of CRP and CXCL10, respectively, than other haplotypes (P = 0.0246, Figure 2B; P = 0.0071, Figure 3C; respectively). Intriguingly, several haplotypes were associated with increased plasma TNF levels (Figure 2D). Thus, because they were found to be associated with elevated or decreased levels of CRP, CXCL10, and TNF, haplotypes may be associated with the outcomes of vivax malaria infection.
Figure 2.

Haplotype influence on inflammatory mediators and outcomes of malaria infection. Each symbol represents a single patient, and the lines represent medians and interquartile range. (A) The figure represents all the possible haplotypes for TNF-308G > A, DDX39B-348C > T, and DDX39B-22C > G polymorphisms. Haplotypes were compared between the 3 categories of vivax malaria infections (uninfected, asymptomatic, mild malaria). (B) CRP levels in participants with GGC haplotype compared to individuals with other haplotypes. (C) CXCL10 plasma concentration in participants with GCC haplotype compared to individuals with other haplotypes. (D) TNF levels in participants with different haplotypes. Each haplotype was compared with participants with other haplotypes. The differences between groups illustrated in (A) were compared using the chi-square test. The data in (B), (C), and (D) were compared using the Mann–Whitney test. P values are shown in each panel.
Figure 3.
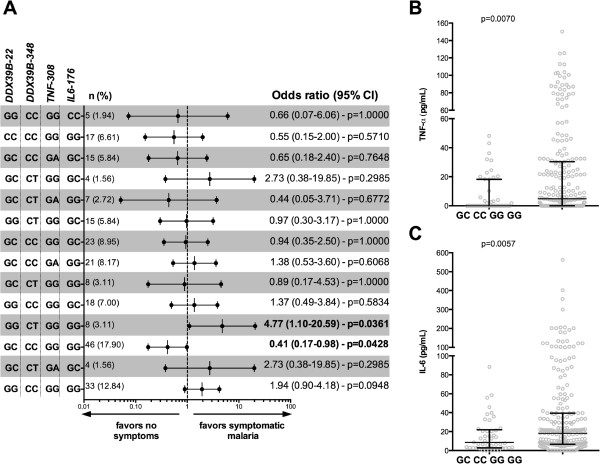
Genotype combinations and outcomes of malaria infection. (A) Univariate linear regression analysis of the different combination of TNF-308G > A, IL6-176G > C, DDX39B-348C > T, and DDX39B-22C > G genotypes. The odds ratios and respective 95% confidence intervals (95% CI) are shown according to each combination of genotypes compared with the other combinations. (B-C) Each symbol represents a single patient, and the lines represent medians and interquartile range. TNF and IL-6 levels in participants with the GC/CC/GG/GG combination compared with the other combinations. The data were compared using the Mann–Whitney test. P values are shown in each panel.
Analysis of combinations of genotypes was performed to determine association with manifestations of vivax malaria. Participants with DDX39B-22/DDX39B-348/TNF-308/IL6-176 genotype combinations GC/CC/GG/GG and GG/CT/GG/GG had decreased and increased risk, respectively, of developing manifestations of malaria relative to asymptomatic participants (OR 0.41, 95% CI 0.17-0.98, P = 0.0428; OR 4.77, 95% CI 1.10-20.59, p = 0.0361; respectively, Figure 3A). Moreover, the genotype combination GC/CC/GG/GG was associated with lower TNF and IL-6 levels than other genotypes (P = 0.0070 and P = 0.0057, respectively; Figure 3B,C), suggesting that this set of genotypes may protect against more severe malaria because of association with reduced levels of inflammatory cytokines.
Second series of study participants: DDX39B polymorphisms and complicated vivax malaria
An additional aim of this combined study was to specifically evaluate the association between DDX39B polymorphisms and complicated P. vivax malaria. Patients screened in a reference hospital from the Brazilian Amazon who manifested complicated malaria were compared with those presenting with mild disease. Participants with mild malaria had a median age of 36 years (IQR 27–46) and 81.16% were male (n = 56). Participants with complicated disease had a similar age (median 31 years, IQR 18–41; P = 0.0563) and there was a slightly lower proportion of males (19 of 31 [61.29%], P = 0.0460). Characteristics of the patients with complicated malaria are shown in Table 1. Interestingly, the patients in this study who were categorized with complicated vivax malaria exhibited more often hyperbilirubinaemia (serum total bilirubin >51.3 μmol/L), which has been reported to be a common complication of patients with vivax malaria [28,29].
