Summary
OsXXT1 showed xyloglucan 6-xylosyltransferase activity and is involved in maintaining cell wall structure and tensile strength in rice. Mutation of OsXXT1 results in abnormal root hair development.
Key words: Rice, root hair, type I cell wall, type II cell wall, xyloglucan, xylosyltransferase.
Abstract
Root hairs are important for nutrient uptake, anchorage, and plant–microbe interactions. From a population of rice (Oryza sativa) mutagenized by ethyl methanesulfonate (EMS), a short root hair2 (srh2) mutant was identified. In hydroponic culture, srh2 seedlings were significantly reduced in root hair length. Bubble-like extrusions and irregular epidermal cells were observed at the tips of srh2 root hairs when grown under acidic conditions, suggesting the possible reduction of the tensile strength of the cell wall in this mutant. Map-based cloning identified a mutation in the gene encoding xyloglucan (XyG) 6-xylosyltransferase (OsXXT1). OsXXT1 displays more than 70% amino acid sequence identity with the previously characterized Arabidopsis thaliana XYG XYLOSYL TRANSFERASE 1 (AtXXT1) and XYG XYLOSYL TRANSFERASE 2 (AtXXT2), which catalyse the transfer of xylose onto β-1,4-glucan chains. Furthermore, expression of the full-length coding sequence of OsXXT1 could complement the root hair defect, and slow growth and XyG synthesis in the Arabidopsis xxt1 xxt2 double mutant. Transgenic plants expressing the β-glucuronidase (GUS) reporter under the control of the OsXXT1 promoter displayed GUS expression in multiple tissues, most prominently in root epidermal cells. These results demonstrate the importance of OsXXT1 in maintaining cell wall structure and tensile strength in rice, a typical grass species that contains relatively low XyG content in cell walls.
Introduction
Plant cell walls are composed of cellulose, hemicellulose, and pectin, in addition to a number of inorganic compounds. Based on the chemical structures, wall architecture, and cell wall biosynthetic processes, primary cell walls of flowering plants are divided into two classes, type I and type II (Carpita and Gibeaut., 1993; Carpita, 1996; Carpita and McCann, 2000). Type I cell walls are typically found in dicots and non-commelinoid monocots. They are characterized by a cellulose–xyloglucan (XyG) network with high pectin and structural proteins content. Type II cell walls are found only in the commelinoid monocots (e.g. grasses, rushes, and gingers) and are composed of cellulose fibres encased in glucuronoarabinoxylans (GAX), high levels of hydroxycinnamates, and very low levels of pectin and structural proteins. Additionally, cell walls of grasses and some related families contain significant amounts of mixed linkage glucans (Carpita, 1996; Carpita and McCann, 2000; Vogel, 2008).
XyG is the most abundant hemicellulosic polysaccharide in type I cell walls, which comprises ~20–25% dry mass of cell walls (Obel et al., 2007; Vogel, 2008). XyG consists of a β-1,4-glucan backbone with α-d-xylose substitution on the oxygen-6 position in a regular pattern. There are two general types of XyG: the poly-XXXG and poly-XXGG; approximately 75% and 50% of their backbone residues are branched, respectively (Vincken et al., 1997). These xylosyl residues can be further substituted at the oxygen-2 position with either a single β-D-galactose or an α-L-fucose-(1,2)-β-d-galactose dimer, depending on the plant species (Obel et al., 2007). The sub-structure of XyG has been investigated in detail using β-1,4-endoglucanase to digest the non-substituted glucose units in the polymer (Hoffman et al., 2005). Hydrolysed Arabidopsis XyG released XXXG, XLXG, XXFG, XLLG, and XLFG subunits (see Figure 1 and 2 in Obel et al. (2007) for a description of XyG nomenclature). In contrast to the observations made in Arabidopsis, the hemicelluloses of primary cell walls in graminaceous monocots are mainly comprised of xylans and mixed linkage glucans. Only ~1–5% of XyG is found in cereals or grasses with a low degree of substitution (Carpita, 1996; Hoffman et al., 2005; Sims et al., 2000).
Fig. 1.
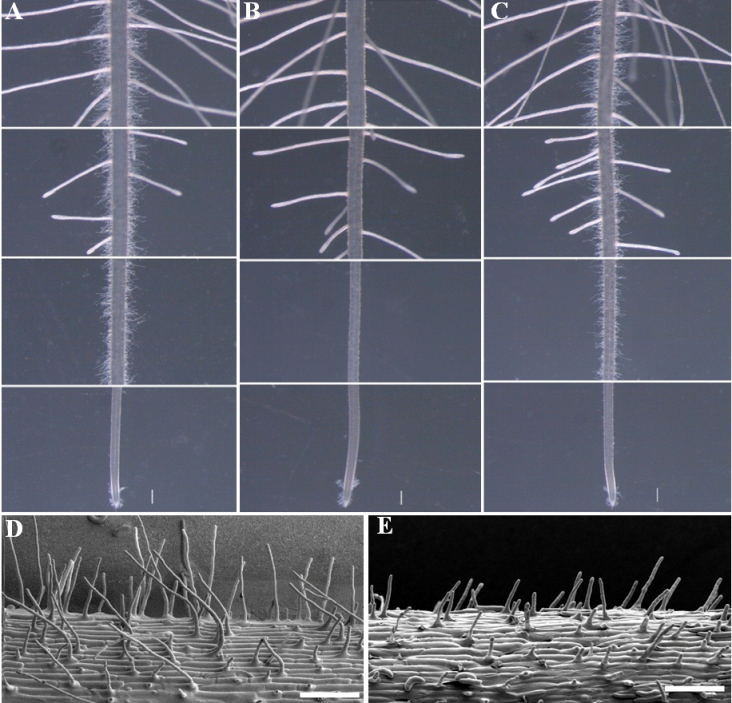
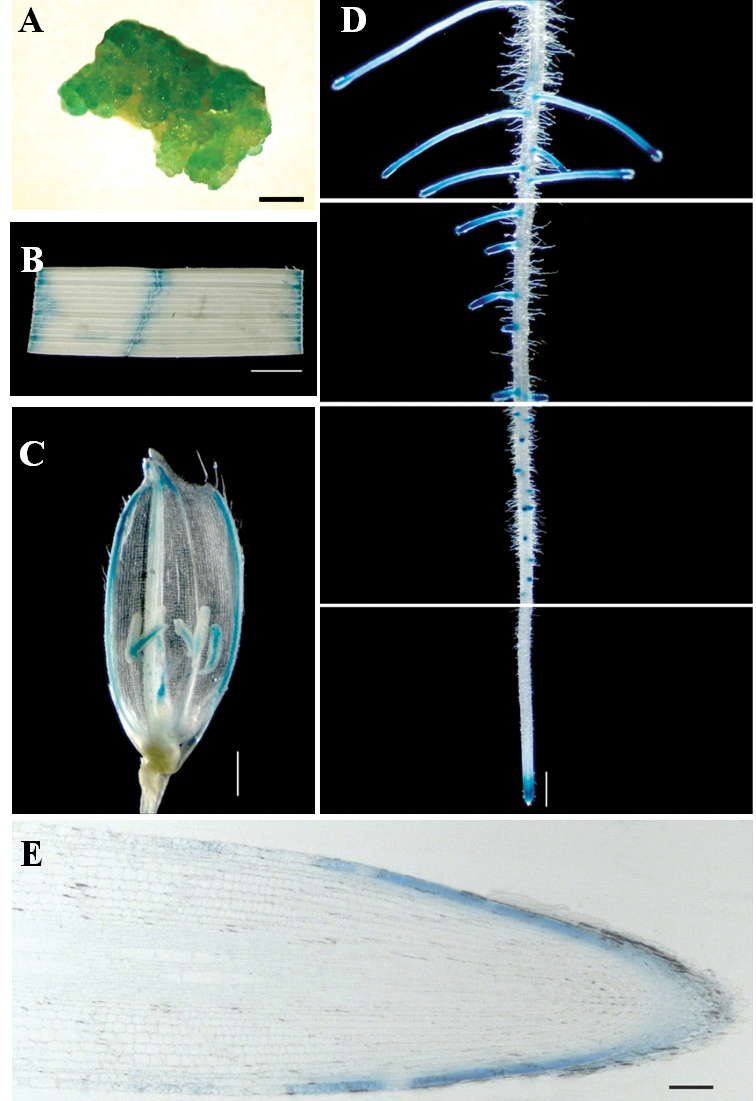
Phenotype of root hairs from wild type (WT, cv Kasalath), srh2, and srh2 complemented by Ubiquitin-1 promoter::OsXXT1. (A and D) root hairs from the WT; (B and E) root hairs from srh2; (C) root hairs from transgenic seedling of srh2 overexpressing OsXXT1. Seedlings of A, B, and C were grown under the same pots with nutrient solution for seven days. Bar=1cm. Seedlings of D and E were grown for three days on Murashige and Skoog medium (pH 5.5) and examined under an electron microscope examination. Bar=200 μm.
