Summary
Increased endogenous NO content modulates ROS accumulation and related antioxidant enzyme activities, osmolyte levels, and the expression of stress-responsive genes, such as AtPYL4/5, resulting in enhanced drought resistance in Arabidopsis.
Key words: Abscisic acid, drought stress, in vivo, neuronal nitric oxide synthase, nitric oxide, physiological, PYL, transcriptomic.
Abstract
Nitric oxide (NO) is involved in plant responses to many environmental stresses. Transgenic Arabidopsis lines that constitutively express rat neuronal NO synthase (nNOS) were described recently. In this study, it is reported that the nNOS transgenic Arabidopsis plants displayed high levels of osmolytes and increased antioxidant enzyme activities. Transcriptomic analysis identified 601 or 510 genes that were differentially expressed as a consequence of drought stress or nNOS transformation, respectively. Pathway and gene ontology (GO) term enrichment analyses revealed that genes involved in photosynthesis, redox, stress, and phytohormone and secondary metabolism were greatly affected by the nNOS transgene. Several CBF genes and members of zinc finger gene families, which are known to regulate transcription in the stress response, were changed by the nNOS transgene. Genes regulated by both the nNOS transgene and abscisic acid (ABA) treatments were compared and identified, including those for two ABA receptors (AtPYL4 and AtPYL5). Moreover, overexpression of AtPYL4 and AtPYL5 enhanced drought resistance, antioxidant enzyme activity, and osmolyte levels. These observations increase our understanding of the role of NO in drought stress response in Arabidopsis.
Introduction
As a gaseous diatomic radical, nitric oxide (NO) is an essential endogenous signalling molecule involved in multiple physiological processes in plants, including growth, development, and response to environmental stresses (Shi et al., 2012b, c ). Interestingly, NO is rapidly induced by multiple hormonal and environmental stimuli, including abscisic acid (ABA) (Guo et al., 2003), hydrogen peroxide (H2O2) (Bright et al., 2006), polyamines (Tun et al., 2006; Shi and Chan, 2013, 2014; Shi et al., 2013a ; Wang et al., 2011), auxin (Kolbert et al., 2007), salicylic acid (SA) (Zottini et al., 2007), brassinosteroids (BRs) (Cui et al., 2011), drought (Fan and Liu, 2012), salt (Zhao et al., 2007; Corpas et al., 2009), cold (Zhao et al., 2009), and heat (Bouchard and Yamasaki, 2008). NO can also act as a secondary messenger in environmental stress signal transduction (Gupta et al., 2011; Gill et al., 2013).
Understanding the complex effects of NO in plants requires a detailed analysis of the physiological and molecular changes. In recent years, transcriptional analyses of plant response to NO have been performed using different techniques (Huang et al., 2002; Polverari et al., 2003; Parani et al., 2004; Grün et al., 2006; Palmieri et al., 2008; Ahlfors et al., 2009; Besson-Bard et al., 2009). These studies have identified thousands of NO-responsive genes, most of which are stress related and serve a variety of functions ranging from plant defence and oxidative stress response to hormonal interplay (Huang et al., 2002; Polverari et al., 2003; Parani et al., 2004; Grün et al., 2006; Ahlfors et al., 2009; Besson-Bard et al., 2009). Further bioinformatics analysis identified several common transcription factor-binding sites (TFBSs) that are enriched in the promoters of these NO-responsive genes, such as WRKY, GBOX, and octopine synthase element-like sequence (OCSE) (Palmieri et al., 2008). However, most of these results were obtained by exogenous application of NO donors such as sodium nitroprusside (SNP), S-nitroso-N-acetyl-d-penicillamine (SNAP), and nitrosoglutathione (GSNO), NO scavengers such as 2-[4-carboxyphenyl]-4,4,5,5-tetramethylimidazoline-1-oxy-3-oxide (c-PTIO), or mammalian-type NO synthase (NOS) or its inhibitors including l-N G-nitro arginine methylester (l-NAME) (Huang et al., 2002; Polverari et al., 2003; Parani et al., 2004; Grün et al., 2006; Palmieri et al., 2008; Besson-Bard et al., 2009). Recent studies sshowed inconsistent findings concerning the effects of these NOS inhibitors, indicating that these chemicals have different or even opposite metabolic effects, and care must be taken in making inferences based on the use of these NO-modulating compounds (Arasimowicz-Jelonek et al., 2011; Gupta et al., 2011).
Several recent reports documented that the constitutive expression of rat nNOS in transgenic Arabidopsis plants resulted in the accumulation of endogenous NO and increased tolerance to abiotic and biotic stresses (Shi et al., 2012b, c ). Similarly, Chun et al. (2013) introduced rat nNOS into tobacco plants and found that nNOS transgenic plants with overproduction of NO exhibited enhanced resistance to bacteria, fungi, and viruses. The use of nNOS transgenic Arabidopsis plant represents a new approach to study the effect of NO. In this system, NO is released in planta as a consequence of the constitutive expression of mammalian nNOS (Shi et al., 2012b, c ; Chun et al., 2013).
To gain insight into NO-mediated stress tolerance, nNOS transgenic Arabidopsis plants with increased in vivo NO content were used for physiological and transcriptomic analyses in the current study. Physiological assays characterized the effects of increased in planta NO production on antioxidant enzyme activities, reactive oxygen species (ROS), and osmolyte accumulation under drought stress conditions. Transcriptomic analysis identified several stress-related genes and revealed related pathways that were significantly changed in the nNOS transgenic plants. Functional analyses of downstream NO-regulated genes, including those for two ABA receptors (PYL4 and PYL5), indicated that they played important roles during drought stress response. This study increases our understanding of the physiological and molecular roles of NO in the response of Arabidopsis to drought stress.
Materials and methods
Plant materials and growth conditions
This study used two transgenic Arabidopsis lines (nNOS-2 and nNOS-25) with the nNOS gene (Shi et al., 2012c ) and also used AtPYL4- and AtPYL5-overexpressing plants under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and Col-0 (wild type, WT). For the overexpression of AtPYL4 and AtPYL5, AtPYL4 and AtPYL5 cDNAs were cloned into the pCAMBIA99-1 vector. Then the constructs were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis WT (Col-0) plants using the floral dip method. Homozygous transgenic plants were selected using hygromycin resistance and were confirmed by PCR analyses.
After stratification in deionized water at 4 °C for 3 d in darkness, the Arabidopsis seeds were sown in soil-filled plastic containers in a growth room. The growth room was maintained at 22–25 °C with an irradiance of 120–150 μmol quanta m–2 s–1, 65% relative humidity, and a 16h light/8h dark cycle. Nutrient solution was added twice each week.
Drought stress treatment
To apply the drought stress treatment (via soil water deficit), water was withheld from 2-week-old WT and transgenic plants in soil for 21 d before the plants were re-watered. The survival rate of the stressed plants was recorded 7 d after re-watering. Leaf samples were harvested at day 7, 14, and 21 (referring to the number of days since the initiation of the drought stress treatment) under control and drought conditions for physiological parameter analyses.
Quantification of NO content and plant growth parameters
The NO content in leaf samples was quantified using the haemoglobin method by examining the conversion of oxyhaemoglobin to methaemoglobin as previously described (Shi et al., 2012c, 2013a). Plant height and dry weight (DW) were measured ~80 d after the seeds were sown.
Determination of electrolyte leakage (EL) and leaf water content (LWC)
EL was determined as described by Shi et al. (2012a , 2013a , b). For determination of EL and LWC, leaf samples were harvested at 0, 7, 14, and 21 d after drought stress and 7 d after re-watering. Fresh weight (FW) was measured immediately after harvest, and the DW was measured after 16h at 80 °C. LWC (%) was measured as (FW–DW)/FW×100.
Determination of proline, sucrose, and total soluble sugar levels
For determination of proline content, a 0.5g aliquot of each leaf sample was ground and extracted in 3% (w/v) sulphosalicylic acid before 2ml of ninhydrin reagent and 2ml of glacial acetic acid were added. The mixed solutions were boiled at 100 °C for 40min and cooled to room temperature. The proline level in the sample was calculated based on absorbance at 520nm and was expressed as μg per g FW of sample (Shi et al., 2012a ). For the determination of sucrose and total soluble sugars, the anthrone method was used as previously described (Shi et al., 2012a ).
Determination of H2O2 content and antioxidant enzyme activities
For determination of H2O2 content and antioxidant enzyme activities, plant extracts were isolated in 50mM sodium phosphate buffer (pH 7.8) using materials harvested from drought-stressed and control plants at 7, 14, and 21 d. H2O2 content and the activities of antioxidant enzymes [superoxide dismutase (SOD; EC 1.15.1.1), catalase (CAT; EC 1.11.1.6), glutathione reductase (GR; EC 1.6.4.2), glutathione peroxidase (GPX; EC 1.11.1.9), and peroxidase (POD; EC 1.11.1.7)] were quantified using previously published protocols (Shi et al., 2012a , 2013a , b). The H2O2 content was expressed as μM per FW. The relative activities of these antioxidant enzymes were expressed as the fold change relative to the WT (Col-0) under control conditions at 7 d.