There was significant difference in the distribution frequency of the polymorphism DDX39B-22C > G in the participants with mild versus complicated malaria (x2 = 6.72, P = 0.0347). The proportion of -22C homozygosity among patients with complicated and mild malaria was 25.80% (n = 8) and 7.25% (n = 5), respectively (Table 5). Participants were categorized based on the presence or absence of the G allele (genotypes GG and GC vs genotype CC) and the G allele was significantly more frequent in patients with mild vivax malaria than in patients with complicated infection (P = 0.0207, Table 5). No differences were seen for the polymorphism DDX39B (-348C > T) regarding the distribution of genotypes and alleles in patients with mild and complicated malaria (Table 5). The distribution of DDX39B-22C > G genotypes in the patients with mild malaria was not under HWE (x2 = 5.22, P = 0.0222). The DDX39B genotypes were under HWE in the patients with complicated malaria.
Table 5.
DDX39B polymorphisms (-22C > G and -348C > T) and complicated vivax malaria infections: second series of participants from Amazonas, Brazil
| |
Genotype |
χ2 |
Allele frequency |
χ2 | CC vs CG + GG χ2 | |||
|---|---|---|---|---|---|---|---|---|
| CC n (%) | CG n (%) | GG n (%) | C (%) | G (%) | ||||
|
DDX39B-22
polymorphism |
|
|
|
|
|
|
|
|
| Mild malaria (n = 69) |
5 (7.25) |
41 (59.42) |
23 (33.33) |
6.72 p = 0.0347 |
51 (36.96) |
87 (63.04) |
p = 0.0628* |
p = 0.0207* |
| Complicated malaria (n = 31) |
8 (25.80) |
16 (51.61) |
7 (22.58) |
32 (51.61) |
30 (48.39) |
|||
| |
Genotype |
|
Allele frequency |
|
CC
vs
TC + TT χ2 |
|||
| |
CC n (%) |
TC n (%) |
TT n (%) |
|
C (%) |
T (%) |
|
|
|
DDX39B-348
polymorphism |
|
|
|
|
|
|
|
|
| Mild malaria (n = 69) |
55 (79.71) |
14 (20.29) |
0 (0.00) |
p = 7,850*# | 124 (89.86) |
14 (10.14) |
p = 0.7965* | p = 7,850* |
| Complicated malaria (n = 31) | 26 (83.87) | 5 (16.13) | 0 (0.00) | 57 (91.93) | 5 (8.07) | |||
*In these cases, Fisher exact test was used. #Analysis excluded genotype TT. χ2: coefficient and P value measured using Chi-square test.
Discussion
Immunity of the human host to malaria is likely to be mediated by T-cell recognition of Plasmodium sp. epitopes on infected host cells via class I and II MHC antigens [30]. Given the importance of the MHC to the immune response, genetic studies of the human MHC I have correlated polymorphisms in this region with susceptibility to malaria. The alleles A*30:01 and A*33:01 of MHC I were found to be associated with malaria severity in Mali [30-32]. The DDX39B (BAT1) gene is located on chromosome 6 near the TNF gene in the same MHC region and seems to influence expression of several immune-related genes [6]. This study found that the C allele of DDX39B-22C > G is a potential risk factor of complicated vivax malaria in the Brazilian Amazon. This finding may be expected, as this allele has been associated with reduced binding to transcription factors and expression of inflammatory cytokines [5]. Haplotype analysis (including DDX39B and TNF polymorphisms) found that genetic alterations in these immune-mediated genes may influence disease outcome by altering TNF plasma levels. In addition, the genotype combinations GC/CC/GG/GG and GG/CT/GG/GG, corresponding to the respective polymorphisms DDX39B-22/DDX39B-348/TNF-308/IL6-176, were associated with a decreased or increased risk, respectively, of developing mild vivax malaria, probably by altering TNF and IL-6 levels. To the best of our knowledge, this is the first report assessing the relationship between DDX39B polymorphisms and malaria outcomes, and also one of the few studies to analyze the manifestations of P. vivax infections in relation to combinations of immune-related genotypes.
