Summary
UGT84A produces a dynamic pool of hydroxycinnamoyl-glucose esters in vegetative tissues and can modulate phenylpropanoid metabolism in response to developmental and environmental cues, such as nitrogen limitation, in Populus.
Key words: Glycosylation, hydroxycinnamate glucose ester, metabolite profiling, phenylpropanoid, Populus, stress, UGT84A.
Abstract
The diversity of phenylpropanoids offers a rich inventory of bioactive chemicals that can be exploited for plant improvement and human health. Recent evidence suggests that glycosylation may play a role in the partitioning of phenylpropanoid precursors for a variety of downstream uses. This work reports the functional characterization of a stress-responsive glycosyltransferase, GT1-316 in Populus. GT1-316 belongs to the UGT84A subfamily of plant glycosyltransferase family 1 and is designated UGT84A17. Recombinant protein analysis showed that UGT84A17 is a hydroxycinnamate glycosyltransferase and able to accept a range of unsubstituted and substituted cinnamic and benzoic acids as substrates in vitro. Overexpression of GT1-316 in transgenic Populus led to plant-wide increases of hydroxycinnamoyl-glucose esters, which were further elevated under N-limiting conditions. Levels of the two most abundant flavonoid glycosides, rutin and kaempferol-3-O-rutinoside, decreased, while levels of other less abundant flavonoid and phenylpropanoid conjugates increased in leaves of the GT1-316-overexpressing plants. Transcript levels of representative phenylpropanoid pathway genes were unchanged in transgenic plants, supporting a glycosylation-mediated redirection of phenylpropanoid carbon flow as opposed to enhanced phenylpropanoid pathway flux. The metabolic response of N-replete transgenic plants overlapped with that of N-stressed wild types, as the majority of phenylpropanoid derivatives significantly affected by GT1-316 overexpression were also significantly changed by N stress in the wild types. These results suggest that UGT84A17 plays an important role in phenylpropanoid metabolism by modulating biosynthesis of hydroxycinnamoyl-glucose esters and their derivatives in response to developmental and environmental cues.
Introduction
Phenylpropanoids play important roles in plant structural integrity (e.g. lignin) and defence against biotic and abiotic stressors (e.g. flavonoids, condensed tannins (CTs), and phenolic glycosides (PGs)). Their composition and abundance thus have significant impact on biomass utilization for pulp, biofuels, forage, or atmospheric carbon sequestration and on ecological interactions (Boerjan et al., 2003; Dixon, 2005; Tsai et al., 2006). Phenylpropanoids also possess nutritive and pharmaceutical value that can be exploited for human health applications (Kammerer et al., 2005; Ververidis et al., 2007).
Hydroxycinnamates derived from phenylalanine are the simplest of the phenylpropanoids and are precursors to other more elaborate phenylpropanoid metabolites, such as lignin and flavonoids. Hydroxycinnamates accumulate in a great variety of ester or amide conjugates with monosaccharides, organic acids, lipids, and amines (Strack, 2001). These hydroxycinnamate conjugates have been implicated in pathogen response (Bernards et al., 1991; Beimen et al., 1992), symbiont interactions (Weiss et al., 1999), and ultraviolet (UV) protection (Landry et al., 1995; Sheahan, 1996; Grace et al., 1998; Kolb et al., 2001). Activated hydroxycinnamates in the forms of CoA or glucose esters are major acyl donors for modification of secondary metabolites (Teusch et al., 1987; Gläßgen and Seitz, 1992; Mock and Strack, 1993) into end products with altered physicochemical properties and hence bioactivities (D’Auria, 2006; Yoshida et al., 2009). Hydroxycinnamoyl esters also cross-link with lignocellulosic polymers, thereby affecting cell-wall strength and biomass utilization (Ralph et al., 2004). Consistent with their multiple roles in phenylpropanoid metabolism, genetic perturbations affecting hydroxycinnamates or hydroxycinnamate conjugates have wide-ranging effects on phenylpropanoid carbon allocation between different branch pathways (Sinlapadech et al., 2007; Lanot et al., 2008; Clauß et al., 2011; Mittasch et al., 2013).
Hydroxycinnamate glucose esters represent the most common form of hydroxycinnamate conjugates in plants (Corner and Swain, 1965). Their synthesis depends on family 1 glycosyltransferases (GT1), which catalyse the transfer of sugars to small acceptor molecules (Bowles et al., 2006). To date, only a handful of GT1s have been shown to catalyse the formation of hydroxycinnamoyl-glucose esters, and all belong to the UGT84A subfamily of group L of plant GT1 proteins (Milkowski et al., 2000a; Lim et al., 2001; Lunkenbein et al., 2006; Mittasch et al., 2007). Arabidopsis UGT84A2 and its Brassica napus (oilseed rape) orthologue UGT84A9 represent the best characterized members. Both enzymes exhibit a specific substrate preference for sinapic acid and produce sinapoyl-glucose as the acyl donor for the biosynthesis of sinapoyl-malate and sinapoyl-choline, the major soluble phenylpropanoids in Brassicaceae (Milkowski et al., 2000a; Lim et al., 2001). Whereas sinapoyl-malate functions as a UV protectant in leaves (Landry et al., 1995), sinapoyl-choline accumulates at high levels in seed of these species (Hüsken et al., 2005). In strawberry (Fragaria × ananassa), the fruit-specific UGT84A6 exhibited a slight substrate preference for cinnamic acid, and the most significant effect of its antisense downregulation was reduced levels of the flavour constituent cinnamoyl-glucose (Lunkenbein et al., 2006). All other characterized UGT84A proteins, such as Arabidopsis UGT84A1, A3, and A4 (Milkowski et al., 2000b; Lim et al., 2001) and oilseed rape UGT84A10 (Mittasch et al., 2007), utilize multiple hydroxycinnamate substrates in vitro, but their in vivo functions remain poorly understood.
This study describes the identification and characterization of UGT84A orthologues from Populus, a species known for its large and diverse reserves of phenylpropanoids (Tsai et al., 2006). Populus harbours three UGT84A members—GT1-315 (UGT84A19), GT1-316a (UGT84A18), and GT1-316 (UGT84A17)—located in a tandem block with high sequence similarity. UGT84A17 exhibited stress-responsive expression and broad in vitro activities toward various hydroxylated and/or methoxylated cinnamic and benzoic acids. Overexpression of UGT84A17 in transgenic Populus led to hyperaccumulation of hydroxycinnamate glucose esters, especially caffeoyl-, 4-coumaroyl-, and cinnamoyl-glucose esters. Widespread changes in phenylpropanoids were also observed, supporting a role of UGT84A17 in modulating phenylpropanoid metabolism.