RNA isolation, array hybridization, and microarray analysis
For RNA isolation, 2-week-old WT and nNOS transgenic plants in pots were well watered (control condition) or subjected to drought conditions by withholding water for 7 d. Each combination of genotype and treatment was represented by two replicate leaf samples, and each sample contained leaves from at least 20 seedlings. Total RNA was extracted with TRIzol reagent (Invitrogen) and was quantified as previously described (Shi et al., 2012c ). RNA quality was determined using a formaldehyde agarose gel and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol.
Array hybridization and microarray analysis were performed by CapitalBio Corporation in China. For array hybridization, 200ng of total RNA was used for first-strand and second-strand cDNA synthesis. An equal amount of RNA from two independent nNOS transgenic lines (line 2 and line 25) was pooled for cRNA labelling. The cRNA was labelled with a biotinylated ribonucleotide analogue and was fragmented with fragmentation buffer using the MessageAmp™ Premier RNA Amplification Kit (Ambion, #1792). After purification, 12.5 μg of labelled and fragmented cRNA probes were hybridized to the Arabidopsis arrays with the Hybridization, Wash and Stain Kit (Affymetrix, #900720) according to the manufacturer’s instructions.
The arrays were scanned using a GeneChip® Scanner 3000 (Affymetrix, #3000). The scanned images were saved as DAT files and were transformed to JPG images. The signal intensities were extracted from the JPG images with Affymetrix® GeneChip® Command Console® Software (AGCC software) and were saved as CEL files. The affylmGUI package (Wettenhall et al., 2006) rooted in R (Gentleman et al., 2004) was used to calculate the intensity ratios and fold changes. All of the differentially expressed genes with P-values <0.05 and log2 fold change >1 or < –1 were chosen for further analysis. The normalized microarray data were submitted to the Gene Expression Omnibus (GEO) database with accession number GSE48474. All of the genes whose expression significantly changed in at least one comparison of genotype or drought treatment (P-value ≤0.05 and log2 fold change ≥1 or log2 fold change ≤ –1) are listed in Supplementary Table S1 available at JXB online.
Quantitative real-time PCR
Total RNA was isolated from 100mg of leaves using 1ml of TRIzol reagent (Invitrogen). The total RNA was treated with RQ1 RNase-free DNase (Promega). RNA samples from two independent nNOS transgenic lines (line 2 and line 25) were pooled for cDNA synthesis. First-strand cDNA was synthesized with the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas) using 2 μg of total RNA according to the manufacturer’s instructions. Reverse transcription products (cDNA) were diluted five times in water, and 2 μl of diluted cDNA was used for quantitative real-time PCR assay with iQ™ SYBR® Green Super mix (Bio-Rad). Quantitative real-time PCR was performed using the CFX96™ Real Time System (Bio-Rad) with SYBR green fluorescence as previously described (Shi et al., 2012c , 2013a , b). The experiment was performed with at least three independent replicates, and the comparative ΔΔCT method was used for comparative gene expression analysis. In total, 30 genes with a ≥2-fold change in expression were randomly selected for real-time PCR assay. The housekeeping gene UBQ10 was used as an endogenous control. The primers used are listed in Table S2 at JXB online.
Biological enrichment and metabolic pathway analyses
All differentially expressed genes with P-values ≤0.05 and log2 fold change ≥1 or ≤ –1 were loaded and annotated in the Classification SuperViewer Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) (Provart and Zhu, 2003). Functional categories of every gene and pathway were assigned using MapMan (http://mapman.mpimp-golm.mpg.de/general/ora/ora.html) as the classification source (Thimm et al., 2004). Additionally, the normalized frequency (NF) of each functional category was calculated as follows: NF=sample frequency of each category in this experiment⁄background frequency of each category in the ATH1 array. For GO term enrichment analysis, differentially expressed genes were input into the agriGO website (http://bioinfo.cau.edu.cn/agriGO/index.php), and the Singular Enrichment Analysis (SEA) tool was used for enrichment analyses (Du et al., 2010).
Hierarchical cluster analysis
The data sets of specific genes were imported for hierarchical cluster analysis, which was performed using an uncentred matrix and complete linkage method with the CLUSTER program (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/) (de Hoon et al., 2004). Resulting tree figures were displayed using the software package Java Treeview (http://jtreeview.sourceforge.net/) as described by Chan et al. (2012) and Chan (2012).
Statistical analysis
All of the experiments in this study were repeated three times, and the values presented are means ±SEs. For each independent experiment, each leaf sample extract was derived from the leaves of at least 15 plants. Asterisks above the columns in figures indicate significant differences relative to the WT at P<0.05 (Duncan’s multiple range test).
Results
Growth of nNOS transgenic and WT plants under control and drought conditions
Under well-watered (control) conditions, growth of nNOS transgenic plants (line 2 and line 25) was generally equivalent to that of the WT (Supplementary Fig. S1a at JXB online). Under water deficit conditions, growth of both nNOS transgenic lines and WT plants was inhibited, as previously reported (Shi et al., 2012c ), but nNOS transgenic plants (line 2 and line 25) had greener leaves than WT plants (Supplementary Fig. S1a). Consistent with previous results (Shi et al., 2012c ), nNOS transgenic plants accumulated higher concentrations of endogenous NO under both control and drought stress conditions (Supplementary Fig. S1b).
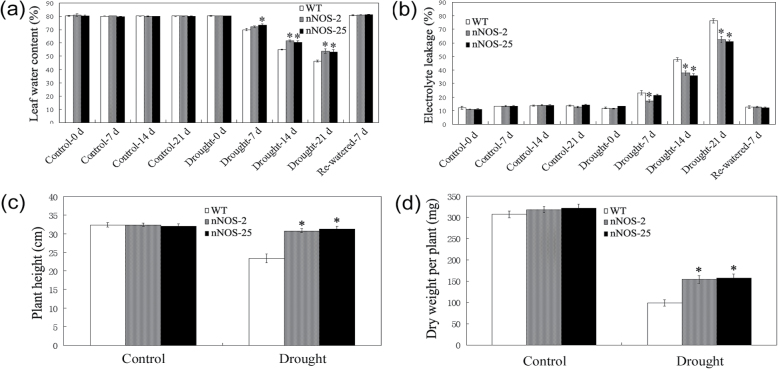
Under control conditions, both nNOS transgenic lines and WT plants maintained LWC at ~80% (Fig. 1a). After drought stress treatment, the LWC gradually declined in all plants, but the decline was greater in the WT than in the nNOS transgenic plants (Fig. 1a). Consistent with the decline in LWC, all plants showed a gradual increase in EL after drought stress treatment, but the increase was less in the nNOS transgenic lines than in the WT (Fig. 1b).
Fig. 1.
Performance of WT and nNOS transgenic Arabidopsis plants under drought stress conditions (soil water deficit). (a and b) LWC (a) and EL (b) of WT and nNOS transgenic plants during control and drought stress conditions. (c and d) Plant height (c) and dry weight (DW) (d) of WT and nNOS transgenic plants under control and drought stress conditions at harvest. Values are means ±SEs (n=4 for a, b, n=20 for c, d). Asterisks indicate significant differences between WT and nNOS transgenic plants (P<0.05).
After re-watering for 7 d, almost all WT plants died, while >55% of the nNOS plants survived (Supplementary Fig. S1a at JXB online). Among the surviving WT and nNOS transgenic plants, LWC and EL were recovered, and there were no significant differences between WT and nNOS transgenic plants in LWC and EL (Fig. 1a, b). At harvest time (i.e. ~45 d after re-watering), plant height and biomass (DW) of plants subjected to water deficit conditions were greater in nNOS transgenic lines than in the WT (Fig. 1c, d).
Osmolyte accumulation and ROS metabolism in nNOS transgenic and WT plants under drought stress
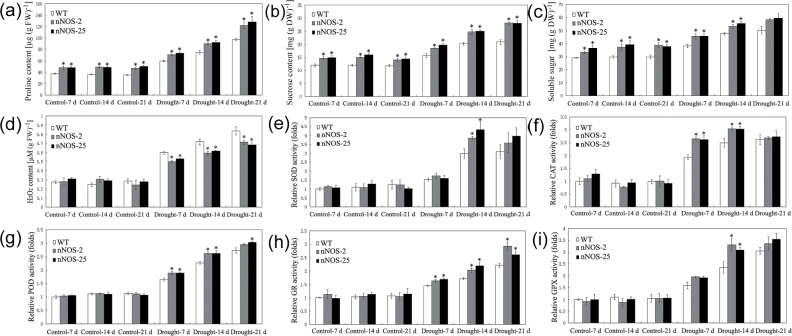
Under control conditions, both nNOS transgenic lines (nNOS-2 and nNOS-25) accumulated significantly higher levels of proline, sucrose, and total soluble sugars than the WT (Fig. 2a–c). Drought stress increased the levels of proline, sucrose, and total soluble sugars in both nNOS transgenic lines and WT plants, but the increase was greater in the nNOS transgenic lines than in the WT (Fig. 2a–c).