Plasma concentrations of IL-6 have been found to be associated with severe disease and death from malaria, and the IL6-176C allele was associated with increased expression of IL-6 in neonates and adults developing acute phase reactions [14,15,33,34]. A study of sympatric ethnic groups in Mali found that the frequency of IL6 CG/GG genotypes was higher in non-Fulani ethnic groups, who have increased susceptibility to malaria, in both symptomatic and asymptomatic falciparum malaria cases [35]. However, in our study, there were no differences in the distribution of IL6-176G > C in the participants making up the different clinical groups of vivax malaria, which may indicate that this polymorphism plays different roles in P. vivax infection and/or in a Brazilian population. Genetic changes in TNF have been described in several studies of different populations throughout the world, and there have sometimes been contradictory results [23]. The SNP TNF-308G > A has been associated with increased susceptibility, resistance or there has been no association with severity to malaria caused by P. falciparum[36-39]. A study of a population in the Brazilian Amazon that was similar to our population found that one TNF haplotype (TNF-1031T/-863A/-857T/-308G/-238G) including the TNF-308G allele was associated with increased susceptibility to mild vivax malaria [40]. Our study did not find an association between TNF-308G and clinical manifestations of malaria. It is noteworthy that TNF-308G > A may not be a causal mutation, and that this polymorphism could be in linkage disequilibrium with other causal mutations located close to the TNF gene [41]. Thus, this polymorphism may have a moderate effect or may have an epistatic effect on DDX39B mutations [42]. Polymorphisms in DDX39B (BAT1) have been described in several diseases with inflammatory profiles, including neuropathy, myasthenia gravis, allergies, Alzheimer disease, myocardial infarction, hepatitis, rheumatoid arthritis, Chagas disease, among others [25,43-48]. In this study it was found that the G allele of DDX39B-22C > G may be a resistance factor to malaria and the C allele a risk factor for disease complications. It has been reported that DDX39B promoter polymorphisms alter the binding of transcription factors (YY1 and Oct1) and may affect the transcription of this gene, and the sequences with -22G and -348 T alleles were expressed more efficiently than sequences containing -22C and-348C alleles [6]. An in vitro study found that BAT1 appeared to decrease the expression of TNF and IL-6 [5]; thus the G allele of DDX39B-22C > G, which enhances the expression of BAT1, may be protective against complicated malaria by decreasing the expression of proinflammatory cytokines.
Although the polymorphisms described in this study are not directly associated with the clinical manifestations of malaria, they can indirectly influence disease by altering the levels of inflammatory mediators involved in disease immunopathology. High plasma levels of TNF are related to the pathogenesis of signs associated with malaria, such as fever, and severe forms of infection, such as cerebral malaria and severe anemia [49]. Our study found that participants heterozygous (AG) for TNF-308G > A had higher plasma concentrations of TNF than homozygous participants with the wild-type (GG) polymorphism. The A allele of this SNP has been associated with increased production of TNF in several studies, and was often associated with the HLA-A1-B8-DR3 haplotype of the MHC region [50-53]. In this study results did not clearly demonstrate increased levels of TNF in participants homozygous for the A mutation, probably because of the small number of participants with this genotype who were recruited for the study. Furthermore, participants heterozygous (CT) for DDX39B-348C > T had higher plasma TNF than homozygous participants with the wild-type (CC) polymorphism, suggesting that an additional genetic mechanism appears to be associated with increased levels of this cytokine, and consequently the clinical outcome of malaria infection. CRP is an acute-phase inflammatory protein, and this study findings indicate that participants homozygous (CC) for DDX39B-22 (G > C) had increased levels of CRP, supporting an association of the C allele with risk of complicated malaria.
A single-point mutation is often not sufficient for predicting the susceptibility or resistance of individuals to malaria [4]. Another approach to investigating the differences in response to malaria infection is haplotype analysis of mutant alleles. DDX39B is situated in the central region of MHC on the short arm of human chromosome 6 and is approximately 150 kb from the TNF gene. The NFKBIL1 gene, which encodes the inhibitor of κB-like protein (IκBL), a protein of unknown function, is situated between DDX39B and TNF[54]. Genetic variations in NFKBIL1 are associated with susceptibility to inflammatory conditions such as periodontitis, chronic thromboembolic pulmonary hypertension, rheumatoid arthritis, and malaria [54-58]. Although DDX39B and TNF genes are near each other on the same chromosome and appear to influence the transcription of its gene products [5], this study did not identify a haplotype with DDX39B (22C > G and 348C > T) and TNF-308G > A polymorphisms that increased the risk of clincal vivax malaria. However, many haplotypes appear to markedly increase TNF levels, indirectly contributing to malaria susceptibility. It is noteworthy that conclusive findings from DDX39B and TNF haplotype analysis are limited by the considerable distance between these genes (approximately 150 kb). Similarly, it is reported here an analysis of susceptibility to clinical manifestations of malaria as a result of genotype combinations found that CG/CC/GG/GG and GG/CT/GG/GG, corresponding to the respective polymorphisms DDX39B-22/DDX39B-348/TNF-308/IL6-176, were associated with decreased and increased risk, respectively, of developing clinical manifestations of P. vivax infection. Intriguingly, TNF-308 and IL6-176 genotypes were wild-type homozygotes (GG for both) among the combinations, and changes were related to DDX39B genotypes. The substitution of DDX39B genotypes (-22 and -348) in the combinations completely changed the risk of developing manifestations of malaria, from susceptibility to resistance to illness from vivax malaria infections. These results lend support to the role of DDX39B as a regulatory gene that can alter transcription factors and inflammatory cytokines and influence the clinical outcome of inflammatory diseases [5,25]. Moreover, study participants with the genotype combination described here that was associated with resistance against manifestations of P. vivax infection (CG/CC/GG/GG) also had lower levels of proinflammatory TNF and IL-6, suggesting that DDX39B confers protection against malaria pathogenesis by reducing the inflammatory response.