Materials and methods
Phylogenetic analysis
Populus GT1 sequences annotated by Geisler-Lee et al. (2006) based on the Populus genome v1.0 were used for initial phylogenetic analysis with Arabidopsis GT1 family (Li et al., 2001) to identify group L orthologues. The gene models were cross-referenced with the Populus genome v3.0 to obtain updated gene models for phylogenetic analysis, along with other experimentally characterized group L members, using the Arabidopsis group E member UGT72B1 as outgroup. Sequence alignment was performed using the MAFFT program housed on the EMBL-EBI server (http://www.ebi.ac.uk/Tools/msa/). The alignment output was imported into MEGA5 (Tamura et al., 2011) for phylogenetic tree reconstruction using the maximum-likelihood method and the Jones–Taylor–Thornton (JTT) substitution matrix with 500 bootstrap iterations.
Recombinant PfaGT1-316 analysis and enzyme assays
A Populus fremontii × angustifolia expressed sequence tag (MTUNUL1.P64.D01, GenBank accession DY801582) matching the predicted GT1-316 in the Populus trichocarpa genome was fully sequenced and used for subcloning. The coding region (with the start codon converted to CTG) was amplified by PCR using gene-specific primers that introduced 5′-BamHI and 3′-SmaI sites (Supplementary Table S1 available at JXB online) and cloned into pCRII-TOPO (Invitrogen/Life Technologies, Grand Island, NY, USA). After sequencing confirmation, the BamHI and SmaI fragment was subcloned into pGEX-2TK (GE Healthcare, Piscataway, NJ, USA) and transformed into Escherichia coli BL21. Recombinant proteins were purified using a glutathione sepharose purification kit (GE Healthcare).
Activity of recombinant GT1-316 was first tested using 5mM UDP-glucose as the sugar donor and a variety of potential acceptor substrates (e.g. phenylpropanoids, terpenoids, indole acetic acid, zeatin) at 1mM. Kinetic analysis was performed using phenylpropanoid substrates ranging from 50 to 500 μM. Reaction conditions were based on Lim et al. (2001) and contained 0.2 μg protein, 100mM Tris (pH 7.5), 1mM DTT, 2mM EGTA, and 0.2mg ml–1 BSA in 50 μl. After prewarming at 30 °C for 75 s, the reaction was started by addition of UDP-glucose and terminated after 5min by snap freezing in liquid nitrogen. Control reactions were stopped immediately after adding UDP-glucose. The protein was denatured by addition of 60 μl acetonitrile with 0.2mM 13C6-cinnamic acid as internal standard. Following centrifugation, the supernatant was dried under vacuum to remove acetonitrile prior to analysis on an Agilent 1200 HPLC, equipped with a diode array detector and a 6220 accurate-mass time-of-flight (TOF) mass spectrometer (Agilent Technologies, Wilmington, DE, USA). Samples (1 μl each) were separated on a reversed-phase column (ZORBAX Eclipse Plus C18, 2.1×150mm, 3.5 µm; Agilent) for quantification of UDP released from UDP-glucose during the reaction, which allowed one quantification method for all glucose acceptor substrates. N,N dimethylhexylamine (DMHA) was included in the mobile phase as an ion-pairing agent (Cordell et al., 2008). The gradient of mobile phase A (95% water, 5% acetonitrile, 5mM DMHA, pH 7) to mobile phase B (95% acetonitrile, 5% water, 5mM DMHA) was linear from 5% B to 30% B over 3min, and then linear up to 40% B over 3.5min, at a flow rate of 0.3ml min–1. The electrospray ionization was set in negative mode, with gas temperature 300 °C, drying gas 11 l min–1, nebulizer pressure 206.8 kPa (30 psig), capillary voltage 3500V, and fragmentor 125V. UDP was detected with a diode array detector at 260nm and by MS using the extracted ion chromatogram at m/z 402.9935 (expected m/z 402.9943). UDP concentration was estimated by a standard curve using authentic UDP (Sigma, St Louis, MO, USA). Glucose ester products were confirmed by MS and a mild alkaline treatment (Lim et al., 2001). Briefly, the assay products were incubated with 0.1M NaOH at room temperature for 1hr, and neutralized with 3M sodium acetate (pH 5.2) prior to HPLC-MS/TOF analysis.
Transgenic Populus production and N stress experiments
The coding region of PfaGT1-316 was PCR amplified using gene-specific primers (Supplementary Table S1 available at JXB online), cloned into pCRII-TOPO (Invitrogen), and sequence-confirmed. The insert was digested by SpeI and EcoRV and subcloned into pCambia1302 behind the CaMV 35S promoter in a sense orientation. The construct was mobilized into Agrobacterium tumefaciens pMP90 and transformed into Populus tremula × alba (clone INRA 717-IB4) using standard methods (Meilan and Ma, 2006). Independent transgenic lines confirmed by PCR were transplanted to soil and maintained in a glasshouse. Plants were fertilized weekly with a 15% solution of Miracle-Gro Water Soluble All Purpose Plant Food (Scotts, Marysville, OH, USA). Leaf tissues (leaf plastochron index LPI-5) from the original transformants were taken for initial screening analyses.
Selected plants were vegetatively propagated for N-stress experiments. Rooted cuttings were transferred to hydroponic culture in perlite pots, with N maintained at 2.5mM (ammonium/nitrate 4:1) as described (Harding et al., 2005). Plants from wild-type (WT) and three transgenic lines were distributed evenly among eight hydroponic tubs. Nutrient solutions were replaced weekly and deionized water was added daily as necessary to maintain volume, with pH maintained at ~5.8. When plants were approximately 1 m tall, they were randomly divided into two groups that received either full (2.5mM) or reduced (0.25mM) N levels with the same ammonium/nitrate molar ratio, and the treatment lasted for 13 days. Plant heights, basal stem diameter (2cm above the perlite surface), and leaf lengths were measured at regular intervals. Leaf (LPI-2 and 7), young stem (internodes between LPI-0 and LPI-4), phloem (bark) and xylem (de-barked stem) from internodes between LPI-7 and LPI-12, and coarse root tissues were harvested, snap frozen, and ground to a fine powder under liquid nitrogen. An aliquot was freeze dried for metabolic analysis and the rest stored at –80 °C until use.