Fig. 2.
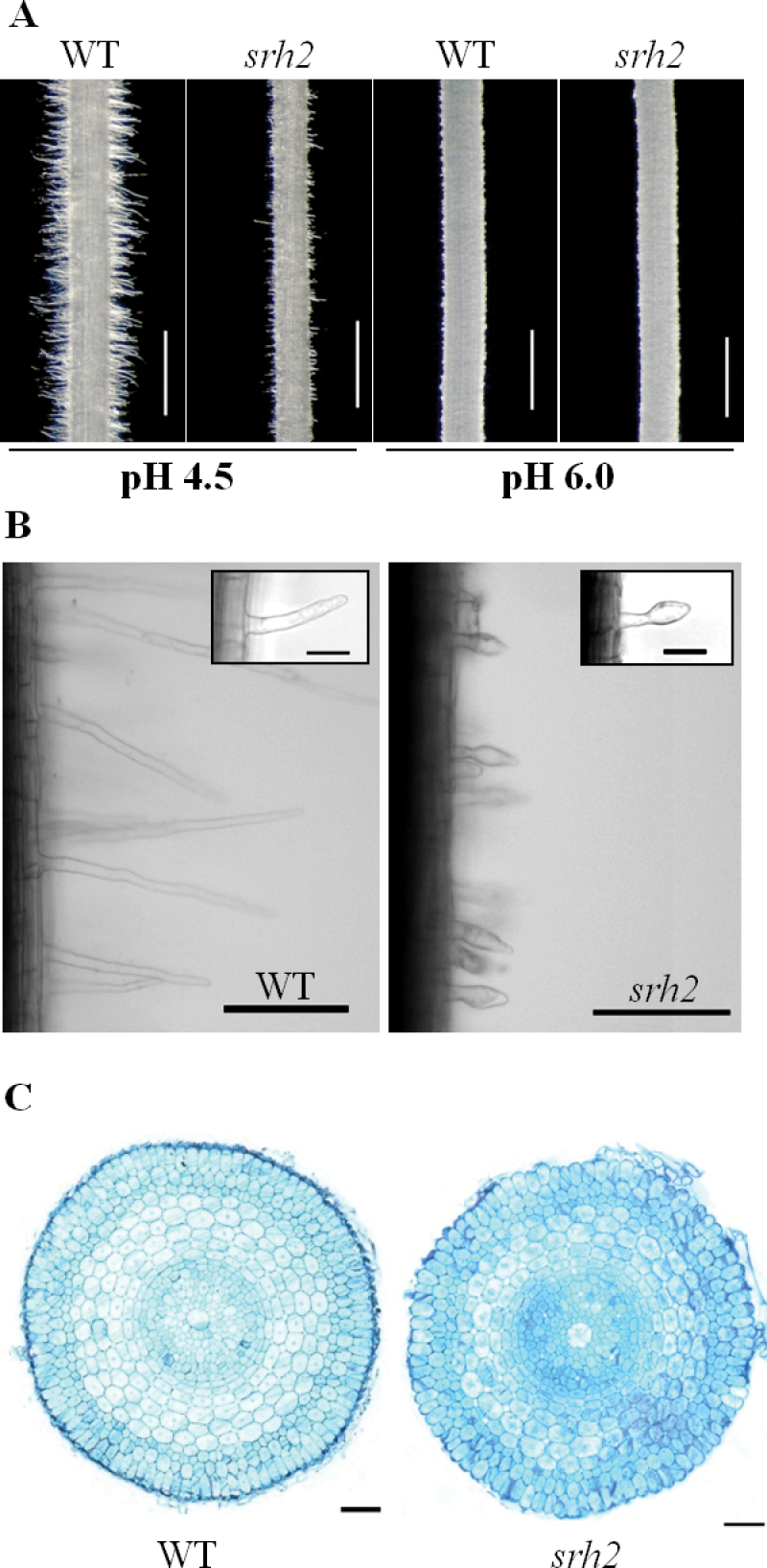
Root hair morphology of rice seedlings grown in different media with various pH. (A) Root hairs of three-day-old seedlings of wild-type (WT) and srh2 grow on Murashige and Skoog medium at pH 4.5 and 6.0. Bar=1mm. (B) The root hairs of the WT and srh2 examined under a microscope (pH 4.5). Bar=100 μm. (C) Cross section of root tip from WT and srh2 seedlings grown at pH 4.5. The section was strained with toluidine blue (TBO). Bar=20 μm. (This figure is available in colour at JXB online.)