This study was limited because of a small numbers of participants, which may have reduced its ability to detect significant differences between study groups. Indeed, the detection of a small difference (effect size) between groups at a significance level of 0.05 ideally would require at least 1,194 study participants. Therefore, our study results require validation in larger studies.
Conclusion
Genetic alterations in the immune response against vivax malaria may predispose individuals to disease complications or protect them from clinical disease. The findings of this study provide support that the C allele of DDX39B-22C > G is a risk factor of complicated vivax malaria, and different haplotypes (including DDX39B and TNF polymorphisms) may influence disease outcomes by altering plasma levels of TNF. Moreover, the results suggest that combinations of genotypes (including IL6-176G > C) are associated with a decreased or increased risk of developing clinical manifestations of malaria and may also influence plasma TNF and IL-6 levels. Further prospective studies should be able to determine if these genetic determinants are critical for protection against the development of clinical P. vivax infection, and also identify individuals who are at risk of developing more complicated forms of malaria.
Abbreviations
DDX39B: DEAD [Asp-Glu-Ala-Asp] box polypeptide 39B; BAT1: HLA-B associated transcript 1; RNA: Ribonucleic acid; TNF: Tumor necrosis factor; IL-6: Interleukin 6; SNP: Single nucleotide polymorphism; CXCL10: C-X-C motif chemokine 10; CRP: C-reactive protein; FUNASA: Brazilian National Foundation of Health; PCR: Polymerase chain reaction; FIOCRUZ: Oswaldo Cruz Foundation; HAV: Hepatitis A virus; HBV: Hepatitis B virus; HCV: Hepatitis C virus; HDV: Hepatitis D virus; HIV: Human immunodeficiency virus; FMT-HVD: Fundação de Medicina Tropical Dr Heitor Vieira Dourado; DNA: Deoxyribonucleic acid; OR: Odds ratio; CI: Confidence interval; HWE: Hardy-Weinberg equilibrium; IQR: Interquartile range; MHC: Major histocompatibility complex; IκBL: Inhibitor of κB-like protein.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VRRM performed the experiments, analyzed the data, and wrote the manuscript together with BBA and MBN. LCLS and GCG helped perform the experiments. BBA performed the field study and sampling in Rondonia and performed the plasma measurements and wrote the manuscript. BMLM was responsible for the field study and sampling in Manaus. MVGL supervised the clinical study and sampling in Manaus. MBN conceptualized the study, supervised the clinical study in Rondonia, and helped with data interpretation and the writing of the manuscript. All authors have read and approved the final version of the manuscript.
Contributor Information
Vitor RR Mendonça, Email: vitorrosaramos@hotmail.com.
Ligia CL Souza, Email: ligiacorreialima@gmail.com.
Gabriela C Garcia, Email: gabibarreto92@hotmail.com.
Belisa ML Magalhães, Email: magalhaesbelisa@gmail.com.
Marcus VG Lacerda, Email: marcuslacerda.br@gmail.com.
Bruno B Andrade, Email: andradebd@niaid.nih.gov.
Marilda S Gonçalves, Email: mari@bahia.fiocruz.br.
Manoel Barral-Netto, Email: mbarral@bahia.fiocruz.br.
Acknowledgements
The authors thank João Gambati (FUNASA, Brazil), Luis Marcelo Camargo (USP, Brazil) and our team at FIOCRUZ-Bahia, Brazil: Jorge Tolentino, Adorielze Leite, Nataly Alexandrino and Andrezza Kariny, who help in logistics of the field study and experiments.