Fig. 2.
Osmolyte levels and ROS metabolism in WT and nNOS transgenic plants during drought stress. (a–i) Proline content (a), sucrose content (b), soluble sugar content (c), H2O2 content (d), and activities of SOD (e), CAT (f), POD (g), GR (h), and GPX (i) in WT and nNOS transgenic plants under control and drought stress conditions. The relative activities were quantified as the fold change relative to the activity in the WT under control conditions at 7 d. Values are the means ±SEs (n=4). Asterisks indicate significant differences between WT and nNOS transgenic plants (P<0.05).
As the major indicator of ROS level and oxidative damage, H2O2 functions as the key stress-related signal, and H2O2 content was assayed in this study. Under control conditions, H2O2 levels did not significantly differ in the nNOS transgenic lines and the WT plants (Fig. 2d). After 7, 14, and 21 d of drought stress treatment, however, H2O2 content was significantly lower in the nNOS transgenic lines than in the WT (Fig. 2d). Under control conditions, the activities of five antioxidant enzymes (SOD, CAT, POD, GR, and GPX) did not significantly differ in nNOS transgenic lines and the WT (Fig. 2e–i). After drought stress treatment, however, the activities of the five enzymes were significantly higher at several time points in nNOS transgenic lines than in the WT (Fig. 2e–i).
Differentially expressed genes in WT and nNOS transgenic plants under control and drought conditions
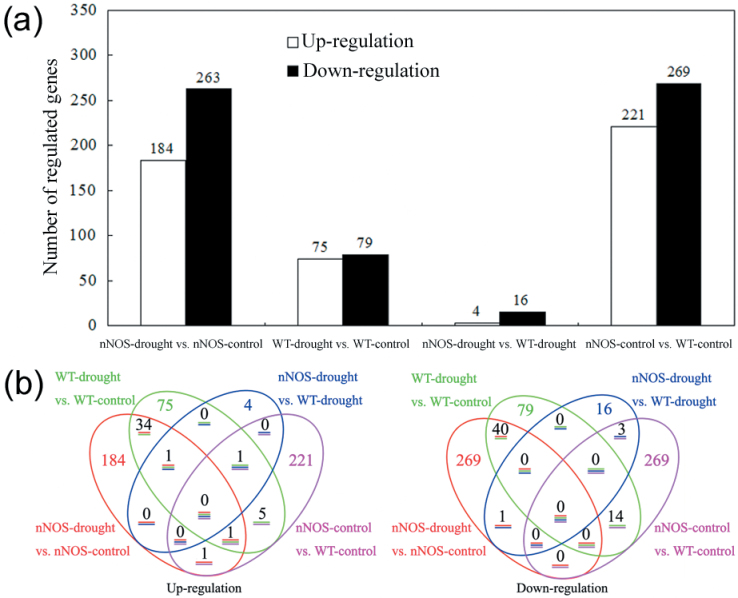
Microarray analysis was performed to gain insight into NO-mediated drought stress responses. In total, the expression levels of 154 genes (75 were up-regulated genes and 79 were down-regulated genes) in WT plants and 447 genes (184 were up-regulated genes and 263 were down-regulated genes) in nNOS transgenic plants were changed by at least 2-fold in response to drought stress treatment (Fig. 3a; Supplementary Table S1 at JXB online). Under control conditions, 490 genes differed in expression level in nNOS transgenic plants compared with the WT, comprising 221 up-regulated and 269 down-regulated genes (Fig. 3a). Under drought conditions, 20 genes differed in expression level in nNOS transgenic plants compared with the WT, comprising four up-regulated and 16 down-regulated genes (Fig. 3a). As shown in Fig. 3b, only a small number of differentially expressed genes were common to all four comparisons, including two comparisons for genotype effect (nNOS control versus WT control, nNOS drought versus WT drought) and two comparisons for stress effect (WT drought versus WT control, nNOS drought versus nNOS control). However, 30% of the genes changed by drought stress in the WT were also changed by drought stress in the nNOS transgenic plants (Fig. 3b). Thirty genes regulated by drought in the WT (WT drought versus WT control) and by the nNOS transgene effect in the absence of drought stress (nNOS control versus WT control) were also identified (Table 1). These genes might play direct roles in nNOS transgene-induced drought stress tolerance. Interestingly, only two genes were regulated in common in nNOS control versus WT control and nNOS drought versus nNOS control (Fig. 3b), while 155 genes were oppositely regulated in these two comparisons (Supplementary Table S1).
Fig. 3.
Number of genes differentially expressed in WT versus nNOS transgenic plants under control and drought conditions. (a) Total number of affected genes in WT and nNOS transgenic plants under control and drought conditions. (b) Venn diagram showing the number of overlapping genes that are differentially expressed between WT and nNOS transgenic plants under control and drought conditions. (This figure is available in colour at JXB online.)
Table 1.
Genes highly regulated by drought stress and by the nNOS transgenic effect in Arabidopsis
| AGI | nNOS drought versus nNOS control | WT drought versus WT control | nNOS drought versus WT drought | nNOS control versus WT control | Description | MapMAN Bin name | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2 | P-value | log2 | P-value | log2 | P-value | log2 | P-value | |||
| At4g25100 | 1.23 | 0.0000 | 2.13 | 0.0000 | 0.21 | 0.2317 | 1.11 | 0.0001 | Fe-superoxide dismutase | Redox. dismutases and catalases |
| At5g53870 | –0.02 | 0.9214 | 1.42 | 0.0001 | 0.12 | 0.5431 | 1.57 | 0.0000 | Early nodulin-like protein 1 | Misc. plastocyanin-like |
| At2g35980 | 0.26 | 0.1609 | 1.30 | 0.0002 | 0.06 | 0.7789 | 1.10 | 0.0002 | NDR1/HIN1-LIKE 10 | Stress. biotic |
| At1g24140 | –0.03 | 0.8622 | 1.20 | 0.0002 | 0.22 | 0.2381 | 1.45 | 0.0000 | Matrixin family protein | Protein. degradation.metalloprotease |
| At4g27654 | –0.18 | 0.5689 | 1.14 | 0.0083 | 1.10 | 0.0138 | 2.41 | 0.0001 | Unknown protein | Not assigned. unknown |
| AtCg00690 | 0.51 | 0.1258 | 1.14 | 0.0125 | 0.78 | 0.0524 | 1.40 | 0.0012 | 5kDa protein subunit PSII-T | PS. Light reaction. photosystem II |
| At1g23040 | 0.05 | 0.7613 | 1.02 | 0.0003 | 0.14 | 0.4071 | 1.10 | 0.0001 | Hydroxyproline-rich glycoprotein | No ontology.hydroxyproline rich proteins |
| At1g75940 | –0.01 | 0.9742 | –1.07 | 0.0087 | –0.28 | 0.3655 | –1.33 | 0.0007 | beta-Glucosidase 20 | Misc. gluco-, galacto- and mannosidases |
| At1g57750 | 0.90 | 0.0002 | –1.11 | 0.0002 | –0.60 | 0.0116 | –2.61 | 0.0000 | CYP96A15 | Not assigned. unknown |
| At3g49620 | –0.06 | 0.7921 | –1.15 | 0.0011 | –0.63 | 0.0245 | –1.72 | 0.0001 | DIN11 | Development. unspecified |
| At1g11580 | –0.17 | 0.3290 | –1.24 | 0.0002 | 0.00 | 0.9973 | –1.07 | 0.0001 | Pectin methylesterase | Cell wall. pectin esterases.PME |
| At5g07550 | 0.40 | 0.0342 | –1.33 | 0.0001 | –0.58 | 0.0177 | –2.31 | 0.0000 | GRP19 | No ontology. glycine rich proteins |
| At2g39330 | 0.15 | 0.6476 | –1.37 | 0.0035 | –0.30 | 0.3505 | –1.82 | 0.0002 | JAL23 | Misc. myrosinases-lectin-jacalin |
| At2g45130 | –0.41 | 0.0273 | –1.39 | 0.0001 | –0.03 | 0.8772 | –1.01 | 0.0002 | SPX3 | Stress. abiotic |
| At3g25050 | –0.03 | 0.9571 | –1.48 | 0.0095 | 0.00 | 0.9992 | –1.45 | 0.0028 | XTH3 | Cell wall. modification |
| At1g17710 | –0.29 | 0.2000 | –1.74 | 0.0001 | 0.07 | 0.7900 | –1.38 | 0.0002 | Phosphocholine phosphatase | Misc. acid and other phosphatases |
| At1g73010 | –0.78 | 0.0012 | –1.85 | 0.0000 | –0.46 | 0.0372 | –1.53 | 0.0000 | Pyrophosphate-specific phosphatase | Misc. acid and other phosphatases |
| At1g54020 | 0.09 | 0.7179 | –1.95 | 0.0001 | –0.22 | 0.3571 | –2.26 | 0.0000 | Myrosinase-associated protein | Secondary metabolism. degradation |