Although the cell wall composition differs between type I and type II cell walls, many cell wall-related genes are conserved between species with both types, presumably to maintain the basic structure of cell walls (Penning et al., 2009; Yokoyama and Nishitani, 2004). The β-glucan backbone of XyG is synthesized by a cellulose synthase-like C family protein, named CSLC4 in Nasturtium and Arabidopsis (Cocuron et al., 2007). AtXXT1, AtXXT2, and AtXXT5 are involved in linking xylose to the β-glucan backbone at different positions (Cavalier and Keegstra, 2006; Cavalier et al., 2008; Faik et al., 2002; Vuttipongchaikij et al., 2012; Zabotina et al., 2008; Zabotina et al., 2012). The galactosyltransferase MUR3 and fucosyltransferase MUR2 further catalyse the UDP–galactose or UDP–fucose to the side chain of a XyG oligosaccharide block (Madson et al., 2003; Vanzin et al., 2002). Plants with mutations affecting XyG structure or content exhibited collapsed trichome papillae or abnormal root hairs (Cavalier et al., 2008; Madson et al., 2003; Pena et al., 2012; Vanzin et al., 2002; Zabotina et al., 2008), and Arabidopsis XyG mutants had reduced tensile strength in primary cell walls (Cavalier et al., 2008; Park and Cosgrove, 2012a; Pena et al., 2004).
Grass cell walls are a major source of dietary fibre for animals and a significant source of renewable energy (Ragauskas et al., 2006). Cell walls in grass species contain significant amounts of GAX and mixed linkage glucans, but relatively low amounts of XyG. It is commonly accepted that XyG is a less important component in type II cell walls compared with type I cell walls (Vogel, 2008). The function and genetic controls of synthesis of XyG is unknown in type II cell walls.
In this study, we isolated and characterized the rice short root hair2 (srh2) mutant and provide evidence for the importance of XyG in cell wall structure and root hair tip growth in a grass species.
Materials and methods
Plant materials and growth conditions
The rice srh2 mutant was identified in an EMS-mutagenized population from the rice cultivar Kasalath. For all experiments, the srh2 mutant and wild-type seeds were germinated in distilled water for two days. The seedlings were then grown in a hydroponic solution (Yoshida et al., 1976). Seedlings were grown in a growth chamber at 30 °C/22 °C day/night temperatures with a 12h light/12h dark regime (450 μmol photons m–2s–1). For the pH treatment experiment, seeds were surface sterilized with 95% (v/v) ethanol for 2min and 15 % (v/v) bleach for 20min. After rinsing in distilled water, seeds were germinated and grown in test tubes (15 cm×3cm) containing Murashige and Skoog medium, 0.059% (w/v) 2-(N-morpholino) ethanesulfonic acid (MES), 1% (w/v) sucrose and 0.3% (w/v) phytagel (Sigma, US). The basic medium contained 2.0mM NH4NO3, 1.9mM KNO3, 0.3mM CaCl2·2H2O, 0.15mM MgSO4·7H2O, 5 μM KI, 25 μM H3BO3, 0.1mM MnSO4·H2O, 0.3mM ZnSO4·7H2O, 1 μM Na2MO4·2H2O, 0.1 μMCuSO4·5H2O, 0.1 μM CoCl2·6H2O, 0.1mM FeSO4·7H2O, and 0.1mM Na2EDTA·2H2O.
Map-based cloning and genetic complementation
An F2 mapping population was generated from crosses between homozygous srh2 mutant plants and the Japonica cultivar Nipponbare. The SRH2 gene was mapped to chromosome 3 between simple sequence repeat (SSR) markers RM232 and RM3280 using 1800 F2 mutant plants. SSR markers were obtained from NCBI database (http://www.ncbi.nlm.nih.gov/unists). The mutation was further mapped to a 36-kb region between STS274-04 and STS274-04-06 using nine newly developed SSR markers. Based on the phenotype of srh2, the OsXXT1 gene was selected as a candidate gene. A derived cleaved amplified polymorphic sequences (dCAPS) marker was developed using the dCAPS finder 2.0 program (http://helix.wustl.edu/dcaps/dcaps.html) to further confirm the mapping result. The genes were amplified by PCR from genomic DNA isolated from srh2 and wild-type plants and sequenced to identify the mutation in the genomic sequence.
For complementation, the full-length open reading frame of OsXXT1 was amplified by reverse transcription PCR and inserted into the modified binary vector pTF101-ubi (Zheng et al., 2010) between the maize Ubiquitin-1 promoter and a nopaline synthase terminator. The resulting transformation plasmid, pXXT1-Oe, was used for the Agrobacterium-mediated rice transformation of srh2 mutant as described (Nishimura et al., 2006).
For the complementation of Arabidopsis xxt1 xxt2 double mutant, the full length coding sequence of OsXXT1 was PCR amplified and cloned into the binary vector (pH7WG2D). The expression of OsXXT1 was driven by the constitutive 35S CaMV promoter. This binary vector was then transformed into Agrobacterium tumefaciens GV3130 strain. Floral dipping was performed with an inoculum medium containing 10% (w/v) sucrose and 0.05% (v/v) Silwet-77 (Clough, 1998). T1 transformants were screened on hygromycin (50mg l–1) (Harrison, 2006). Successful transformants, were age matched and Columbia-0 and Arabidopsis xxt1 xxt2 double mutant plants were used for imaging using a Nikon Eclipse 80i Microscope (Nikon, Japan).
Microscopic analysis
The root hairs were examined by a Leica MZ95 stereomicroscope with a colour CCD camera (Leica Instrument, Nusslosh, Germany). For cryo-scanning electron microscopy seeds were plated on Murashige and Skoog medium and grown for three days (pH 5.7). Root samples were placed on moist nitrocellulose paper mounted on a stub and immersed in liquid nitrogen slush, then transported under vacuum to a cryo preparation chamber. Ice was sublimed at –90 °C and the specimens were sputter-coated with gold and observed using a Hitachi S-3000N scanning electron microscope (Hitachi, Naka, Japan) with a Gatan Alto 2100 cryo preparation system (Gatan UK, Abingdon, UK).
For light microscopic analysis, root tips were fixed overnight in 2.5% (v/v) glutaraldehyde with 0.1M sodium phosphate buffer (pH 7.2) and washed three times for 30min in the same buffer. Root samples were then re-fixed for 4h in 1% (v/v) OsO4 with 0.1M sodium phosphate buffer (pH 7.2) and washed for 30min in the same buffer. The samples were dehydrated in a gradient ethanol and embedded in Spurr resin. Semi-thin sections (2 μm) were made using glass knives on a Power Tome XL (RMC-Boeckeler Instruments, Arizona, USA) microtome and stained in 0.1% (w/v) methylene blue for 3min at 70 °C. The samples were rinsed with distilled water and visualized with a Zeiss Axiovert 200 microscope (Zeiss, Jena, Germany).
For GUS staining, tissues samples were fixed overnight in FAA (constituted of 5% (v/v) formalin, 5% (v/v) acetic acid, 75% (v/v) alcohol) and washed twice for 30min in 70% (v/v) ethanol. The tissues were dehydrated in gradient acetone and embedded in Spurr resin. The sections and visualizations were carried out as described above.
Sequence alignments and phylogenic analysis
The putative XXT sequences for alignment were extracted from NCBI (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment of XXT proteins was conducted using the ClustalX 1.83 program (Thompson et al., 1997) with default multiple alignment parameters and viewed by GeneDoc 3.2. A phylogenic tree of the gene family was constructed using the Neighbor–Joining method by MEGA5.