Quantitative real-time PCR
Various tissues from Populus tremuloides (clone 271) and P. tremula × alba (clone 717-1B4) as described in Payyavula et al. (2011) or from the N stress experiments were used for RNA extraction by the CTAB method (Chang et al., 1993). DNase treatment, cDNA synthesis, and quantitative real-time PCR (qRT-PCR) were conducted as described (Tsai et al., 2006), using the ABsolute qRT-PCR SYBR Green Mix (ABgene/Thermo Fisher Scientific, Pittsburgh, PA, USA) and a Mx3005P Real-Time PCR system (Stratagene, La Jolla, CA, USA). Relative transcript abundance was estimated by the ΔCT method (Tsai et al., 2006), using the geometric mean of three stable housekeeping genes (elongation factor 1-β, α-tubulin 4, and ubiquitin-conjugating enzyme E2) for normalization (see Supplementary Table S1, available at JXB online, for gene-specific primers).
Metabolite extraction and HPLC-MS/TOF analysis
Freeze-dried tissues (10mg) were extracted in ice-cold methanol in an ultrasonicator bath for 5min. The extracts were clarified by centrifugation and stored at –80°C or analysed directly (1 μl) by HPLC-MS/TOF for nontargeted profiling as described in Xue et al. (2013). Metabolite data were processed by MassHunter Qualitative Analysis software (Agilent) using the ‘Molecular Feature Extraction’ function. After removing noise (signal-to-noise <150 and absolute ion counts <500), the software groups chemically related ions (isotopes, formide adducts, and dimers) to identify putative compounds (ion groups) in negative mode and generate molecular formulas (mass error≤10 ppm). The resultant files were aligned using Mass Profiler (Agilent) to generate a list of target compounds, which was filtered to retain those that were present in at least half of the samples from either WT or the transgenic group. The list was then converted to XML format using a custom script and imported into the MassHunter Quantitative Analysis software (Agilent) for peak fitting and integration to obtain abundance values. Internal standard (13C6-cinnamic acid)-normalized and tissue mass-corrected abundance values were used for statistical analysis. Separate comparisons for genotypic effects within each N regime, and N effects within genotype were made for each tissue using SLIM in R (Wang et al., 2011), with statistical significance determined by either a P-value or a Q-value cut off of 0.05.
For metabolites that were significantly affected by the transgene expression, their m/z and tentative molecular formulas obtained from the MassHunter Qualitative Analysis software were used to search the mass spectral databases KNApSAcK (Afendi et al., 2012), Metabolome Tomato Database (MoToDB, Moco et al., 2006), and Dictionary of Natural Products (http://dnp.chemnetbase.com) to assign putative identity. Where possible, compound identities were confirmed by authentic standards (Supplementary Table S3), including the phenolic glucose esters obtained from in vitro GT1-316 assays.
GC-MS analysis of wall-bound phenolics
An aliquot (20mg) of freeze-dried LPI-7 was extracted three times in 1ml methanol/chloroform (33:67, v/v), followed by 100% methanol and then water, 15min each by sonication at room temperature. The pellet was resuspended in 1ml 2M NaOH with methoxybenzoic acid as internal standard, and incubated on an orbital shaker (800rpm) overnight at room temperature. After centrifugation, the supernatant was adjusted to pH ~5 using 8M HCl and extracted three times with 500 µl water-saturated ethyl acetate. The pooled ethyl acetate fractions were evaporated to dryness and resuspended into 200 µl acetonitrile. A portion (150 µl) of the sample was used for derivitization and GC-MS analysis following conditions detailed in Xue et al. (2013). The identity of hydroxycinnamic acids was confirmed by authentic standards.
Lignin and CT analyses
Lignin content and syringyl-to-guaiacyl monolignol (S/G) ratio were determined by pyrolysis molecular beam mass spectrometry at the Complex Carbohydrate Research Center according to Sykes et al. (2009), using freeze-dried stem xylem samples. Condensed tannins (CTs) were analysed by a modified n-butanol-HCl method (Porter et al., 1986), using purified CTs from Populus leaves as standards according to Harding et al. (2005).
Statistics
Unless otherwise noted, one-way or two-way ANOVA was performed for comparisons between treatments and/or genotypes using SigmaPlot v.12.3 (Systat Software, San Jose, CA, USA). The Tukey multiple comparison correction was used where appropriate. Gene expression data from qRT-PCR were log transformed prior to statistical comparison to approximate a normal distribution, as indicated by the Shapiro–Wilk test.
Results
Identification of a stress-responsive GT1-315/316 gene cluster
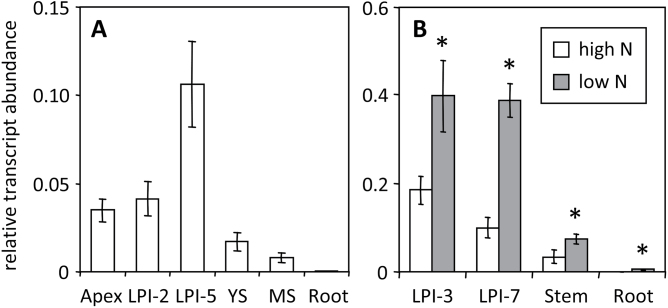
Two Affymetrix probe-sets (PtpAffx.125962.1.S1_at and PtpAffx.18005.2.A1_a_at) corresponding to previously annotated Populus GT1-315 and GT1-316 (Geisler-Lee et al., 2006) were found to exhibit elevated expression in response to multiple stress treatments (e.g. N limitation, wounding, and detopping) and in multiple genotypes (Supplementary Fig. S1 available at JXB online; Yuan et al., 2009; Tuominen et al., 2011). In the current (v3.0) genome release, these probes match three gene models—Potri.009G095100 (GT1-316), Potri.009G095300 (GT1-316a, not previously annotated), and Potri.009G095400 (GT1-315)—that share a high degree of sequence identity (hereafter referred to as the GT1-315/316 cluster). Thus, the stress-responsive expression that the current study observed likely reflected the collective response of this gene cluster. To verify their expression, qRT-PCR was conducted in two separate experiments, using primers designed to amplify all members of the GT1-315/316 cluster. In P. tremula × alba clone 717-1B4, transcript levels of GT1-315/316 were highest in leaves, lower in stems, and very low in roots (Fig. 1A). A similar tissue expression pattern was found in P. tremuloides clone 271. Consistent with the microarray findings, transcript levels of GT1-315/316 were elevated (two-way ANOVA P N-treatment<0.001, P tissue<0.001) in P. tremuloides plants that were grown under N-limiting conditions (Fig. 1B).