| At1g66850 | 0.77 | 0.0010 | –2.03 | 0.0000 | –0.73 | 0.0094 | –3.54 | 0.0000 | Lipid-transfer protein | Misc. protease inhibitor |
| At5g20790 | –0.93 | 0.0010 | –2.39 | 0.0000 | 0.09 | 0.6912 | –1.36 | 0.0001 | Unknown protein | Not assigned. unknown |
| At1g72260 | –0.08 | 0.7225 | –2.73 | 0.0000 | 0.38 | 0.1165 | –2.26 | 0.0000 | THI2.1 | Stress. biotic.receptors |
| At5g45890 | 1.75 | 0.0000 | 1.27 | 0.0009 | –0.92 | 0.0103 | –1.41 | 0.0002 | SAG12 | Protein. degradation.cysteine protease |
| At2g17880 | –1.93 | 0.0000 | –1.03 | 0.0006 | 0.12 | 0.5189 | 1.01 | 0.0002 | DNAJ heat shock protein, | Stress. abiotic.heat |
| At3g28310 | –2.22 | 0.0000 | –1.15 | 0.0012 | 0.03 | 0.9057 | 1.10 | 0.0004 | Unknown protein | Not assigned. no ontology |
| At5g56100 | –2.12 | 0.0000 | –1.31 | 0.0001 | 0.52 | 0.0225 | 1.34 | 0.0001 | Glycine-rich protein | Lipid metabolism. TAG synthesis |
| At4g12500 | –3.42 | 0.0000 | –1.61 | 0.0000 | –0.20 | 0.2350 | 1.62 | 0.0000 | Lipid-transfer protein | Misc. protease inhibitor |
| At1g15010 | –2.32 | 0.0000 | –1.67 | 0.0000 | 0.69 | 0.0103 | 1.34 | 0.0001 | Unknown protein | Not assigned. unknown |
| At2g38310 | –0.95 | 0.0002 | –0.33 | 0.0699 | 0.50 | 0.0187 | 1.12 | 0.0000 | PYL4/RCAR10 | Stress. abiotic |
| At5g05440 | –2.40 | 0.0000 | –1.75 | 0.0000 | 0.55 | 0.0166 | 1.21 | 0.0001 | PYL5/RCAR8 | Stress. abiotic |
| At1g19610 | –4.01 | 0.0000 | –1.78 | 0.0000 | –0.14 | 0.4867 | 2.08 | 0.0000 | Pathogenesis-related protein | Stress. biotic |
Genes highly regulated by drought stress and by the nNOS transgenic effect (i.e. with a P-value ≤0.05 and log2 fold change ≥1 or log2 fold change ≤ –1) were classified using MapMan.
Values in bold indicate significant up-regulation, and those in italics indicate significant down-regulation.
To confirm the reliability of the microarray data, the expression of 30 genes that were differentially expressed between WT and nNOS transgenic plants was assessed via quantitative real-time PCR. Although several genes (only four genes) showed a <2-fold change in real-time PCR, the results of the real-time PCR assay exhibited the same trend and were correlated with the microarray data (R 2=0.8951) (Supplementary Fig. S2a–c at JXB online), confirming the reproducibility of microarray analysis.
Cluster and pathway enrichment analyses of differentially expressed genes changed by nNOS transgene or by drought
Pathway analysis revealed that the nNOS transgene and drought affected the expression of many genes involved in photosynthesis, hormone metabolism, redox, stress, mitochondrial electron transport, oxidative pentose phosphate (OPP), and lipid metabolism (Table 2, group I). Further analyses showed that many redox- and phytohormone metabolism-related genes were extensively regulated by the nNOS transgene effect (Supplementary Fig. S3a, b at JXB online). Interestingly, genes involved in ABA, indole-3-acetic acid (IAA), benzylaminopurine (BA), and ethylene metabolism were constitutively up-regulated in nNOS transgenic plants (Supplementary Fig. S3a, b). Moreover, ectopic expression of the nNOS gene in Arabidopsis also resulted in activation of the light reaction and sugar biosynthesis pathways (Supplementary Fig. S4). Several other pathways including development, major carbohydrate (CHO) metabolism, protein, signalling, and cell wall were over-represented in nNOS trangenic versus WT plants under control conditions (nNOS control versus WT control) but under-represented in nNOS transgenic versus WT plants under drought conditions (nNOS drought versus WT drought) (Table 2, group II).
Table 2.
Pathway enrichment analysis of genes whose expression was significantly affected by drought stress and by the nNOS transgenic effect in Arabidopsis
| Groups | Pathways | nNOS drought versus nNOS control | WT drought versus WT control | nNOS drought versus WT drought | nNOS control versus WT control | ||||
|---|---|---|---|---|---|---|---|---|---|
| NF | P-value | NF | P-value | NF | P-value | NF | P-value | ||
| I | PS | 5.49 | 0.0000 | 2.11 | 0.1740 | 8.15 | 0.1090 | 6.68 | 0.0000 |
| Metal handling | 4.54 | 0.0041 | 15.77 | 0.0000 | – | – | – | – | |
| Hormone metabolism | 4.18 | 0.0000 | 2.41 | 0.0260 | – | – | 1.52 | 0.0440 | |
| Redox | 2.15 | 0.0390 | 11.4 | 0.0000 | – | – | 1.96 | 0.0520 | |
| Stress | 2.09 | 0.0000 | 3.37 | 0.0000 | 2.73 | 0.1300 | 1.45 | 0.0140 | |
| Miscellaneous | 1.66 | 0.0011 | 3.7 | 0.0000 | 4.22 | 0.0110 | 1.34 | 0.0200 | |
| Mitochondrial electron transport | 1.50 | 0.1820 | 1.44 | 0.3480 | – | – | 2.28 | 0.0470 | |
| Oxidative Pentose phosphate | 2.43 | 0.2750 | 7.03 | 0.1240 | – | – | 2.22 | 0.2900 | |
| Lipid metabolism | 1.05 | 0.1610 | 1.01 | 0.2730 | – | – | 1.12 | 0.1440 | |
| II | Secondary metabolism | 1.53 | 0.0650 | 0.98 | 0.2730 | – | – | 2.48 | 0.0006 |
| RNA | 1.48 | 0.0005 | 0.64 | 0.0450 | 1.65 | 0.1690 | 1.01 | 0.0620 | |
| Transport | 1.40 | 0.0330 | 1.7 | 0.0520 | – | – | 0.53 | 0.0200 | |
| Development | 0.95 | 0.1260 | 1.38 | 0.1400 | 4.26 | 0.0680 | 1.39 | 0.0440 | |
| Cofactor and vitamin metabolism | 0.93 | 0.3700 | – | – | – | – | 3.4 | 0.0240 | |
| Major CHO metabolism | 0.74 | 0.3530 | – | – | – | – | 2.72 | 0.0440 | |
| Protein | 0.7 | 0.0016 | 0.54 | 0.0049 | 0.69 | 0.2400 | 1.08 | 0.0380 | |
| Signalling | 0.66 | 0.0350 | 0.64 | 0.1220 | 1.23 | 0.3700 | 1.11 | 0.0770 | |
| Amino acid metabolism | 0.58 | 0.1890 | 0.83 | 0.3640 | – | – | 1.58 | 0.0930 | |
| Cell wall | 0.41 | 0.0440 | 1.19 | 0.2170 | – | – | 1.88 | 0.0081 | |
| Cell | 0.36 | 0.0095 | 0.52 | 0.1600 | 6.07 | 0.0110 | 0.66 | 0.0630 | |
| III | Not assigned | 0.93 | 0.0240 | 0.53 | 0.0000 | 0.56 | 0.0730 | 0.89 | 0.0096 |
| DNA | 0.07 | 0.0000 | 0.13 | 0.0000 | – | – | 0.11 | 0.0000 | |
Differentially expressed genes (i.e. with P value ≤0.05 and log2 fold-change ≥1 or log2 fold-change ≤ –1) were annotated using the Classification SuperViewer Tool and MapMan. shading scales of NF are as follows:
≥3 2–3 1–2 0.5–1 ≤0.5
Not surprisingly, GO term enrichment analysis showed that many metabolic-, ion homeostasis-, and transport-related pathways were extensively changed after drought stress treatment, resulting in enrichment of stress-responsive GO terms (Supplementary Fig. S5a at JXB online). Comparatively, transformation of Arabidopsis with the nNOS gene changed nitrogen metabolism as expected, as well as photosynthesis, energy-producing, and phytohormone metabolism pathways. These changes might contribute to the enrichment of stress-related GO terms (Supplementary Fig. S5b).