Matrix-assisted laser desorption/ionizationd time-of-flight (MALDI-TOF) mass spectrometry analysis of xyloglucan oligosaccharides
Cell wall was extracted from leaves that were ground into powder in liquid nitrogen. The homogenate was washed three times with hot 70% (v/v) ethanol and extracted with a mixture of chloroform and methanol (1:1). The pellet was suspended in acetone and air-dried overnight. The alcohol-insoluble residues (AIRs) were de-starched with α-amylase (Bacillus sp). The XyG enriched KOH-soluble fraction was prepared by neutralizing 50mg of de-starched AIRs in 4M KOH solution, samples were then dialysed and finally lyophilized. Then, 0.5mg of AIRs or KOH fraction was incubated in 100ml of 50mM ammonium formate, pH 5.0, with one unit of xyloglucanase (EXEGP; Megazyme) for 18h at 37 °C. The supernatants were recovered, and 1ml of aqueous sample plus 10ng xylopentaose was spotted with an equal volume of matrix solution (10mg ml–1 2,5-dihydroxbenzoic acid). After being dried on the MALDI target plate, spectra were analysed on a Bruker Autoflex MALDI-TOF mass spectrometry instrument (Bruker) in the positive reflection mode with an acceleration voltage of 20kV. The relative height of each generated oligosaccharide ion peak was counted to determine their relative abundance as described previously (Zhang et al., 2012).
Generation of OsXXT1::GUS transgenic lines
The 1.8-kb region upstream of the start codon of OsXXT1 gene was amplified from the genomic DNA of Kasalath using primers shown in Table S1. The PCR product was then cloned into the pBIGUS-plus vector, in which the original GUS gene in binary vector pBI101.3 was replaced by a GUS-Plus sequence from pCAMBIA1305.1. The resultant vector was introduced into Nipponbare rice using Agrobacterium-mediated rice transformation as described (Nishimura et al., 2006).
GUS histochemical analysis
Histochemical GUS staining was performed as described in Jefferson et al. (1987). Plant tissues from GUS transgenic lines were immediately submerged in GUS staining solution after harvest and placed under a vacuum for 10min. The samples were incubated overnight in darkness at 37 °C. Chlorophyll was removed by submerging the stained tissues in 70% (v/v) ethanol. Plant material was placed on glass slides using 20% chloral (w/v) in 25% glycerol (v/v) for 10min. GUS staining was visualized using a Leica MZ95 stereomicroscope with a colour CCD camera (Leica Instrument, Nusslosh, Germany). The GUS staining solution contained 100mM sodium phosphate buffer (pH 7.0), 10mM Na2EDTA, 1mM K3[Fe(CN6)], 1mM K4[Fe(CN6)], 0.5% (v/v) TritonX-100, 20% (v/v) methanol, and 0.5mg ml–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc).
Expression analysis
Publically available Affymetrix Arabidopsis and rice microarray CEL files were downloaded from the Gene Expression Omnibus within the National Centre for Biotechnology Information database or from the MIAME ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/). The CEL files were imported and quantile normalized together using Partek Genomics Suite version 6.5 (St. Louis, Missouri, USA) as carried out in previous studies (Narsai et al., 2011). The accession numbers for the Arabidopsis studies were GSE30223 and E-AFMX-9, and for rice several were combined, including E-MEXP-1766, E-MEXP-2267, GSE6908, GSE11966, GSE7951, and GSE6893.
Results
Isolation of the srh2 mutant
Seeds from the M2 generation of an EMS-mutagenized population of Indica cultivar Kasalath, were germinated and grown in nutrient solution to screen for mutants with abnormal root hair phenotype. Seven days after germination, a mutant with significantly reduced length in root hairs was identified (Fig. 1A, B). No obvious difference was observed in leaf or root growth between wild type and mutant (Fig. S1). The mutant was designated as srh2. Scanning electron microscopy of the wild-type and srh2 root surface showed that the length of the root hairs of srh2 were shorter than those of wild type. However, no significant difference was found in root hair density or distribution pattern between srh2 and wild type (Fig. 1D, E).
It is reported that extracellular pH can regulate root hair growth by modification to cell wall rigidity (Monshausen et al., 2007). To investigate whether the root hair growth of srh2 is affected by extracellular pH, wild-type and mutant seeds were germinated on Murashige and Skoog medium under pH 4.5 and 6.0. The medium pH was stabilised by MES. At pH 6.0, root hair growth in both wild type and the srh2 mutant was significantly inhibited (Fig. 2A). The acidic conditions (pH 4.5) induced root hair growth in wild-type seedlings, but not in the srh2 mutant (Fig. 2A). Bubble-like extrusions were observed at the tip of the hairs in the srh2 mutants grown under acidic conditions (Fig. 2B). Transverse section of the wild-type and mutant roots showed that the shape of epidermal cells in the srh2 root meristem zone was irregular (Fig. 2C).
Gene cloning of the srh2 mutant
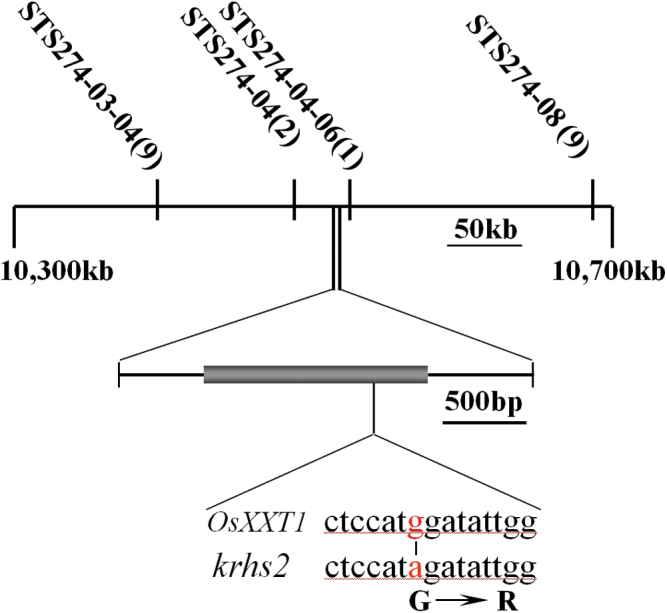
Genetic analysis showed that a single recessive gene was responsible for the mutant phenotype. In 1800 mutant seedlings from the F2 population derived from a cross between the mutant (from the Indica cultivar Kasalath) and the Japonica cultivar Nipponbare, the roots hairs of 1362 seedlings grew normally, whereas 438 seedlings showed short root hairs. Using this population, the mutation was mapped to a 36-kb region between SSR markers STS274-04 and STS274-04-06 on chromosome 3 (Fig. 3). This region contains seven open reading frames, including a putative OsXXT1 gene (LOC_Os03g18820). Altered XyG structure or content of cell walls in Arabidopsis caused collapsed trichome papillae or a short root hair phenotype (Cavalier et al., 2008; Madson et al., 2003; Vanzin et al., 2002; Zabotina et al., 2012). Therefore, the phenotype of srh2 suggested that the OsXXT1 gene is a tentative candidate gene for the mutation. To confirm this, the full-length cDNA sequence of the OsXXT1 gene was amplified from both the wild-type and srh2 mutant genomic DNA. Comparison of these two sequences revealed the presence of a single point mutation (G to A) at the nucleotide position 1009bp from the start codon of OsXXT1 (Fig. 3). The nucleotide substitution of the srh2 mutant sequence resulted in an amino acid change of a glycine (G) into an arginine (R) (Fig. 3). To further verify the positional cloning of the mutant, a dCAPS marker was developed using the NcoI restriction endonuclease (Fig. S2). The enzyme cuts the PCR product of the wild-type OsXXT1 gene into four fragments: 456bp, 73bp, 244bp, and 1210bp. In contrast, the PCR product amplified from the srh2 mutant produced only three fragments when digested with NcoI: 529bp, 244bp, and 1210bp (Fig. S2).