Fig. 1.
Relative transcript abundance of Populus GT1-315/316 gene cluster in different tissues. (A) Populus tremula × alba clone 717-1B4. (B) P. tremuloides clone 271 grown under N-replete (high N) or N-limited (low N) conditions. Asterisks indicate statistically significant N treatment effects.
Phylogenetic analysis
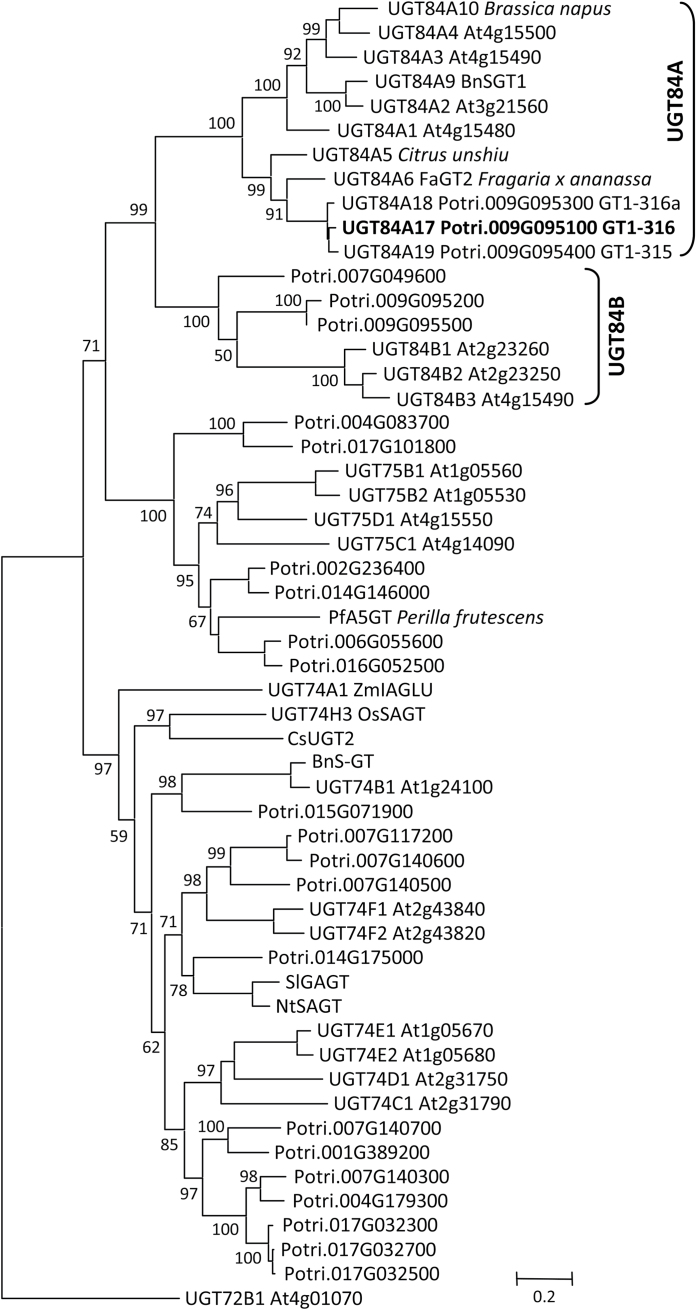
Phylogenetic analysis showed that the deduced GT1-315/316 proteins are most closely related to the UGT84A subfamily (Fig. 2), belonging to group L of plant GT1s (Li et al., 2001). They were assigned UGT84A17 (GT1-316), UGT84A18 (GT1-315), and UGT84A19 (GT1-316a) by the UDP Glucuronosyltransferase Nomenclature Committee. The UGT84A clade included several biochemically characterized members known to catalyse the formation of hydroxycinnamate glucose esters. Examples are the sinapate-specific UGT84A9 (oilseed rape BnSGT1, Milkowski et al., 2000a) and UGT84A2 (Arabidopsis At3g21560; Lim et al., 2001), the cinnamic acid-biased UGT84A6 (FaGT2) from strawberry (Lunkenbein et al., 2006), and several other members with a broad substrate specificity: UGT84A1 (At4g15480), UGT84A3 (At4g15490), and UGT84A4 (At4g15500) from Arabidopsis (Milkowski et al., 2000b; Lim et al., 2001) and UGT84A10 from oilseed rape (Mittasch et al., 2007). This strongly supported UGT84A clade was sister to another strongly supported branch that includes the Arabidopsis indole-3-acetic acid glucosyltransferase (At2g23260) of the UGT84B subfamily (Jackson et al., 2001). Taken together, the strong phylogenetic association of GT1-315/316 with UGT84A members supports their potential involvement in phenylpropanoid metabolism.
Fig. 2.
Maximum-likelihood tree of group L glycosyltransferases from Populus and Arabidopsis, along with experimentally characterized members from other species. An Arabidopsis group E member (UGT72B1) was used as outgroup. Bootstrap support for the branches is shown. Populus gene models were from the genome release v3.0 (Phytozome), whereas the Arabidopsis sequences were from TAIR10. GenBank accession numbers for the other sequences are: Brassica napus UGT84A9/BnSGT1 (AF87143), UGT84A10 (CAJ77650), and BnS-GT (AAL09350); Crocus sativus CsUGT2 (Q6X1C0); Fragaria × ananassa FaGT2/UGT84A6 (Q66PF4); Citrus unshiu UGT84A5 (BAA93039); Nicotiana tabacum NtSAGT (AF190634); Oryza sativa UGT74H3/OsSAGT (BAD34358); Perilla frutescens var. crispa PfA5GT (BAA36421); Solanum lycopersicum SlGAGT (CAI62049); and Zea mays UGT74A1/ZmIAGLU (AAA59054). UGT code assigned by the UDP Glucuronosyltransferase Nomenclature Committee is included when available.