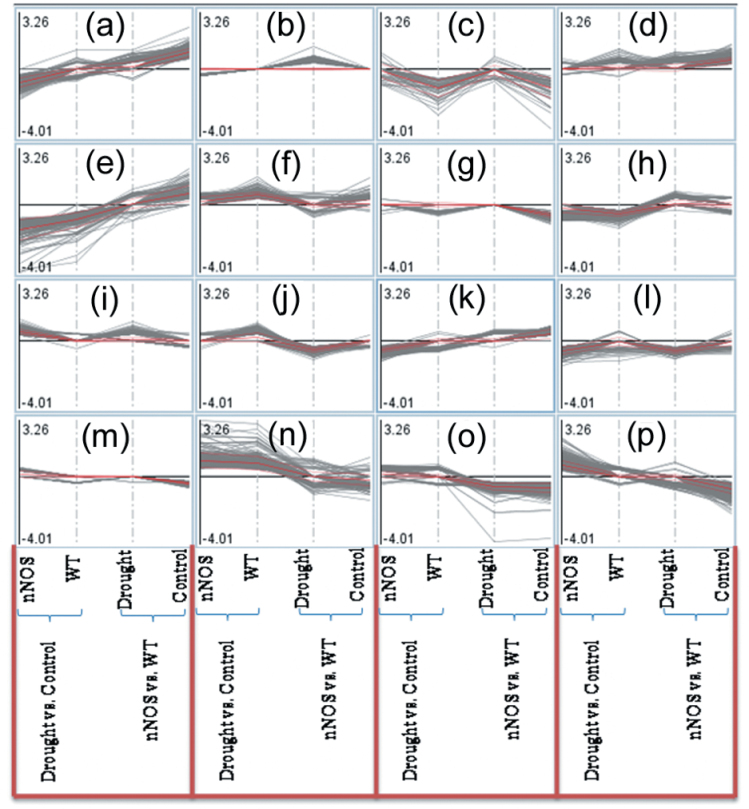
Genome-wide cluster analysis indicated that genes in cluster d were mainly up-regulated, while those in cluster l were mainly down-regulated by both the nNOS transgene and drought stress treatment. In addition, genes in clusters e and h were mainly up-regulated by the nNOS transgene but down-regulated by the drought stress treatment, and genes in clusters n and o were mainly up-regulated by drought stress treatment but down-regulated by the nNOS transgene (Fig. 4; Table S3 at JXB online).
Fig. 4.
Cluster analysis of microarray data using MapMan software. All microarray data were divided into 16 clusters that were labelled from (a) to (p). Detailed information for each cluster is provided in Supplementary Table S3 at JXB online. (This figure is available in colour at JXB online.)
Comparison of genes whose expression is altered by the nNOS transgene and NO donor treatment
Genes whose expression was affected by both the nNOS transgene and by NO donor (SNP) treatment were also identified (Ahlfors et al., 2009). Among the 28 genes whose expression was affected by both factors, 21 were up-regulated and one was down-regulated by both treatments, while six others differed in expression pattern due to the nNOS transgene and SNP treatment (Table 3).
Table 3.
Genes that were differentially expressed in response to both the nNOS transgenic effect and the NO donor (SNP) effect in Arabidopsis
| ID | Description | nNOSa versus WT | SNPb versus control | MapMAN | |
|---|---|---|---|---|---|
| Bin code | Bin name | ||||
| At3g44720 | ADT4 (arogenate dehydratase 4) | 1.18 | 1.21 | 13.1.6 | Amino acid metabolism. synthesis |
| At4g35630 | PSAT; O-phospho-l-serine:2-oxoglutarate aminotransferase | –1.11 | 0.98 | 13.1.5 | Amino acid metabolism. synthesis |
| At4g09030 | AGP10 (ARABINOGALACTAN PROTEIN 10) | 1.11 | 1.12 | 10.5.1 | Cell wall. cell wall proteins. AGPs |
| At1g19300 | Polygalacturonate 4-alpha-galacturonosyltransferase | 1.34 | 1.04 | 10.3.2 | Cell wall. hemicellulose synthesis. glucuronoxylan |
| At2g38360 | PRA1.B4 (PRENYLATED RAB ACCEPTOR 1.B4) | 1.14 | 0.96 | 31.4 | Cell. vesicle transport |
| At5g65870 | ATPSK5 (PHYTOSULPHOKINE 5 PRECURSOR) | 1.20 | 1.64 | 33.99 | Development. unspecified |
| At3g15210 | RAP2.5/ERF4 | 1.39 | 1.74 | 17.5.2 | Hormone metabolism. ethylene.signal transduction |
| At1g23440 | Pyrrolidone-carboxylate peptidase family protein | –0.54 | 1.03 | 29.5 | Protein. degradation |
| At1g24140 | Matrixin family protein | 1.45 | 1.88 | 29.5.7 | Protein. degradation. metalloprotease |
| At5g27420 | Zinc finger (C3HC4-type RING finger) family protein | 1.49 | 2.98 | 29.5.11 | Protein. degradation. ubiquitin. E3. RING |
| At4g35480 | RING-H2 finger protein RHA3b | 1.24 | 1.59 | 29.5.11 | Protein. degradation. ubiquitin. E3. RING |
| At5g47610 | Zinc finger (C3HC4-type RING finger) family protein | 1.21 | –1.01 | 29.5.11 | Protein. degradation. ubiquitin. E3. RING |
| At5g66070 | Zinc finger (C3HC4-type RING finger) family protein | 1.02 | 1.83 | 29.5.11 | Protein. degradation. ubiquitin. E3. RING |
| At5g47070 | Protein kinase, putative | –1.08 | 1.39 | 29.4.1 | Protein. postranslational modification. kinase |
| At1g28480 | GRX480; electron carrier/protein disulfide oxidoreductase | 1.15 | 1.46 | 21.4 | Redox. glutaredoxins |
| At5g22250 | CCR4-NOT transcription complex protein, putative | 1.31 | 1.53 | 27.1.19 | RNA. processing. ribonucleases |
| At1g27730 | ZAT10/STZ (salt tolerance zinc finger) | 1.15 | 2.66 | 27.3.11 | RNA. regulation of transcription. zinc finger family |
| At5g54490 | PBP1 (PINOID-BINDING PROTEIN 1) | 1.11 | 2.39 | 30.3 | Signalling. calcium |
| At3g01830 | Calmodulin-related protein, putative | 1.00 | 2.95 | 30.3 | Signalling. calcium |
| At4g36040 | DNAJ heat shock N-terminal domain-containing protein (J11) | 1.00 | 0.70 | 20.2.1 | Stress. abiotic.heat |
| At1g72940 | Disease resistance protein (TIR-NBS class) | –1.05 | 0.77 | 20.1.7 | Stress. biotic. PR-proteins |
| At5g52760 | Heavy-metal-associated domain-containing protein | 1.01 | 1.33 | 35.1 | Not assigned. no ontology |
| At3g04640 | Glycine-rich protein | 1.22 | 1.55 | 35.1.40 | Not assigned. no ontology. Glycine-rich proteins |
| At1g56060 | Unknown protein | 1.07 | 2.48 | 35.2 | Not assigned. unknown |
| At2g25735 | Unknown protein | 1.05 | 1.84 | 35.2 | Not assigned. unknown |
| At2g28400 | Unknown protein | 1.02 | 2.38 | 35.2 | Not assigned. unknown |
| At5g53420 | Unknown protein | –1.02 | 0.77 | 35.2 | Not assigned. unknown |
| At1g13650 | 18S pre-ribosomal assembly protein gar2-related | –1.18 | –1.04 | 35.2 | Not assigned. unknown |
a The data for nNOScontrol versus WT control are from this study.
b The data for log2 fold change of SNP 3h versus 0h are from Ahlfors et al. (2009).
Values in bold indicate significant up-regulation, and those in italics indicates significant down-regulation.