Fig. 3.
Molecular identification of srh2 by positional cloning. Physical map of the chromosomal region encompassing the srh2 gene was defined by high-resolution mapping. srh2 was mapped between the simple sequence repeat (SSR) markers STS274-04 and STS274-04-06 with the number of recombinants given in parentheses. (This figure is available in colour at JXB online.)
A genetic complementation test was carried out to confirm that the point mutation in OsXXT1 was responsible for the mutant phenotype. The full-length open reading frame of the wild-type OsXXT1 gene was inserted into the binary vector pTF101.1 under the control of a maize Ubiquitin-1 promoter. The resulting construct was used to introduce a full-length OsXXT1 gene into the srh2 mutant genome via Agrobacterium-mediated transformation. Four positive transgenic lines were identified. The root hair from the T2 transgenic seedlings displayed normal root hair growth (Fig. 1C), indicating overexpression of the OsXXT1 gene could complement the mutant phenotype completely.
Protein structure and phylogenetic analysis of OsXXT1
The OsXXT1 gene encodes a predicted protein of 448 amino acids that has been classified as a member of glycosyltransferase family 34 (GT34) in CAZy (http://www.cazy.org). The OsXXT1 protein is predicted to have a transmembrane domain near the N-terminus and a glycosyltransferase domain near the C-terminus (Fig. 4A). The base substitution of the srh2 mutant sequence resulted in an amino acid change of a glycine to an arginine in the highly conserved glycosyltransferase domain (Fig. S3).
Fig. 4.
Schematic diagram of protein domain structure and phylogenetic analysis of OsXXT1. (A) Predicted schematic of OsXXT1 protein domain structure. TM, predicted transmembrane domain; GT, predicted glycosyltransferase domain. (B) Neighbor–joining phylogenic tree of putative xylosyltransferases in Arabidopsis and rice using MEGA5 program. CAB52246 is accession number of fenugreek α-(1,6) galactosyltransferase. The gene locus of rice GT34 genes are: OsXXT1, LOC_Os03g18820; OsGT2, LOC_Os02g32750; OsGT3, LOC_Os12g05380; OsGT4, LOC_Os03g19310; OsGT5, LOC_Os03g19330; OsGT6, LOC_Os11g34390; OsGT7, LOC_Os02g49140.
Phylogenetic analysis of 14 putative xylosyltransferases in Arabidopsis and rice revealed that these genes could be divided into two classes (Fig. 4B). Five genes from rice, and five genes from Arabidopsis—AtXXT1 to AtXXT5, which have demonstrated XXT activity—are grouped into class I. Class II contains two Arabidopsis genes (AtGT6 and AtGT7) and two rice genes (OsGT6 and OsGT7), which are clustered with the fenugreek α-(1,6) galactosyltransferase (CAB52246). Although there are 8, 10, and 18 RNA variants encoding GT34 protein members in Arabidopsis, rice, and maize (Penning et al., 2009), respectively, only class I proteins contained both the transmembrane and glycosyltransferase domains in Arabidopsis and rice (Fig. S3, and Fig. 4B).
Functional verification of OsXXT1 by complementation of the Arabidopsis xxt1 xxt2 double mutant
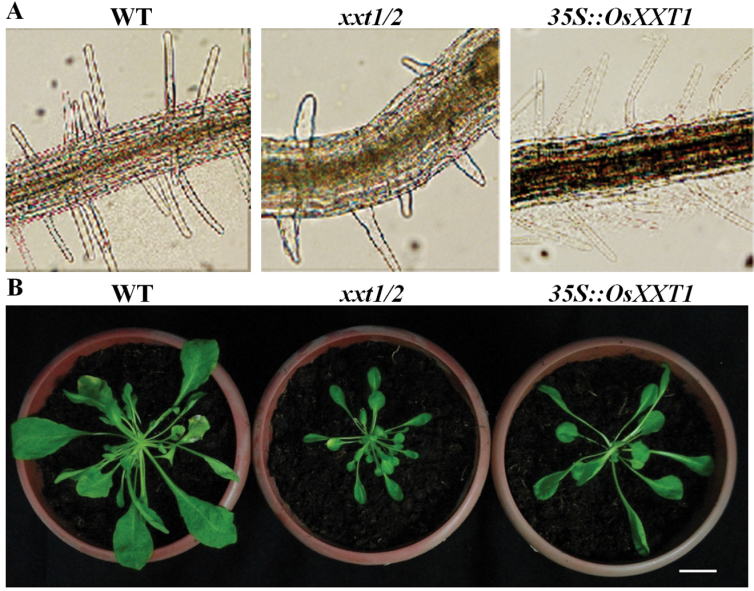
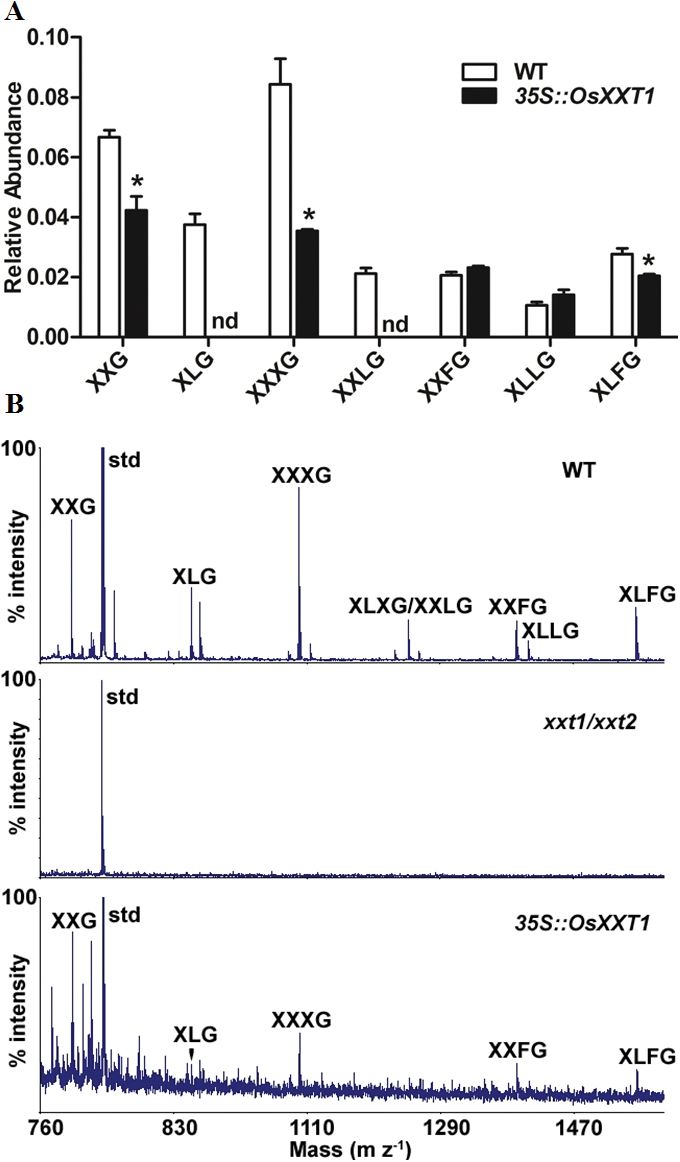
The amino acid sequence of OsXXT1 is 70% and 72% identical to that of AtXXT1 and AtXXT2 in Arabidopsis, respectively. This suggests that OsXXT1 functions as an XXT gene in rice, as AtXXT1 and AtXXT2 function in Arabidopsis. To test if OsXXT1 possesses xyloglucan 6-xylosytransferase activity, a functional complementation test of the Arabidopsis xxt1 xxt2 double mutant, which displays a root hair development phenotype (Cavalier et al., 2008), was carried out. Expression of OsXXT1 under the 35S CaMV promoter was measured by reverse-transcription PCR (RT-PCR) in