Biochemical characterization of GT1-316
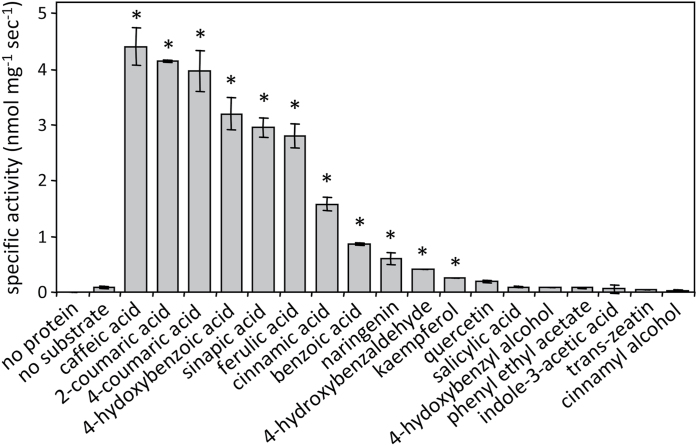
Data mining of Populus expressed sequence tag collections (Ranjan et al., 2004; unpublished data) identified one candidate full-length GT1-315/316 clone (DY801582) derived from P. fremontii × angustifolia (genotype NUL). This clone was fully sequenced and found to share 99% coding sequence identity with GT1-316, followed by GT1-315 and GT1-316a (~97%) of the P. trichocarpa genome (Tuskan et al., 2006). The clone was therefore named PfaGT1-316 (GenBank accession KF552072). The coding region of PfaGT1-316 was expressed in E. coli for in vitro protein characterization. The purified recombinant PfaGT1-316 protein exhibited activity for 11 out of the 18 potential glucose acceptor substrates tested (Fig. 3, Supplementary Fig. S2, Supplementary Table S2). The PfaGT1-316 activity was higher for substituted hydroxycinnamic acids and 4-hydroxybenzoic acid than for unsubstituted cinnamic and benzoic acids. The activity with flavonoid substrates was very low or below detection. Kinetic analysis was performed for eight phenolic acids and two flavonoids (naringenin and kaempferol). PfaGT1-316 exhibited the highest catalytic activities and turnover rates toward caffeic acid, 4-coumaric acid, 4-hydroxybenzoic acid, 2-coumaric acid, ferulic acid, and sinapic acid as glucose-acceptors, while flavonoids were relatively poor substrates (Table 1). The hydroxycinnamate conjugates were hydrolysable by a mild alkaline treatment, suggesting that PfaGT1-316 preferentially catalyses the formation of glucose esters rather than O-glucosides (Supplementary Fig. S3), similarly to the other UGT84A orthologues (Lim et al., 2001).
Fig. 3.
In vitro enzyme activity of recombinant PfaGT1-316 with various substrates. Specific activity was measured as UDP released from UDP-glucose during the glycosylation reaction. Asterisks inidicate reaction products verified by HPLC-MS/TOF (Supplementary Fig. S2 and Table S1).
Table 1.
Enzyme kinetics of recombinant PfaGT1-316 as determined by Lineweaver-Burke plotData represent mean and SD from three independent batches of protein purification, each with at least three technical replicates. ND, K m for kaempferol could not be determined due to poor activity.
| Substrate | V max (pkat) | K m (mM) | k cat (s–1) | k cat/K m (mM–1 s–1) |
|---|---|---|---|---|
| Caffeic acid | 15.85±4.50 | 0.22±0.09 | 6.49±1.84 | 31.3±4.3 |
| 4-Coumaric acid | 15.07±2.55 | 0.62±0.12 | 6.17±1.05 | 10.1±0.3 |
| 4-Hydroxybenzoic acid | 10.61±2.95 | 0.72±0.33 | 4.35±1.21 | 6.6±2.0 |
| 2-Coumaric acid | 8.99±3.44 | 0.11±0.05 | 3.68±1.41 | 35.9±3.9 |
| Ferulic acid | 7.93±1.27 | 0.15±0.02 | 3.25±0.52 | 22.1±1.6 |
| Sinapic acid | 7.55±2.12 | 0.13±0.05 | 3.09±0.87 | 24.7±3.0 |
| Benzoic acid | 3.68±0.89 | 1.59±0.41 | 1.51±0.36 | 1.0±0.1 |
| Cinnamic acid | 3.42±0.59 | 0.30±0.07 | 1.40±0.24 | 4.7±0.9 |
| Naringenin | 1.99±0.31 | 0.22±0.10 | 0.81±0.13 | 4.3±1.6 |
| Kaempferol | 0.55±0.13 | ND | 0.23±0.05 | ND |
Overexpression of PfaGT1-316 in Populus
To investigate the in vivo role of GT1-316, transgenic P. tremula × alba (717-1B4) that overexpressed PfaGT1-316 under the CaMV 35S promoter were generated. Based on qRT-PCR screening of 16 independent transgenic lines, three (A, D, and H) with >50-fold elevated GT1-316 transcript levels (Supplementary Fig. S4A) were selected for metabolite analysis. The expanding leaves (LPI-5) of PfaGT1-316 transgenic plants accumulated ~15-fold higher levels of caffeoyl-glucose compared to WT (Supplementary Fig. S4B). Levels of 4-coumaroyl-glucose, feruloyl-glucose, and cinnamoyl-glucose also increased, but to a much lesser extent (1.5–3-fold) (Supplementary Fig. S4C–E).