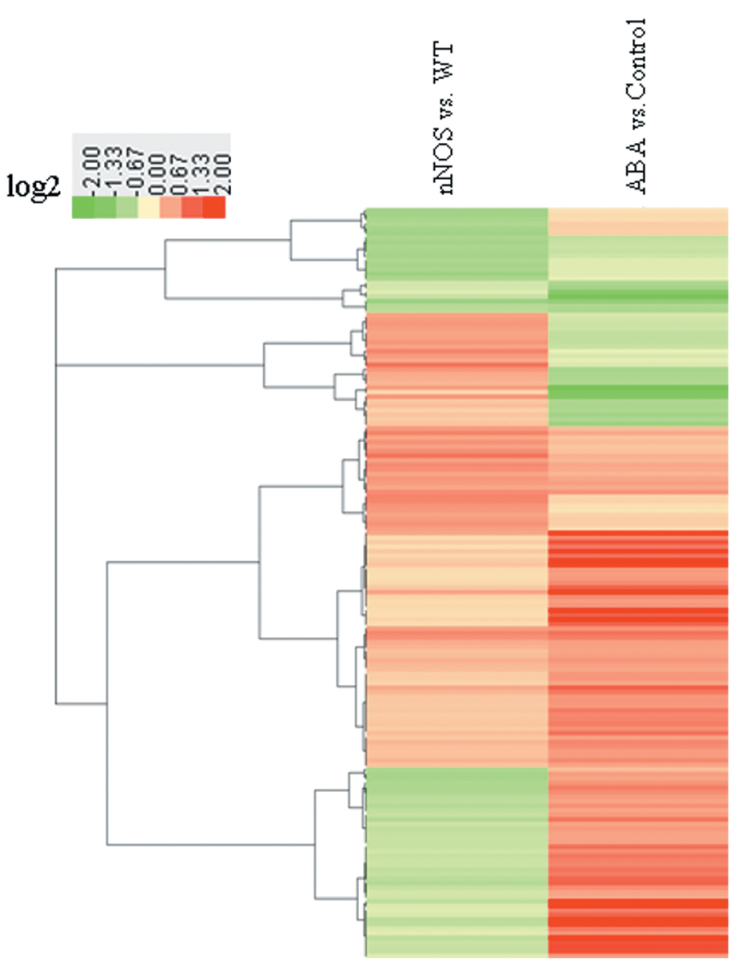
Additionally, based on published data (Goda et al., 2008), 165 genes that were regulated by both the nNOS transgenic effect and ABA treatment were found (Supplementary Table S4 at JXB online). Cluster analysis revealed that most of these genes were up-regulated or down-regulated by both the nNOS transgene and ABA effect (Fig. 5). Since ABA plays a critical role in plant drought stress response, further experiments were carried out to characterize putative connections between ABA and NO signalling pathways.
Fig. 5.
Hierarchical cluster analysis of genes regulated both by the nNOS transgene and by ABA treatment in Arabidopsis. The expression data for genes commonly regulated by nNOS and ABA were imported for cluster analysis, and the resulting tree figure was displayed using the software package and Java Treeview. The detailed information is provided in Supplementary Table S4 at JXB online. (This figure is available in colour at JXB online.)
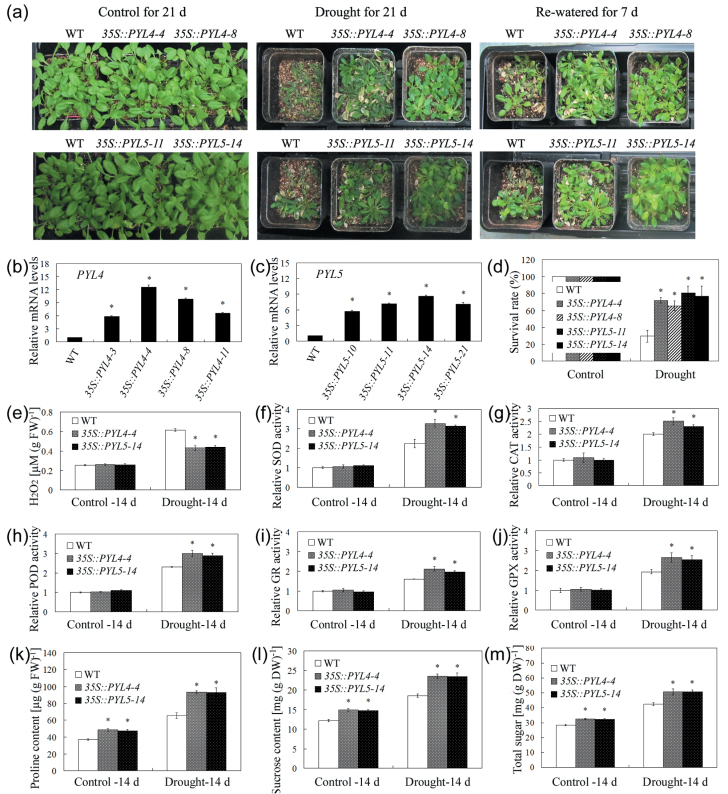
Overexpression of AtPYL4 and AtPYL5 enhances drought tolerance
To characterize further the in vivo roles of some differentially expressed genes in the nNOS transgenic lines, two ABA receptor genes (AtPYL4/RCAR10 and AtPYL5/RCAR8), which act upstream of the ABA pathway and directly modulate many downstream genes (Table 1), were constitutively overexpressed in Arabidopsis (Fig. 6a–c). AtPYL4- and AtPYL5-overexpressing transgenic plants exhibited enhanced drought resistance (Fig. 6a–d). Under drought stress conditions, H2O2 levels were lower and activities of antioxidant enzymes (SOD, CAT, POD, GR, and GPX) were higher in AtPYL4- and AtPYL5-overexpressing plants than in the WT (Fig. 6e–j). Additionally, AtPYL4- and AtPYL5-overexpressing plants accumulated higher levels of proline, sucrose, and soluble sugars than the WT under both control and drought stress conditions (Fig. 6l, m). These results indicated that AtPYL4 and AtPYL5 enhanced drought tolerance, in part by modulating ROS metabolism and osmolyte levels.
Fig. 6.
Enhanced drought resistance of plants overexpressing AtPYL4 and AtPYL5. (a) Two-week-old plants were subjected to drought stress conditions (or to well-watered, control conditions) for 21 d before they were re-watered; the plants were photographed 7 d after watering was resumed. (b and c) Gene expression of AtPYL4- (b) and AtPYL5- (c) overexpressing plants. The relative mRNA level of WT plants was set at 1.0. (d) Survival of AtPYL4- and AtPYL5-overexpressing plants under control and drought stress conditions. (e–m) H2O2 content (e), SOD activity (f), CAT activity (g), POD activity (h), GR activity (i) GPX activity (j), proline content (k), sucrose content (l), and soluble sugar content (m) of WT, AtPYL4, and AtPYL5 transgenic plants during control and drought stress conditions. The relative activities were quantified as fold change relative to the WT under control conditions for 14 d. Values are means ±SEs (n=3). Asterisks indicate significant differences between WT and nNOS transgenic plants (P<0.05). (This figure is available in colour at JXB online.)
Discussion
In this study, comparative physiological and transcriptomic analyses (Huang et al., 2011; Chan et al., 2012) were used to assess the drought stress response of transgenic Arabidopsis plants that ectopically expressed the rat nNOS gene, which resulted in the specific release of NO in planta. Although mutants with increased NO content are available, some (such as cue1/nox1 and gsnor) negatively affect plant development and yield (He et al., 2004; Chen et al., 2009), while others have a non-specific role in NO synthesis; argah1/2 mutants, for example, modulate not only NO accumulation but also polyamine accumulation and arginine metabolism (Shi and Chan, 2013; Shi et al., 2013a ). In addition to investigating NO-mediated physiological responses, this study also partially revealed the transcriptomic modulations caused by constitutive NO release. Importantly, this study avoided the side effects caused by the use of NO-modulator compounds (Arasimowicz-Jelonek et al., 2011; Gupta et al., 2011), and characterized functions of some genes (such as PYL4 and PYL5) induced in the nNOS transgenic plants, but not by SNP application (Ahlfors et al., 2009).