complemented transgenic lines (35S::OsXXT1). Two transgenic lines showed a high expression level of OsXXT1 (Fig. S4), and in these two lines the short root hair growth was restored (Fig. 5A). Furthermore, the expression of OsXXT1 also partially complemented the slow growth phenotype of the Arabidopsis xxt1 xxt2 double mutant (Fig. 5B). Thus, OsXXT1 may have xyloglucan 6-xylosytransferase similar to AtXXT1 and AtXXT2. To measure the effect of OsXXT1 on XyG synthesis, cell walls from the leaves of wild type, xxt1 xxt2 double mutant and 35S::OsXXT1-complemented Arabidopsis plants were digested with a XyG-specific endoglucanase (XEG) followed by extraction with 4M KOH. Xyloglucan oligosaccharides released after XEG digestion were analysed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Fig. 6B). As previously reported, we could not detect any signature of XyG fragment in the Arabidopsis xxt1 xxt2 mutant (Fig. 6B). In the 35S::OsXXT1-complemented Arabidopsis lines, we detected several XyG oligosaccharides, including normal wild-type levels of XXFG, XLLG, and XLFG, and relatively lower level of XXG, XLG, XXXG, and XXLG compared with wild-type plants (Fig. 6A, B).
Fig. 5.
Complementation of root hair and slow growth phenotype of Arabidopsis xxt1 xxt2 double mutant with OsXXT1 in ten-day-old seedlings. Representative examples of root hair and growth are shown. (A) Roots hairs of wild type (WT, Columbia-0), Arabidopsis xxt1 xxt2 double mutant, and complemented Arabidopsis xxt1 xxt2 double mutant with 35S::OsXXT1 (35S::OsXXT1). Bars=100 μm. (B) Seedlings of wild type (WT, Columbia-0), Arabidopsis xxt1xxt2 double mutant and 35S::OsXXT1. Bars=2cm. (This figure is available in colour at JXB online.)
Fig. 6.
MALDI-TOF mass spectrometry analysis of the relative abundance of xyloglucan oligosaccharides released by xyloglucan-specific endoglucanase. (XEG). (A) Relative proportions of xyloglucan subunits generated from cell wall preparations of wild-type (WT, white bar) and 35S::OsXXT1 Arabidopsis (black bar) leaves digested with XEG. Results are expressed as the percentile of the areas of the corresponding peaks for each subunit on high-performance anion exchange chromatography (HPAEC). Arabidopsis were grown in growth chamber for two months and leaves of each of the plants were sampled as a biological replicate. All analyses were performed on three plant preparations. Significant differences are indicated with an asterisk. nd, not detected. (B) MALDI-TOF mass spectrometry of XEG-generated xyloglucan fragments from the hemicellulosic fractions of wild-type (WT, Columbia-0), Arabidopsis xxt1 xxt2 double mutant, and complemented Arabidopsis xxt1 xxt2 double mutant with 35S::OsXXT1 (35S::OsXXT1). Xyloglucan subunit is described from the nomenclature introduced by Fry et al. (1993). WT (top), Arabidopsis xxt1 xxt2 (middle) and 35S::OsXXT1 (bottom). Xylopentaose was used as a standard control (std).
Expression pattern of OsXXT1
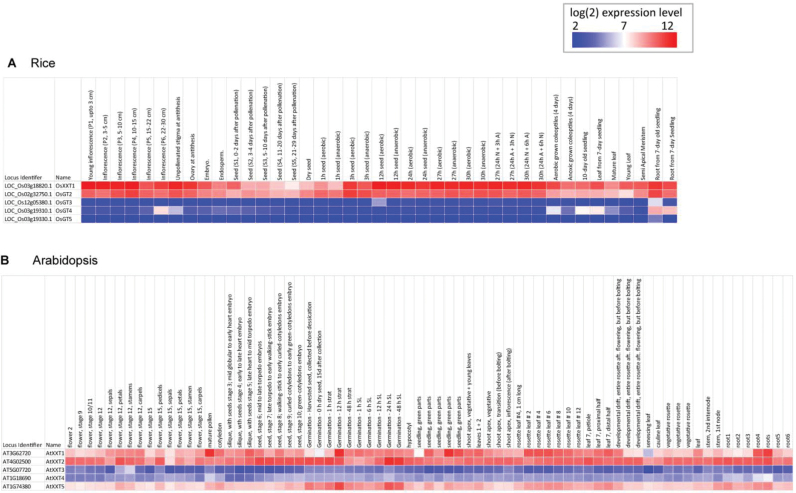
Expression patterns of rice XXT subfamily genes were determined by examining expression across development using publically available microarray datasets. The expression data are colour coded according to the RNA expression level (Fig. 7). OsXXT1 and OsGT2 have similar expression patterns and are expressed across a variety of tissues. In contrast, other members of this family (OsGT3 to OSGT5) display a more restricted expression pattern, suggesting that OsXXT1 and OsGT2 are the predominantly expressed genes (Fig. 7A). A comparison to the expression in Arabidopsis displays a similar pattern, with AtXXT1 and AtXXT2 being predominantly expressed, followed by AtXXT5 (Fig. 7B).