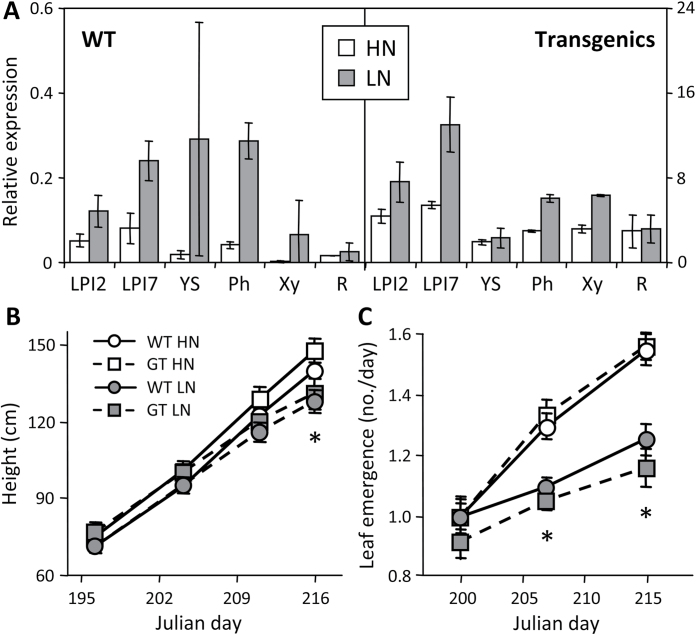
WT and transgenic PfaGT1-316 plants were subjected to hydroponic N manipulation to perturb plant growth and phenylpropanoid metabolism. Low N availability clearly stressed the plants, causing leaf yellowing and reduced shoot growth and leaf emergence rates (Fig. 4; see also Supplementary Fig. S5 available at JXB online). Transcript levels of endogenous GT1-315/316 were upregulated by 1.5–30-fold, depending on the tissue, in response to N stress in WT (Fig. 4A). In comparison, the magnitude of PfaGT1-316 overexpression (relative to the endogenous GT1-315/316) in transgenic plants was much greater, by 67–1500-fold at high N or by 8–121-fold at low N (Fig. 4A). There was little or no morphological phenotype of PfaGT1-316 transgenic plants regardless of N status (Fig. 4B, C). However, under N-limited conditions, 53% of the PfaGT1-316 plants ceased growth and set buds by the end of the experiment, but only 20% of the WT plants did so (data not shown).
Fig. 4.
Molecular and growth characterization of transgenic Populus. (A) Relative transcript abundance of the GT1-315/316 cluster in WT and transgenic plants under N-replete (HN) or N-limited (LN) treatments. Leaves (LPI2, LPI7), young stem (YS), phloem (Ph), xylem (Xy), and coarse roots (R) were analysed. Values are mean and SD of three biological replicates from line D. (B and C) Height growth (B) and leaf emergence (C) rates of WT and transgenic (GT) plants. Values are mean and SE of 15 biological replicates, pooled from lines A, D, and H. Asterisks indicate significant LN treatment effects.
Metabolic consequences of PfaGT1-316 overexpression in Populus
Several (hydroxyl)cinnamate/benzoate glucose esters were detected in the P. tremula × alba tissues examined, including six of the PfaGT1-316 glycosylation products in vitro: caffeoyl-, 4-coumaroyl-, 4-hydroxybenzoyl-, feruloyl-, cinnamoyl-, and benzoyl-glucose (Fig. 5). All six glucose esters were more abundant in transgenic plants than in WT across all tissues and N regimes (Fig. 5A, B). Also elevated in transgenic plants was a putative hydroxycinnamoyl-glucose with a matching m/z but a different retention time as compared to 2- or 4-hydroxycinnamoyl-glucose esters. In contrast, the in vitro PfaGT1-316 glycosylation products of naringenin and kaempferol were not detected in Populus tissues. The results were consistent with the observed broad in vitro substrate preference of PfaGT1-316 toward various cinnamic and benzoic acid derivatives, but not flavonoids.
Fig. 5.
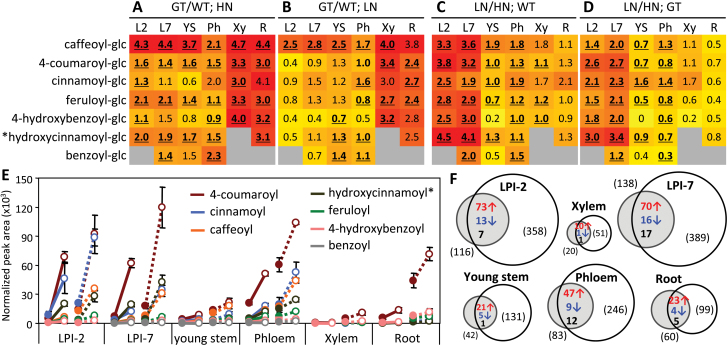
Effects of PfaGT1-316 overexpression and N limitation on hydroxycinnamate glucose ester accumulation. (A and B) Transgenic effects (GT/WT) under high-N (A) or low-N (B) conditions. (C and D) N stress response (LN/HN) in WT (C) and transgenic (D) plants. Shown are log2-transformed ratios in heatmaps. Values in bold-face with underline denote statistical significance (Q <0.05). Grey indicates below detection. (E) The same data plotted by normalized peak areas to illustrate the relative abundance of the glucose esters (solid symbols, HN; open symbols, LN; solid lines, WT; dashed lines, transgenic). Values are mean and SD of three biological replicates. The asterisk indicates an unknown hydroxycinnamoyl-glucose ester with a m/z of 325.0921. (F) Venn diagrams showing overlap of significantly changed metabolites (P<0.05) due to transgenic (GT/WT under HN, grey circles) or N (LN/HN in WT, white circles) manipulation. The numbers of metabolites with increased (red) or decreased abundance (blue), or inconsistent changes (black) are shown in the overlap, with the total number of significantly changed metabolites noted in parentheses.
Across all tissues and N treatments, caffeoyl-glucose exhibited the greatest fold-increase in transgenic plants (Fig. 5A, B), due partly to its relatively low abundance in WT plants (Fig. 5E, Supplementary Table S3). Levels of the more abundant 4-coumaroyl-glucose and cinnamoyl-glucose (LPI-2) also showed large absolute increases in transgenic plants, although the fold-change was lower than that of caffeoyl-glucose (Fig. 5A, B, E). Most of the hydroxycinnamoyl- and benzoyl-conjugates were present at low abundance in xylem and roots, and overexpression of PfaGT1-316 resulted in large fold changes from near absence in WT (Fig. 5A, B, E, Supplementary Table S3 available at JXB online). Under N-limited conditions, accumulation of most of these glucose conjugates was stimulated, particularly in leaves, regardless of genotype (Fig. 5C–E). Some hydroxycinnamates, such as 4-coumaric, caffeic, and ferulic acids are known to accumulate in Populus as wall-bound phenolics (Gou et al., 2008). No consistent transgenic or N treatment effects on the abundance of wall-bound phenolics were observed (Supplementary Fig. S6).