Consistent with the results reported previously (Shi et al., 2012b, c ), nNOS transgenic plants exhibited significantly improved drought stress resistance (Fig. 1; Supplementary Fig. S1 at JXB online), which might be attributed to the increased in vivo NO content. Additionally, the effect of re-watering (Xu et al., 2010; Ziogas et al., 2013) was also determined in this study. After re-watering for 7 d, almost all WT plants died, while >55% of the nNOS plants survived (Supplementary Fig. S1a). Although no significant differences of LWC and EL were observed between the surviving WT and nNOS transgenic plants after re-watering for 7 d, the drought stress effect was fully recovered, as nNOS transgenic lines exhibited higher plant height and more biomass (DW) at harvest time (~45 d after re-watering) (Fig. 1). These results indicated that re-watering recovered the water status and cell damage caused by drought stress, and nNOS transgenic plants exhibited improved drought resistance with a higher survival rate. Under control and drought stress conditions, nNOS transgenic plants accumulated higher levels of osmolytes (proline, sucrose, and total soluble sugars) relative to WT plants (Fig. 2a–c), which would protect Arabidopsis plants by increasing cell membrane stability and balancing osmotic pressure in response to drought stress. Additionally, nNOS transgenic plants had lower levels of H2O2, the major indicator of ROS accumulation, and higher activities of antioxidant enzymes (SOD, CAT, POD, GR, and GPX) than WT plants under drought stress conditions (Fig. 2d–i). Previous studies in various plant species have shown that NO could modulate the activities of several antioxidant enzymes such as CAT, Fe-SOD, and dehydroascorbate reductase (DHAR) via S-nitrosylation modification (Shi and Chan, 2014). Based on the microarray data, although there were no significant changes in proline biosynthesis-related genes (Supplementary Table S1), many genes involved in redox metabolism (Supplementary Fig. S3a, b) and sugar metabolism (Supplementary Fig. S4a, b) were extensively changed by the nNOS transgene. As expected, some genes were up-regulated and others were down-regulated, indicating that NO had significant effects on the metabolism of these compounds partially through gene transcriptional modulation and protein post-translational modification. The results obtained with nNOS transgenic plants were consistent with those obtained with arginase1/2 mutants, which exhibited increased in vivo NO content, enhanced activities of antioxidant enzymes, reduced water loss and EL, and thus increased stress tolerance relative to the WT (Shi et al., 2013a). The reduced ROS accumulation and increased antioxidant enzyme activities confirmed that in vivo NO reduces drought stress-triggered oxidative stress and thereby reduces drought stress-triggered cell damage. These results, together with previously published observations (Gupta et al., 2011; Shi et al., 2012b , c, 2013a ; Tanou et al., 2012; Shi and Chan, 2013), indicated that osmolyte accumulation, ROS accumulation, and antioxidant enzyme activities were important in drought stress tolerance. Additionally, Tanou et al. (2012) characterized some potentially carbonylated, nitrated, and nitrosylated proteins with distinct and overlapping signatures that belong to metabolic categories linked to ROS and NO acclimation signalling.
To gain insight into the NO-regulated defence response at the molecular level, comparative transcriptomic analysis was performed, and 490 and 20 genes that were differentially expressed in WT and nNOS transgenic plants under control and drought stress conditions, respectively, were identified (Fig. 3). Quantitative real-time PCR of 30 genes supported the reliability of the microarray analysis (Supplementary Fig. S2a–c at JXB online). Interestingly, far fewer genes were changed by drought stress in the WT (WT drought versus WT control, 154) than by the nNOS transgene under normal conditions (nNOS control versus WT control, 490). Based on physiological analyses, nNOS transgenic plants accumulated a high level of NO- and stress-related metabolites (proline, sugars, and antioxidants), which functioned as stress signals and therefore might affect a wide range of transcripts. Pathway enrichment analysis indicated that nine pathways were over-represented among differentially expressed genes in nNOS transgenic plants, including photosynthesis, hormone metabolism, redox, stress, mitochondrial electron transport, OPP, and lipid metabolism (Table 2). Additionally, 24 stress-related genes, and in particular 16 of them with log2 fold change ≥1, were significantly regulated by the nNOS transgene effect under control conditions (Supplementary Table S1). Among these stress-related genes, At5g66590 is involved in unspecified abiotic stress; At3g05890, At2g24040, and At2g45130 are involved in drought and salt stresses; At2g17880 and At4g36040 are involved in heat stress; and other genes are involved in biotic stress. All of these genes might contribute to the enhanced stress tolerance of nNOS transgenic plants, indicating that nNOS transgenic lines might be pre-conditioned to be resistant to abiotic stresses.
Previous studies have identified genes regulated by treatment with the NO donor SNP (Ahlfors et al., 2009). Comparative analysis identified 22 genes that were regulated both by the nNOS transgene effect and by the SNP treatment, while six other genes showed the opposite expression pattern due to nNOS and SNP effects (Table 3). These 28 co-regulated genes might play essential roles in NO-mediated plant stress responses. A total of 165 genes that were regulated by both nNOS transgenic and ABA effects were also identified (Goda et al., 2008), and most of them displayed the same pattern of up- and down-regulation by nNOS transgenic and ABA effects (Supplementary Table S4 at JXB online). The cross-talk between ABA and NO has been studied in depth, especially in terms of how it relates to abiotic stress tolerance (Guo et al., 2003; Bright et al., 2006; Lozano-Juste and León, 2010). Lozano-Juste and León (2010) found several ABA-related phenotypes as well as enhanced dehydration stress tolerance in NO-deficient plants, which might be due to the function of NO as an endogenous negative regulator of the sensitivity to ABA, thus leading to NO-independent inhibition of stomatal opening and enhanced closure by ABA. Among these 165 genes, the expression of two ABA receptor genes (AtPYL4 and AtPYL5), which act upstream of the ABA pathway and directly modulate many downstream genes, was decreased by ABA and drought stress treatments (Chan, 2012), but increased in nNOS transgenic lines (Table 1; Supplementary Table S4), indicating a possible connection between NO and ABA pathways. Recent reports indicated that the constitutive overexpression of specific PYL genes in several plants enhanced their resistance to drought stress (Santiago et al., 2009; Sun et al., 2011). In this study, overexpression of AtPYL4 and AtPYL5 increased antioxidant enzyme activities and osmolyte levels and enhanced drought tolerance (Fig. 6; Supplementary Fig. S6). Up-regulation of AtPYL4 and AtPYL5 might contribute to the increased resistance of nNOS transgenic plants to drought stress. One possible mechanism might involve stomatal regulation because NO is involved in the stomatal closure triggered by ABA (Guo et al., 2003). The increased NO content in nNOS transgenic plants (Supplementary Fig. S1b) might promote stomatal closure and result in the previously reported decrease in water loss (Shi et al., 2012c ). Additionally, the up-regulation of AtPYL4 and AtPYL5 by the endogenous NO content as a consequence of nNOS gene transformation or stress treatment might also contribute to improved drought stress resistance. Therefore, the co-regulation of genes by ABA and NO provides new clues regarding the interaction between ABA and NO signal transduction pathways.
Additionally, 14 members of the zinc finger family of proteins, a family that regulates the transcription of several stress-responsive genes (Miller et al., 2008; Kodaira et al., 2011; Liu et al., 2012), were differentially expressed in WT versus nNOS transgenic plants (Supplementary Table S1 at JXB online). AtZAT10 was commonly up-regulated by the nNOS transgenic effect and by SNP treatment (Table 3). The overexpression of zinc finger family protein genes such as AZF1, AZF2, AZF3, ZAT6, ZAT7, ZAT10, and ZAT12 increased the tolerance of Arabidopsis to high light, high salt, drought, osmotic, and oxidative stresses (Miller et al., 2008; Kodaira et al., 2011; Liu et al., 2012). Although the mechanisms by which different zinc finger proteins affect stress tolerance may be diverse and complex, the modulation of the expression of genes that encode several zinc finger family proteins by the nNOS transgene might contribute to the enhanced drought tolerance in nNOS transgenic plants. Moreover, the important role of some zinc finger proteins (AtZAT7, AtZAT10, and AtZAT12) in ROS signalling also suggests an important interaction among NO, zinc finger proteins, and ROS signalling in plant responses to stress. Additionally, CBF/DREB are known to increase the tolerance to abiotic stress (Seki et al., 2001; Haake et al., 2002; Sakuma et al., 2006; Achard et al., 2008; Novillo et al., 2012), and their expression was greater in nNOS transgenic plants than in WT plants (Table 2; Supplementary Fig. S2a); it follows that the enhanced expression of CBF1 and CBF2 might also contribute to the enhanced stress tolerance of nNOS transgenic plants.
Taken together, the present comparative physiological and transcriptomic analyses revealed that WT and nNOS transgenic plants differed greatly in their physiological and molecular responses to drought stress. It is reasonable to infer that these differences might partially explain the difference in resistance to drought stress in WT versus nNOS transgenic plants. It is inferred that increased endogenous NO content resulting from nNOS transformation or from stress treatment modulates ROS accumulation, the activities of antioxidant enzymes, osmolyte levels, and the expression of stress-responsive genes (such as AtPYL4 and AtPYL5), resulting in enhanced drought resistance. These finding increased our understanding of the physiological and molecular mechanisms by which NO mediates the drought stress response in Arabidopsis.
Supplementary data
Supplementary data are available at at JXB online.
Table S1. Spreadsheet of all genes whose expression was changed by the nNOS transgene or drought stress treatment.
Table S2. Primers used for quantitative real-time PCR and vector construction.
Table S3. Gene lists of the 16 clusters analysed by MapMan software.