Fig. 7.
Tissue-specific expression of rice XXT genes from rice (A) and Arabidopsis (B). The averaged normalized publically available expression data across development in both species is shown. Data are reported as log2 average intensities on a heatmap, whereby low expression is indicated by blue shading and higher expression indicated by red shading.
To further analyse the expression of OsXXT1, a 1.8-kb fragment upstream of the OsXXT1start codon was amplified and fused to a GUS reporter gene to make the OsXXT1::GUS construct. Using Agrobacterium-mediated rice transformation, six independent transgenic lines were generated. Analysis of the T1 plants of these transgenic lines showed a similar GUS expression patterns. Expression of GUS was observed in most tissues, including callus, leaves, panicles, inflorescences, and root tips (Fig. 8A–D). Strong GUS staining was observed in the initiating lateral roots (Fig. 8D). Upon development of the lateral roots, the GUS staining was progressively restricted to the tip area (Fig. 8D). Similar expression patterns could be observed in adventitious roots. No GUS staining was detected in the mature zones or root hairs. Horizontal sections of roots showed that the GUS expression was mainly in epidermal cells (Fig. 8D and Fig. S5).
Fig. 8.
GUS reporter gene expression in transgenic plants expressing the OsXXT1 promoter-GUS constructs. (A) Callus; (B) Leaf (2-week-old seedlings); (C) Flower (3-month-old seedlings); (D) Root (2-week-old seedlings); (E) Longitudinal section of root tip (2-week-old seedlings). (A–D) Bar=1mm; (E) Bar=20 μm.
Discussion
Short root hair phenotypes caused by decreasing XyG content in Arabidopsis have been previously reported (Cavalier et al., 2008; Pena et al., 2012; Zabotina et al., 2008; Zabotina et al., 2012). Additionally, it has been previously observed that mutations in the XXT gene causes a reduction or elimination of XyG content in Arabidopsis cell walls, and could result in a reduced resistance of protoplast pressure and tensile strength of cell walls, and thus lead to a defect in root hair development (Cavalier et al., 2008; Park and Cosgrove, 2012a; Zabotina et al., 2008). In comparison to the XyG content of 20–25% in type I cell walls, cell walls of grass species contain less than 5% XyG. Thus, XyG is considered to be a less important component in type II cell walls (Vogel, 2008). However, our study showed that the mutation of rice OsXXT1 resulted in a short root hair phenotype, which is similar to the Arabidopsis xxt1 xxt2 double mutants (Fig. 1). The bubble-like extrusion of root hairs and irregular epidermis cells observed in srh2 roots under an acidic condition suggested that the tensile strength of cell walls is reduced in the mutant (Fig. 1). OsXXT1 shares high sequence and structural similarity with AtXXT1 and AtXXT2 (Fig. 4, S2). The mutation resulting from EMS mutagenesis is in a conserved domain of XXT proteins and probably resulted in the loss of xyloglucan 6-xylosyltransferase activity in srh2, confirmed by the observations that OsXXT1 gene can complement an Arabidopsis xxt1 xxt2 double mutant in both root hair growth and XyG biosynthesis. This demonstrates the importance of XyG in type II cell walls.
Although cell walls of grasses only contain a very low amount of XyG, genes encoding XXT and xyloglucan endotransglucosylase/hydrolase (XET/XTH) in rice, barley, and maize genomes are similar to those in Arabidopsis (Penning et al., 2009; Strohmeier et al., 2004; Yokoyama et al., 2004). It has been shown that these XET/XTH genes displayed a similar XET/XTH activity in rice as in Arabidopsis (Hara et al., 2014). AtXXT1, AtXXT2, and AtXXT5 are the major XXT genes responsible for XyG biosynthesis (Vuttipongchaikij et al., 2012; Zabotina et al., 2012), which is in accordance with the high expression level of these three genes in most tissues (Fig 7B). Interestingly, the rice OsXXT1 and OsGT2 were homologues of AtXXT1/AtXXT2 and AtXXT5, respectively, and displayed similar expression patterns with the Arabidopsis genes (Fig 7A, B). Therefore, OsXXT1 may have a similar function as AtXXT1/AtXXT2 and XyG has an important structural function in both type I and type II cell walls, even though the XyG content is different.
In Arabidopsis, the AtXXT1 and AtXXT2 display functional redundancy and phylogenic analysis indicate they form a distinct cluster (Vuttipongchaikij et al., 2012). Interestingly, OsXXT1 is the only gene in rice that branches with this cluster (Fig. 4). This probably explains why a single mutation of OsXXT1 exhibited a similar short root hair phenotype as the Arabidopsis xxt1 xxt2 double mutant. Moreover, constitutive expression of OsXXT1 in the Arabidopsis xxt1 xxt2 double mutant partially complemented the growth phenotype and XyG synthesis, which demonstrates that OsXXT1 possess xyloglucan 6-xylosyltransferase activity. However, the Arabidopsis xxt1 xxt2 double mutant complemented with a 35S::OsXXT1 transgenic line contained a relatively low abundance of XyG oligosaccharides, especially lacking galactose modified XyG (XLG and XXLG). This result suggests that the xyloglucan 6-xylosyltransferase activity of OsXXT1 may not be exactly the same as AtXXT1 and AtXXT2.
Although the proportions and structure of XyG vary between type I and type II cell walls, all flowering plants studied to date contain XyG in their primary cell walls (Hsieh and Harris, 2009). It is has been observed that the XET activity of epidermal cells in root elongation zones and trichoblasts of diverse species of vascular plants is high (Vissenberg et al., 2003). It is possible that the active form of XET is involved in the restructuring of XyG to regulate root hair development in all vascular plants. OsXXT1 is preferentially expressed in epidermal cells of primary, adventurous, and lateral roots (Fig. 7E, S3). The expression pattern supports the hypothesis that XyG synthesis and modification in root epidermal cells is critical for cell wall development in the root hairs of all plants. Recently, a root hair specific galacturonic acid containing XyG has been discovered in Arabidopsis and the lack of this galacturonic acid containing XyG resulted in a short hair phenotype (Pena et al., 2012). Therefore, XyG is a pivotal component in the regulation of root hair tip growth in both Arabidopsis and rice.
XyG is the most abundant hemicellulose in type I primary cell walls. This polysaccharide has been depicted as a binding surface of cellulose microfibrils, forming a load-bearing network (Thompson, 2005). However, the important role of XyG in this model has been challenged by the discovery that a lack of XyG in Arabidopsis manifests as a short root hair phenotype and an ability to grow normally, despite the defect in root hair morphology (Cavalier et al., 2008). Based on the biomechanical properties of the Arabidopsis xxt1 xxt2 double mutant and wild-type Arabidopsis, a revised architecture of primary cell wall was proposed: a minor XyG component was found in wall mechanics (Park and Cosgrove, 2012b). The analysis of the microarray data and GUS expression patterns under the OsXXT1 promoter suggest that OsXXT1 is expressed in a variety of tissues, yet like in Arabidopsis (Cavalier et al., 2008; Vuttipongchaikij et al., 2012), a phenotypic defect is only observed in root hair development. If only a minor, inaccessible XyG component works as a load-bearing connection between microfibrils in type I cell walls, it is reasonable to suggest that a relatively low abundance of XyG has a similar function in type II cell walls. Thus, it seems that XyG plays a similar role in the cell walls of both plants with type I and type II cell walls, and it seems that under normal situations, this role is only observed to be non-redundant in root hair development.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Growth performance of srh2 mutant and wild-type (cv Kasalath, kas) plants. The seedlings were growth at nutrient solution (pH 5.5) for 7 d and examined under an electron microscope.