Nontargeted HPLC-MS/TOF profiling of secondary metabolites revealed additional metabolic changes due to PfaGT1-316 overexpression or to N-limitation. In general, leaves had the most complex metabolite profiles, while xylem extracts had the lowest number of detectable metabolites. Statistical analysis showed an overall stronger effect of N-limitation than PfaGT1-316 overexpression on Populus metabolism (Fig. 5F). The leaf and phloem metabolomes were affected the most by either perturbation, based on P≤0.05 (Fig. 5F) or Q≤0.05 (Supplementary Table S3). Consistent with the stress-responsive nature of GT1-316 (Figs 1 and 4A, Supplementary Fig. S1), metabolite changes due to GT1-316 overexpression overlapped substantially with those induced by N stress, accounting for 62–74% of significantly changed metabolites in green tissues and 45–55% in xylem and root of transgenic plants (Fig. 5F). A majority of the significantly affected metabolites showed increased abundance in PfaGT1-316 transgenic plants (Fig. 5F), and most of them were predicted to be phenylpropanoid derivatives, including conjugates of various flavonoids and di- and tri-glycosides of hydroxycinnamates (Supplementary Table S3). The latter included several putative caffeic acid derivatives (e.g. dicaffeoylquinates, caffeoyl-salicin, hydroxycinnamoyl-salicin), in addition to the hydroxycinnamoyl-glucose esters discussed above. Relatively fewer metabolites showed decreased abundance in transgenic plants, especially in leaves. Among compounds that decreased in concentration were rutin (quercetin-3-O-rutinoside) and kaempferol-3-O-rutinoside, the two most abundant flavonoids in leaves (Supplementary Table S3). Together, these results suggested that UGT84A-mediated hydroxycinnamate glycosylation plays an important role in phenylpropanoid metabolism during Populus stress response.
Major phenylpropanoids were not affected in PfaGT1-316 transgenic plants
The effects of PfaGT1-316 overexpression on accumulation of major phenylpropanoid end products (PGs, CTs, and lignin) were examined. PGs such as salicortin and tremulacin were most abundant in leaves, while CTs and lignin were present at highest levels in roots and xylem, respectively, of clone 717-1B4 (Supplementary Fig. S7). Any transgenic effects on PGs and lignin (both content and S/G ratio) were minor and inconsistent across tissues and N status (Supplementary Fig. S7A, C, D). CTs were also largely unaffected by PfaGT1-316 overexpression, but unlike PGs, increased significantly in response to N-limitation, except in xylem where CTs were barely detected (Supplementary Fig. S7B). Overall, the data suggested that PfaGT1-316 overexpression had little effect on PGs, CTs, and lignin in Populus.
Transcript levels of phenylpropanoid genes were not affected in transgenic plants
qRT-PCR was conducted to examine the transcriptional response, if any, of representative phenylpropanoid genes in transgenic plants with increased hydroxycinnamoyl-glucose esters. These included two isoforms each of the phenylalanine ammonia-lyase (PAL), 4-coumarate:CoA ligase (4CL), and caffeoyl-CoA 3-O-methyltransferase families. N-sensitive expression responses were observed for PAL and 4CL in an isoform- and tissue-dependent manner, but no transgenic effects were detected for any of the phenylpropanoid genes tested (Supplementary Fig. S8). The results suggested that elevated hydroxycinnamoyl-glucose accumulation was driven by PfaGT1-316 overexpression and redirection of phenylpropanoid pathway intermediates, without stimulating phenylpropanoid biosynthesis at the transcriptional level.
Discussion
PfaGT1-316 encodes a hydroxycinnamate glycosyltransferase
Phylogenetic, biochemical, and transgenic analyses provided strong support that PfaGT1-316 encodes a hydroxycinnamate glycosyltransferase. The ability of PfaGT1-316 to accept multiple hydroxycinnamate/benzoate substrates in vitro was corroborated in vivo, as PfaGT1-316 overexpression in transgenic poplars increased accumulation of caffeoyl-glucose, 4-coumaroyl-glucose, cinnamoyl-glucose, and several other less abundant hydroxycinnamoyl/benzoyl-glucose esters. A similar multisubstrate utilization pattern has been noted for several UGT84A orthologues (Lim et al., 2001; Lunkenbein et al., 2006; Mittasch et al., 2007), consistent with the propensity of many GT1 members to exhibit regioselectivity rather than true specificity (Vogt and Jones, 2000; Lim et al., 2003). However, this differs from the Arabidopsis UGT84A2 and oilseed rape UGT84A9 that exhibit a much more restricted substrate preference for sinapic acid (Lim et al., 2001; Mittasch et al., 2007). The Populus GT1-315/316 transcripts were detected in a wide range of tissues, especially leaves, which contrasts with the seed-, seedling-, and/or flower-preferential expression of Arabidopsis UGT84A2 (Schmid et al., 2005), oilseed rape UGT84A10 (Mittasch et al., 2007), and strawberry UGT84A6 (Lunkenbein et al., 2006). Thus, while the UGT84A family appears evolutionarily conserved (Fig. 2), variation in tissue expression and substrate utilization preference exists among isoforms and may confer species- or tissue-specific roles. The broad expression and substrate utilization patterns of PfaGT1-316 suggest that it may play a more general role of modulating phenylpropanoid metabolism.
PfaGT1-316 overexpression affected phenylpropanoid metabolism
Overexpression of PfaGT1-316 resulted in increases of hydroxycinnamoyl- and benzoyl-glucose esters in all Populus tissues examined, regardless of N status. Levels of various phenylpropanoid derivatives and conjugates were also increased. However, rutin and kaempferol-3-O-rutinoside, the two most abundant flavonoid glycosides in leaves, decreased. As expression of phenylpropanoid pathway genes was unaffected in transgenic plants, the metabolic effects observed appear to be direct consequences of elevated PfaGT1-316 glycosylation activity. The findings that hydroxycinnamoyl-glucose esters were increased at the expense of the abundant flavonoid rutinosides are consistent with both metabolite pools being dependent on aglycone hydroxycinnamates and UDP-glucose for their synthesis. Reduced flavonoid rutinoside accumulation likely led to secondary trade offs within the flavonoids, resulting in increased accrual of many other less abundant flavonoid conjugates. Metabolic trade offs between distinct phenylpropanoid pools have been frequently reported (Xie et al., 2003; Tattini et al., 2004; Clauß et al., 2011; Kosonen et al., 2012) and, in several cases, the trade offs have been associated with altered phenylpropanoid glycosylation (Sinlapadech et al., 2007; Griesser et al., 2008; Lanot et al., 2008; Payyavula et al., 2009). For instance, mutation of UGT84A2 in Arabidopsis resulted in reduced sinapoyl-glucose and its malate and choline esters, while an unusual flavonoid, sinapic acid-derived polyketide, hyperaccumulated in the trichomes (Sinlapadech et al., 2007). Together, these findings are in line with the highly plastic nature of the phenylpropanoid network in response to genetic or environmental perturbations (Vogt, 2010) and suggest that glycosylation of small phenolics can modulate a multitude of cellular and metabolic responses to affect phenylpropanoid pool composition.