Table S4. Spreadsheet of genes commonly regulated by both the nNOS transgene effect and the ABA effect in Arabidopsis.
Figure S1. Improved drought stress resistance in nNOS transgenic Arabidopsis plants.
Figure S2. Verification of the microarray data by quantitative real-time PCR.
Figure S3. Metabolic pathway analyses of differentially expressed genes.
Figure S4. Effects of the nNOS transgene (a) and the drought stress treatment (b) on the plant photosynthesis pathway using MapMan software.
Figure S5. Enriched GO terms resulting from the nNOS transgene and the drought stress treatment.
Figure S6. Hierarchical cluster analysis of physiological parameters differentially expressed in WT, nNOS-, AtPYL4- and AtPYL5-overexpressing plants.
Acknowledgements
We are grateful to Rebecca Ann Stevenson for language editing. We thank Profesor Yingtang Lu and Dr Rongjun Li for their help in this research, and we thank Professor Pingfang Yang for help with quantitative real-time PCR. This research was supported by the National Natural Science Foundation of China (grant no. 31370302) and the Knowledge Innovative Key Program of the Chinese Academy of Sciences (grant no. 54Y154761O01076 and Y329631O0263) to ZC, by the Chinese Academy of Sciences and US National Institutes of Health Grant R01GM059138 to J-K, and by the National Natural Science Foundation of China (grant no. 31200194), Youth Innovation Promotion Association of the Chinese Academy of Sciences (grant no. Y429371O04), and the Outstanding Young Talent Program of Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture (grant no. Y352811O03 and Y452331O03) to HS.
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. 2008. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. The Plant Cell 20, 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors R, Brosché M, Kollist H, Kangasjärvu J. 2009. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana . The Plant Journal 58, 1–12 [DOI] [PubMed] [Google Scholar]
- Arasimowicz-Jelonek M, Floryszak-Wieczorek J, Kosmala A. 2011. Are nitric oxide donors a valuable tool to study the functional role of nitric oxide in plant metabolism? Plant Biology 13, 747–756 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Astiera J, Rasula S, Wawera I, Dubreuil-Maurizia C, Jeandrozb S, Wendehennea D. 2009. Current view of nitric oxide-responsive genes in plants. Plant Science 177, 302–309 [Google Scholar]
- Bouchard JN, Yamasaki H. 2008. Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant and Cell Physiology 49, 641–652 [DOI] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. 2006. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent H2O2 synthesis. The Plant Journal 45, 113–122 [DOI] [PubMed] [Google Scholar]
- Chan Z. 2012. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100, 110–115 [DOI] [PubMed] [Google Scholar]
- Chan Z, Bigelow P, Loescher W, Grumet R. 2012. Comparison of salt stress resistance genes in transgenic Arabidopsis thaliana indicates that extent of transcriptomic change may not predict secondary phenotypic or fitness effects. Plant Biotechnology Journal 10, 284–300 [DOI] [PubMed] [Google Scholar]
- Chen R, Sun S, Wang C, et al. 2009. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Research 19, 1377–1387 [DOI] [PubMed] [Google Scholar]
- Chun HJ, Park HC, Koo SC, et al. 2013. Constitutive expression of mammalian nitric oxide synthase in tobacco plants triggers disease resistance to pathogens. Molecules and Cells 34, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Hayashi M, Mano S, Nishimura M, Barroso JB. 2009. Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiology 151, 2083–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JX, Zhou YH, Ding JG, Xia XJ, Shi K, Chen SC, Asami T, Chen Z, Yu JQ. 2011. Role of nitric oxide in hydrogen peroxide-dependent induction of abiotic stress tolerance by brassinosteroids in cucumber. Plant, Cell and Environment 34, 347–358 [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, and Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QJ, Liu JH. 2012. Nitric oxide is involved in dehydration/drought tolerance in Poncirus citri between kumquat and sweet orange with contrasting canker tolerance. Plant Cell Reports 31, 145–154 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Hasanuzzaman M, Nahar K, Macovei A, Tuteja N. 2013. Importance of nitric oxide in cadmium stress tolerance in crop plants. Plant Physiology and Biochemistry 63, 254–261 [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, et al. 2008. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. The Plant Journal 55, 526–542 [DOI] [PubMed] [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J. 2006. Nitric oxide and gene regulation in plants. Journal of Experimental Botany 57, 507–516 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM. 2003. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. 2011. On the origins of nitric oxide. Trends in Plant Science 16, 160–168 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. 2002. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology 130, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, et al. 2004. Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971 [DOI] [PubMed] [Google Scholar]
- Huang X, von Rad U, Durner J. 2002. Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215, 914–923 [DOI] [PubMed] [Google Scholar]
- Huang XS, Luo T, Fu XZ, Fan QJ, Liu JH. 2011. Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliate whose ectopic expression confers dehydration/drought tolerance in transgenic tobacco. Journal of Experimental Botany 62, 5191–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LSP, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology 157, 742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Bartha B, Erdei L. 2007. Exogenous auxin-induced NO synthesis in nitrate reductase-associated in Arabidopsis thaliana root primordia. Journal of Plant Physiology 165, 967–975 [DOI] [PubMed] [Google Scholar]
- Liu XM, Nguyen XC, Kim KE, Han HJ, Lee K, Yun DJ, Chung WS. 2012. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochemical and Biophysical Research Communications 430, 1054–1059 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. 2010. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiology 152, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. 2008. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum 133, 481–489 [DOI] [PubMed] [Google Scholar]
- Novillo F, Medina J, Rodríguez-Franco M, Neuhaus G, Salinas J. 2012. Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. Journal of Experimental Botany 63, 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri MC, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C. 2008. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. Journal of Experimental Botany 59, 177–186 [DOI] [PubMed] [Google Scholar]
- Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman DW, Goldman SL. 2004. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnology Journal 2, 359–366 [DOI] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M. 2003. Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana . Molecular Plant-Microbe Interactions 16, 1094–1105 [DOI] [PubMed] [Google Scholar]
- Provart N, Zhu T. 2003. A browser-based functional classification superviewer for Arabidopsis genomics. Current Computer Molecular Biology 2003, 271–272 [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell 18, 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. 2009. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. The Plant Journal 60, 575–588 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. 2001. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. The Plant Cell 13, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HT, Chan ZL. 2013. In vivo role of Arabidopsis arginase in arginine metabolism and abiotic stress response. Plant Signaling and Behavior 5, e24138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chan Z. 2014. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. Journal of Integrative Plant Biology 56, 114–121 [DOI] [PubMed] [Google Scholar]
- Shi H, Wang Y, Chen Z, Ye T, Chan Z. 2012a. Analysis of natural variation in bermudagrass (Cynodon dactylon) reveals physiological responses underlying drought tolerance. PLoS One 7, e53422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, Zhang Y, Chan Z. 2013a. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. Journal of Experimental Botany 64, 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Wang Y, Chan Z. 2013b. Arabidopsis ALTERED MERISTEM PROGRAM 1 negatively modulates plant responses to abscisic acid and dehydration stress. Plant Physiology and Biochemistry 67, 209–216 [DOI] [PubMed] [Google Scholar]
- Shi HT, Li RJ, Cai W, Liu W, Fu ZW, Lu YT. 2012b. In vivo role of nitric oxide in plant response to abiotic and biotic stress. Plant Signaling and Behavior 7, 438–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HT, Li RJ, Cai W, Liu W, Wang CL, Lu YT. 2012c. Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant and Cell Physiology 53, 344–357 [DOI] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. 2011. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany 62, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanou G, Filippou P, Belghazi M, Job D, Diamantidis G, Fotopoulos V, Molassiotis A. 2012. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. The Plant Journal 72, 585–599 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939 [DOI] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EIS, Scherer GFE. 2006. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant and Cell Physiology 47, 346–354 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun PP, Chen CL, Wang Y, Fu XZ, Liu JH. 2011. An arginine decarboxylase gene PtADC from Poncirus trifoliate confers abiotic stress tolerance and promotes primary root growth. Journal of Experimental Botany 62, 2899–2914 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. 2006. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22, 897–899 [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhou G, Shimizu H. 2010. Plant responses to drought and rewatering. Plant Signaling and Behavior 5, 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH. 2007. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiology 144, 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH. 2009. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiology 151, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziogas V, Tanou G, Filippou P, Diamantidis G, Vasilakakis M, Fotopoulos V, Molassiotis A. 2013. Nitrosative responses in citrus plants exposed to six abiotic stress conditions. Plant Physiology and Biochemistry 68, 118–126 [DOI] [PubMed] [Google Scholar]
- Zottini M, Costa A, Michele RD, Ruzzene M, Carimi F, Schiavo FL. 2007. Salicylic acid activates nitric oxide synthesis in Arabidopsis. Journal of Experimental Botany 58, 1397–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.