This work was supported by FIOCRUZ and FMT-HVD. MBN and MSG are senior investigators from CNPq. MVGL is scientific director at FMT-HVD. VRRM, LCLS, GCG, and BMLM are supported by CNPq. The work of BBA is supported by the intramural research programme of the National Institute of Allergy and Infectious Diseases, National Institute of Health. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- WHO. World Malaria Report 2013. Geneva: World Health Organization; 2013. [Google Scholar]
- Andrade BB, Barral-Netto M. Biomarkers for susceptibility to infection and disease severity in human malaria. Mem Inst Oswaldo Cruz. 2011;106(Suppl):70–78. doi: 10.1590/s0074-02762011000900009. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN. Heritability of malaria in Africa. PLoS Med. 2005;2:e340. doi: 10.1371/journal.pmed.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mendonça VRR, Goncalves MS, Barral-Netto M. The host genetic diversity in malaria infection. J Trop Med. 2012;2012:940616. doi: 10.1155/2012/940616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allcock RJ, Williams JH, Price P. The central MHC gene, BAT1, may encode a protein that down-regulates cytokine production. Genes Cells. 2001;6:487–494. doi: 10.1046/j.1365-2443.2001.00435.x. [DOI] [PubMed] [Google Scholar]
- Price P, Wong AM-L, Williamson D, Voon D, Baltic S, Allcock RJN, Boodhoo A, Christiansen FT. Polymorphisms at positions -22 and -348 in the promoter of the BAT1 gene affect transcription and the binding of nuclear factors. Hum Mol Genet. 2004;13:967–974. doi: 10.1093/hmg/ddh113. [DOI] [PubMed] [Google Scholar]
- Andrade BB, Reis-Filho A, Souza-Neto SM, Raffaele-Netto I, Camargo LMA, Barral A, Barral-Netto M. Plasma superoxide dismutase-1 as a surrogate marker of vivax malaria severity. PLoS Negl Trop Dis. 2010;4:e650. doi: 10.1371/journal.pntd.0000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M, Kaul A, Bali P, Raziuddin M, Singh MP, Singh OP, Dash AP, Adak T. Alleles -308A and -1031C in the TNF-alpha gene promoter do not increase the risk but associated with circulating levels of TNF-alpha and clinical features of vivax malaria in Indian patients. Mol Immunol. 2008;45:1682–1692. doi: 10.1016/j.molimm.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Aubouy A, Deloron P, Migot-Nabias F. Plasma and in vitro levels of cytokines during and after a Plasmodium falciparum malaria attack in Gabon. Acta Trop. 2002;83:195–203. doi: 10.1016/s0001-706x(02)00094-3. [DOI] [PubMed] [Google Scholar]
- Böttger E, Grangeiro de Carvalho E, Meese S, Kun JFJ, Esen M. Expression of interleukin-6 family receptors in NK92 cells is regulated by cytokines and not through direct interaction with Plasmodium falciparum-infected erythrocytes. J Interferon Cytokine Res. 2013;33:65–71. doi: 10.1089/jir.2012.0094. [DOI] [PubMed] [Google Scholar]
- Gonçalves RM, Scopel KKG, Bastos MS, Ferreira MU. Cytokine balance in human malaria: does Plasmodium vivax elicit more inflammatory responses than Plasmodium falciparum? PLoS One. 2012;7:e44394. doi: 10.1371/journal.pone.0044394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyke KE, Burges R, Cissoko Y, Sangare L, Dao M, Diarra I, Kone A, Harley R, Plowe CV, Doumbo OK, Sztein MB. Serum levels of the proinflammatory cytokines interleukin-1 beta (IL-1beta), IL-6, IL-8, IL-10, tumor necrosis factor alpha, and IL-12(p70) in Malian children with severe Plasmodium falciparum malaria and matched uncomplicated malaria or healthy controls. Infect Immun. 2004;72:5630–5637. doi: 10.1128/IAI.72.10.5630-5637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562–566. doi: 10.1002/jlb.66.4.562. [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Hulkkonen J, Wang XY, Hurme M. The promoter polymorphism of the interleukin-6 gene regulates interleukin-6 production in neonates but not in adults. Eur Cytokine Netw. 2001;12:62–68. [PubMed] [Google Scholar]