Figure S2. Confirmation the single nucleotide mutation of srh2 by dCAPS marker. A, The PCR fragment of WT contained three Nco I site, whereas mutation of srh2 eliminate one NcoI site (red colour). B, Electrophoresis of NcoI-digested PCR product. The red arrows indicated specific digested fragment of wild-type and mutant samples.
Figure S3. Protein sequence alignment of putative xyloglucan 6-xylosyltransferase in Arabidopsis and rice. The transmembrane and glycosyltransferase domains were indicated by red box and black line, respectively.
Figure S4. The expression of OsXXT1 in complementation Arabidopsis by RT-PCR.
Figure S5. A, Transverse section of root mature region of OsXXT1::promoter GUS plants. Bar=20 μm.
Table S1. Primers used in this research.
Acknowledgements
This work was supported by the Key Basic Research Special Foundation of China (2011CB100303), National Science Foundation of China (31172024, 31201675), the Ministry of Science and Technology of China (2010DFA31080, 2014ZX08009328-002), and an Australian Research Council Centre of Excellence grants to JW. (CEO561495 and CE140100008). We would like to thank Reena Narsai for help with the digital expression analysis. The xxt1 xxt2 double mutant was obtained from the Arabidopsis Biological Resource Center (ABRC).
Glossary
Abbreviations:
- dCAPS
derived cleaved amplified polymorphic sequence
- GAX
glucuronoarabinoxylans
- GT34
glycosyltransferase family 34
- GUS
β-glucuronidase
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- MLG
mixed linkage glucans
- MS
Murashige and Skoog
- srh2
short root hair 2
- SSR
simple sequence repeat
- Ubi-1
ubiquitin-1
- XET/XTH
xyloglucan endotransglucosylase/hydrolase
- XXT1
xyloglucan xylosyltransferase1
- XyG
xyloglucan.
References
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal 3, 1–30 [DOI] [PubMed] [Google Scholar]
- Carpita NC. 1996. Structure and biogenesis of the cell walls of grasses. Annual Review of Plant Physiology Plant Molecular Biology 47, 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann M. 2000. The cell wall. In: Buchanan BB, Wilhelm G, Jones RL, eds. In Biochemistry and Molecular Biology of Plants . American Society of Plant Physiologists, 52–108 [Google Scholar]
- Cavalier DM, Keegstra K. 2006. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. The Journal of Biology Chemistry 281, 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, et al. 2008. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. The Plant Cell 20, 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. 2007. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proceedings of the National Academy of Sciences, USA 104, 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. 2002. An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proceedings of the National Academy of Sciences, USA 99, 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, et al. 1993. An unambigous nomenclature for xyloglucan derived oligosaccharides. Physiology Plant 89, 1–3 [Google Scholar]
- Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K. 2014. Function of xyloglucan endotransglucosylase/hydrolases in rice. Annals of Botany (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A. 2006. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Jia Z, Pena MJ, Cash M, Harper A, Blackburn AR, 2nd, Darvill A, York WS. 2005. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydrate Research 340, 1826–1840 [DOI] [PubMed] [Google Scholar]
- Hsieh YS, Harris PJ. 2009. Xyloglucans of monocotyledons have diverse structures. Molecular Plant 2, 943–965 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD. 2003. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. The Plant Cell 15, 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. 2007. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proceedings of the National Academy of Sciences, USA 104, 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT. 2011. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytologist 190, 472–487 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M. 2006. A protocol for Agrobacterium-mediated transformation in rice. Nature Protocols 1, 2796–2802 [DOI] [PubMed] [Google Scholar]
- Obel N, Neumetzler L, Pauly M. 2007. Hemicellulose and cell expansion. Berlin: Springer-Verlag [Google Scholar]
- Park YB, Cosgrove DJ. 2012a. Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis . Plant Physiology 158, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. 2012b. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiology 158, 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena MJ, Ryden P, Madson M, Smith AC, Carpita NC. 2004. The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiology 134, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena MJ, Kong Y, York WS, O’Neill MA. 2012. A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. The Plant Cell 24, 4511–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning BW, Hunter CT, Tayengwa R, et al. 2009. Genetic resources for maize cell wall biology. Plant Physiology 151, 1703–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas AJ, Williams CK, Davison BH, et al. 2006. The path forward for biofuels and biomaterials. Science 311, 484–489 [DOI] [PubMed] [Google Scholar]
- Sims IM, Middleton K, Lane AG, Cairns AJ, Bacic A. 2000. Characterisation of extracellular polysaccharides from suspension cultures of members of the Poaceae. Planta 210, 261–268 [DOI] [PubMed] [Google Scholar]
- Strohmeier M, Hrmova M, Fischer M, Harvey AJ, Fincher GB, Pleiss J. 2004. Molecular modeling of family GH16 glycoside hydrolases: potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Science 13, 3200–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DS. 2005. How do cell walls regulate plant growth? Journal of Experimental Botany 56, 2275–2285 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. 2002. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proceedings of the National Academy of Sciences, USA 99, 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, York WS, Beldman G, Voragen AG. 1997. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiology 114, 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen JP. 2003. Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays . Journal of Experimental Botany 54, 335–344 [DOI] [PubMed] [Google Scholar]
- Vogel J. 2008. Unique aspects of the grass cell wall. Current Opinion in Plant Biology 11, 301–307 [DOI] [PubMed] [Google Scholar]
- Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ. 2012. Arabidopsis GT34 family contains five xyloglucan alpha-1,6-xylosyltransferases. New Phytologist 195, 585–595 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. 2004. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis . Plant Cell Physiology 45, 1111–1121 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JK, Nishitani K. 2004. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiology 134, 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, KA G. 1976. Laboratory manual for physiological studies of rice, Ed 3. Manila: The International Rice Research Institute [Google Scholar]
- Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. 2008. Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. The Plant Journal 56, 101–115 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG. 2012. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiology 159, 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Song XQ, Yu BS, Zhang BC, Sun CQ, Knox JP, Zhou YH. 2012. Identification of quantitative trait loci affecting hemicellulose characteristics based on cell wall composition in a wild and cultivated rice species. Molecular Plant 5, 162–175 [DOI] [PubMed] [Google Scholar]
- Zheng L, Cheng Z, Ai C, et al. 2010. Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS One 5, e10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.