- Brull DJ, Montgomery HE, Sanders J, Dhamrait S, Luong L, Rumley A, Lowe GD, Humphries SE. Interleukin-6 gene -174 g > c and -572 g > c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler Thromb Vasc Biol. 2001;21:1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- Andrade BB, Rocha BC, Reis-Filho A, Camargo LMA, Tadei WP, Moreira LA, Barral A, Barral-Netto M. Anti-Anopheles darlingi saliva antibodies as marker of Plasmodium vivax infection and clinical immunity in the Brazilian Amazon. Malar J. 2009;8:121. doi: 10.1186/1475-2875-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade BB, Reis-Filho A, Barros AM, Souza-Neto SM, Nogueira LL, Fukutani KF, Camargo EP, Camargo LMA, Barral A, Duarte A, Barral-Netto M. Towards a precise test for malaria diagnosis in the Brazilian Amazon: comparison among field microscopy, a rapid diagnostic test, nested PCR, and a computational expert system based on artificial neural networks. Malar J. 2010;9:117. doi: 10.1186/1475-2875-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade BB, Reis-Filho A, Souza-Neto SM, Clarêncio J, Camargo LMA, Barral A, Barral-Netto M. Severe Plasmodium vivax malaria exhibits marked inflammatory imbalance. Malar J. 2010;9:13. doi: 10.1186/1475-2875-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade BB, Araújo-Santos T, Luz NF, Khouri R, Bozza MT, Camargo LMA, Barral A, Borges VM, Barral-Netto M. Heme impairs prostaglandin E2 and TGF-beta production by human mononuclear cells via Cu/Zn superoxide dismutase: insight into the pathogenesis of severe malaria. J Immunol. 2010;185:1196–1204. doi: 10.4049/jimmunol.0904179. [DOI] [PubMed] [Google Scholar]
- Andrade BB, Santos CJN, Camargo LM, Souza-Neto SM, Reis-Filho A, Clarêncio J, Mendonça VRR, Luz NF, Camargo EP, Barral A, Silva AAM, Barral-Netto M. Hepatitis B infection is associated with asymptomatic malaria in the Brazilian Amazon. PLoS One. 2011;6:e19841. doi: 10.1371/journal.pone.0019841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça VRR, Luz NF, Santos NJG, Borges VM, Gonçalves MS, Andrade BB, Barral-Netto M. Association between the haptoglobin and heme oxygenase 1 genetic profiles and soluble CD163 in susceptibility to and severity of human malaria. Infect Immun. 2012;80:1445–1454. doi: 10.1128/IAI.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozzelino R, Andrade BB, Larsen R, Luz NF, Vanoaica L, Seixas E, Coutinho A, Cardoso S, Rebelo S, Poli M, Barral-Netto M, Darshan D, Kühn LC, Soares MP. Metabolic Adaptation to tissue iron overload confers tolerance to malaria. Cell Host Microbe. 2012;12:693–704. doi: 10.1016/j.chom.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Mendonça VRR, Queiroz ATL, Lopes FM, Andrade BB, Barral-Netto M. Networking the host immune response in Plasmodium vivax malaria. Malar J. 2013;12:69. doi: 10.1186/1475-2875-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Guidelines for the treatment of malaria. Second edition. Geneva: World Health Organization; 2010. [Google Scholar]
- Ramasawmy R, Cunha-Neto E, Faé KC, Müller NG, Cavalcanti VL, Drigo SA, Ianni B, Mady C, Kalil J, Goldberg AC. BAT1, a putative anti-inflammatory gene, is associated with chronic Chagas cardiomyopathy. J Infect Dis. 2006;193:1394–1399. doi: 10.1086/503368. [DOI] [PubMed] [Google Scholar]
- Yalçın S, Kayaaltı Z, Söylemezoğlu T. Role of interleukin-6–174 G/C promoter polymorphism in trace metal levels of autopsy kidney and liver tissues. Int J Hyg Environ Health. 2011;214:219–224. doi: 10.1016/j.ijheh.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Wang T-N, Chen W-Y, Wang T-H, Chen C-J, Huang L-Y, Ko Y-C. Gene-gene synergistic effect on atopic asthma: tumour necrosis factor-alpha-308 and lymphotoxin-alpha-NcoI in Taiwan’s children. Clin Exp Allergy. 2004;34:184–188. doi: 10.1111/j.1365-2222.2004.01867.x. [DOI] [PubMed] [Google Scholar]
- Anand AC, Puri P. Jaundice in malaria. J Gastroenterol Hepatol. 2005;20:1322–1332. doi: 10.1111/j.1440-1746.2005.03884.x. [DOI] [PubMed] [Google Scholar]
- Fabbri C, de Cássia Mascarenhas-Netto R, Lalwani P, Melo GC, Magalhães BML, Alexandre MAA, Lacerda MVG, Lima ES. Lipid peroxidation and antioxidant enzymes activity in Plasmodium vivax malaria patients evolving with cholestatic jaundice. Malar J. 2013;12:315. doi: 10.1186/1475-2875-12-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyke KE, Fernández-Viňa MA, Cao K, Hollenbach J, Coulibaly D, Kone AK, Guindo A, Burdett LA, Hartzman RJ, Wahl AR, Hildebrand WH, Doumbo OK, Plowe CV, Sztein MB. Association of HLA alleles with Plasmodium falciparum severity in Malian children. Tissue Antigens. 2011;77:562–571. doi: 10.1111/j.1399-0039.2011.01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flori L, Sawadogo S, Esnault C, Delahaye NF, Fumoux F, Rihet P. Linkage of mild malaria to the major histocompatibility complex in families living in Burkina Faso. Hum Mol Genet. 2003;12:375–378. doi: 10.1093/hmg/ddg033. [DOI] [PubMed] [Google Scholar]
- Diakite M, Clark TG, Auburn S, Campino S, Fry AE, Green A, Morris AP, Richardson A, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP, Rockett KA. A genetic association study in the Gambia using tagging polymorphisms in the major histocompatibility complex class III region implicates a HLA-B associated transcript 2 polymorphism in severe malaria susceptibility. Hum Genet. 2009;125:105–109. doi: 10.1007/s00439-008-0597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day NP, Hien TT, Schollaardt T, Loc PP, Chuong LV, Chau TT, Mai NT, Phu NH, Sinh DX, White NJ, Ho M. The prognostic and pathophysiologic role of pro- and antiinflammatory cytokines in severe malaria. J Infect Dis. 1999;180:1288–1297. doi: 10.1086/315016. [DOI] [PubMed] [Google Scholar]
- Kern P, Hemmer CJ, Van Damme J, Gruss HJ, Dietrich M. Elevated tumor necrosis factor alpha and interleukin-6 serum levels as markers for complicated Plasmodium falciparum malaria. Am J Med. 1989;87:139–143. doi: 10.1016/s0002-9343(89)80688-6. [DOI] [PubMed] [Google Scholar]
- Israelsson E, Maiga B, Kearsley S, Dolo A, Homann MV, Doumbo OK, Troye-Blomberg M, Tornvall P, Berzins K. Cytokine gene haplotypes with a potential effect on susceptibility to malaria in sympatric ethnic groups in Mali. Infect Genet Evol. 2011;11:1608–1615. doi: 10.1016/j.meegid.2011.05.021. [DOI] [PubMed] [Google Scholar]
- McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- Clark TG, Diakite M, Auburn S, Campino S, Fry AE, Green A, Richardson A, Small K, Teo YY, Wilson J, Jallow M, Sisay-Joof F, Pinder M, Griffiths MJ, Peshu N, Williams TN, Marsh K, Molyneux ME, Taylor TE, Rockett KA, Kwiatkowski DP. Tumor necrosis factor and lymphotoxin-alpha polymorphisms and severe malaria in African populations. J Infect Dis. 2009;199:569–575. doi: 10.1086/596320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CG, May J, Luty AJ, Lell B, Kremsner PG. TNFalpha-308A associated with shorter intervals of Plasmodium falciparum reinfections. Tissue Antigens. 2002;59:287–292. doi: 10.1034/j.1399-0039.2002.590406.x. [DOI] [PubMed] [Google Scholar]
- Wattavidanage J, Carter R, Perera KL, Munasingha A, Bandara S, McGuinness D, Wickramasinghe AR, Alles HK, Mendis KN, Premawansa S. TNFalpha*2 marks high risk of severe disease during Plasmodium falciparum malaria and other infections in Sri Lankans. Clin Exp Immunol. 1999;115:350–355. doi: 10.1046/j.1365-2249.1999.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sortica VA, Cunha MG, Ohnishi MDO, Souza JM, Ribeiro-Dos-Santos AKC, Santos NPC, Callegari-Jacques SM, Santos SEB, Hutz MH. IL1B, IL4R, IL12RB1 and TNF gene polymorphisms are associated with Plasmodium vivax malaria in Brazil. Malar J. 2012;11:409. doi: 10.1186/1475-2875-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC, Keating BJ, Kwiatkowski DP. Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat Genet. 2004;36:394–399. doi: 10.1038/ng1331. [DOI] [PubMed] [Google Scholar]
- Flori L, Delahaye NF, Iraqi FA, Hernandez-Valladares M, Fumoux F, Rihet P. TNF as a malaria candidate gene: polymorphism-screening and family-based association analysis of mild malaria attack and parasitemia in Burkina Faso. Genes Immun. 2005;6:472–480. doi: 10.1038/sj.gene.6364231. [DOI] [PubMed] [Google Scholar]
- Cherry CL, Rosenow A, Affandi JS, McArthur JC, Wesselingh SL, Price P. Cytokine genotype suggests a role for inflammation in nucleoside analog-associated sensory neuropathy (NRTI-SN) and predicts an individual’s NRTI-SN risk. AIDS Res Hum Retroviruses. 2008;24:117–123. doi: 10.1089/aid.2007.0168. [DOI] [PubMed] [Google Scholar]
- Degli-Esposti MA, Leelayuwat C, Dawkins RL. Ancestral haplotypes carry haplotypic and haplospecific polymorphisms of BAT1: possible relevance to autoimmune disease. Eur J Immunogenet. 1992;19:121–127. doi: 10.1111/j.1744-313x.1992.tb00051.x. [DOI] [PubMed] [Google Scholar]
- Gnjec A, D’Costa KJ, Laws SM, Hedley R, Balakrishnan K, Taddei K, Martins G, Paton A, Verdile G, Gandy SE, Broe GA, Brooks WS, Bennett H, Piguet O, Price P, Miklossy J, Hallmayer J, McGeer PL, Martins RN. Association of alleles carried at TNFA -850 and BAT1–22 with Alzheimer’s disease. J Neuroinflammation. 2008;5:36. doi: 10.1186/1742-2094-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W, Hoppmann P, Michou E, Jung V, Pfeufer A, Mueller JC, Gieger C, Wichmann H-E, Meitinger T, Schömig A, Kastrati A. Association of variants in the BAT1-NFKBIL1-LTA genomic region with protection against myocardial infarction in Europeans. Hum Mol Genet. 2007;16:1821–1827. doi: 10.1093/hmg/ddm130. [DOI] [PubMed] [Google Scholar]
- Oliveira LC, Porta G, Marin MLC, Bittencourt PL, Kalil J, Goldberg AC. Autoimmune hepatitis, HLA and extended haplotypes. Autoimmun Rev. 2011;10:189–193. doi: 10.1016/j.autrev.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Quiñones-Lombraña A, Lopez-Soto A, Ballina-Garcia FJ, Alperi-López M, Queiro-Silva R, Lopez-Vazquez A, Lopez-Larrea C, Gonzalez S. BAT1 promoter polymorphism is associated with rheumatoid arthritis susceptibility. J Rheumatol. 2008;35:741–744. [PubMed] [Google Scholar]
- Karunaweera ND, Grau GE, Gamage P, Carter R, Mendis KN. Dynamics of fever and serum levels of tumor necrosis factor are closely associated during clinical paroxysms in Plasmodium vivax malaria. Proc Natl Acad Sci U S A. 1992;89:3200–3203. doi: 10.1073/pnas.89.8.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AG, de Vries N, Pociot F, di Giovine FS, van der Putte LB, Duff GW. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. J Exp Med. 1993;177:557–560. doi: 10.1084/jem.177.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob CO, Fronek Z, Lewis GD, Koo M, Hansen JA, McDevitt HO. Heritable major histocompatibility complex class II-associated differences in production of tumor necrosis factor alpha: relevance to genetic predisposition to systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1990;87:1233–1237. doi: 10.1073/pnas.87.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candore G, Cigna D, Gervasi F, Colucci AT, Modica MA, Caruso C. In vitro cytokine production by HLA-B8, DR3 positive subjects. Autoimmunity. 1994;18:121–132. doi: 10.3109/08916939409007985. [DOI] [PubMed] [Google Scholar]
- Bouma G, Crusius JB, Oudkerk Pool M, Kolkman JJ, von Blomberg BM, Kostense PJ, Giphart MJ, Schreuder GM, Meuwissen SG, Peña AS. Secretion of tumour necrosis factor alpha and lymphotoxin alpha in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand J Immunol. 1996;43:456–463. doi: 10.1046/j.1365-3083.1996.d01-65.x. [DOI] [PubMed] [Google Scholar]
- An J, Nakajima T, Shibata H, Arimura T, Yasunami M, Kimura A. A novel link of HLA locus to the regulation of immunity and infection: NFKBIL1 regulates alternative splicing of human immune-related genes and influenza virus M gene. J Autoimmun. 2013;47:25–33. doi: 10.1016/j.jaut.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Atkinson SH, Rockett KA, Morgan G, Bejon PA, Sirugo G, O’Connell MA, Hanchard N, Kwiatkowski DP, Prentice AM. Tumor necrosis factor SNP haplotypes are associated with iron deficiency anemia in West African children. Blood. 2008;112:4276–4283. doi: 10.1182/blood-2008-06-162008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami S, Tanabe N, Ota M, Naruse TK, Katsuyama Y, Nakanishi N, Tomoike H, Sakuma M, Shirato K, Takahashi M, Shibata H, Yasunami M, Chen Z, Kasahara Y, Tatsumi K, Kuriyama T, Kimura A. HLA-DPB1 and NFKBIL1 may confer the susceptibility to chronic thromboembolic pulmonary hypertension in the absence of deep vein thrombosis. J Hum Genet. 2009;54:108–114. doi: 10.1038/jhg.2008.15. [DOI] [PubMed] [Google Scholar]
- Chiba T, Matsuzaka Y, Warita T, Sugoh T, Miyashita K, Tajima A, Nakamura M, Inoko H, Sato T, Kimura M. NFKBIL1 confers resistance to experimental autoimmune arthritis through the regulation of dendritic cell functions. Scand J Immunol. 2011;73:478–485. doi: 10.1111/j.1365-3083.2011.02524.x. [DOI] [PubMed] [Google Scholar]
- Kallio KAE, Marchesani M, Vlachopoulou E, Mäntylä P, Paju S, Buhlin K, Suominen AL, Contreras J, Knuuttila M, Hernandez M, Huumonen S, Nieminen MS, Perola M, Sinisalo J, Lokki M-L, Pussinen PJ. Genetic variation on the BAT1-NFKBIL1-LTA region of major histocompatibility complex class III associates with periodontitis. Infect Immun. 2014;82:1939–1948. doi: 10.1128/IAI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