Hydroxycinnamate glycosylation as a mediator of stress response
The initial identification of GT1-316 from poplar stress transcriptomes (Supplementary Fig. S1 available at JXB online), its N-sensitive expression in multiple genotypes (Figs 1 and 4A), and the large overlap of metabolic response between N-stressed and GT1-316-overexpressing poplars (Fig. 5F) provide multiple lines of evidence to support a role of GT1-316 in stress response. Populus, more than many other species, depends on large, constitutive, yet dynamic phenylpropanoid pools for stress response (reviewed in Tsai et al., 2006; Douglas et al., 2011). Given the central position of free hydroxycinnamates that support multiple phenylpropanoid branchways, GT1-316 activity can potentially modulate stress-induced shifts in carbon partitioning through its action on aglycone hydroxycinnamate pools. This is consistent with elevated expression of several UGT84A genes in response to UV-B or a pharmacologically induced oxidative burst (Lunkenbein et al., 2006; Meißner et al., 2008) and with the roles of hydroxycinnamoyl-glucoses as UV protectants (Landry et al., 1995; Lehfeldt et al., 2000; Meißner et al., 2008) and radical scavengers (Braham et al., 2005; Kylli et al., 2008; D’Abrosca et al., 2010). Recent studies showed that hydroxycinnamoyl-glucose esters serve as acyl donors in anthocyanin acylation, as Arabidopsis mutant and transgenic oilseed rape defective in UGT84A2 and UGT84A9, respectively, exhibited reduced accrual of not only sinapoyl-esters, but also sinapoylated anthocyanins (Wolfram et al., 2010; Yonekura-Sakakibara et al., 2012). Acylation with hydroxycinnamates is a common modification of phenylpropanoids (D’Auria, 2006; Tsai et al., 2006), known to alter the bioactivity, stability, and/or absorbance of the acceptor substrates (reviewed in Yoshida et al., 2009). Aromatic acylation of phenylpropanoids depends on either hydroxycinnamoyl-CoA or hydroxycinnamoyl-glucose esters as the high-energy acyl donors (Teusch et al., 1987; Gläßgen and Seitz, 1992; Mock and Strack, 1993). This places UGT84A in a position to modulate the availability of hydroxycinnamoyl donors for phenylpropanoid acylation. Thus, GT1-316/UGT84A17 could play both direct and indirect roles in modulating phenylpropanoid synthesis, modification, bioactivity, and/or stability in response to stress and glucose availability cues.
In summary, overexpression of GT1-316 caused changes in phenylpropanoid composition in Populus, suggesting an important role of glycosylation in phenylpropanoid metabolism. Populus GT1-316, like its UGT84A orthologues, is developmentally and environmentally regulated. Given the propensity of phenylpropanoids to exhibit taxon-specific diversity (Tsai et al., 2006), the current results suggest that the UGT84A subfamily, while evolutionarily conserved, may serve species-specific functions in modulating phenylpropanoid metabolism in response to developmental and environmental cues. The work presented here opens up new prospects to explore the physiological roles of diverse hydroxycinnamate derivatives in stress responses of the GT1-316-overexpressing Populus.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primer information.
Supplementary Table S2. Characteristics of PfaGT1-316 glycosylation products confirmed by HPLC-MS/TOF.
Supplementary Table S3. List of LC-MS/TOF-identified metabolites significantly changed in transgenic plants.
Supplementary Fig. S1. Expression response of two GT1-315/316 probe-sets to various stress treatments in multiple Populus genotypes.
Supplementary Fig. S2. HPLC-MS/TOF confirmation of PfaGT1-316 enzyme assay products.
Supplementary Fig. S3. Alkaline hydrolysis of PfaGT1-316 assay products to confirm glucose-ester linkage.
Supplementary Fig. S4. Screening of independent GT1-316 transgenic lines.
Supplementary Fig. S5. Additional growth data.
Supplementary Fig. S6. Analysis of wall-bound phenolics in xylem of WT and transgenic Populus grown under different N regimes.
Supplementary Fig. S7. Effects of PfaGT1-316 overexpression on major phenylpropanoid products.
Supplementary Fig. S8. Relative transcript abundance of representative phenylpropanoid pathway genes in WT and transgenic Populus grown under different N regimes.
Acknowledgements
The authors would like to thank Sandra Hubscher for assistance with plant transformation, Steve Pettis for greenhouse plant care, Fielding Callaway for lab assistance, Mark Wilson for scripts that facilitated HPLC-MS file format conversion, Batbayar Nyamdari for analysis of wall-bound phenolics by GC-MS, Vanessa Michelizzi for help with qRT-PCR, and Liang-Jiao Xue for help with microarray data mining. This work was funded by the US National Science Foundation (grants DBI-0421756 and 0836433).
References
- Afendi FM, Okada T, Yamazaki M, et al. 2012. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant and Cell Physiology 53, e1. [DOI] [PubMed] [Google Scholar]
- Beimen A, Bermpohl A, Meletzus D, Eichenlaub R, Barz W. 1992. Accumulation of phenolic compounds in leaves of tomato plants after infection with Clavibacter michaganense supsp michiganense strains differing in virulence. Zeitschrift für Naturforschung C: Journal of Biosciences 47, 898–909 [Google Scholar]
- Bernards MA, Strack D, Wray V, Ellis BE. 1991. Caffeoyl glucosides in fungal challenged tomato suspension cultures. Phytochemistry 30, 497–499 [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54, 519–546 [DOI] [PubMed] [Google Scholar]
- Bowles D, Lim E-K, Poppenberger B, Vaistij FE. 2006. Glycosyltransferases of lipophilic small molecules. Annual Review of Plant Biology 57, 567–597 [DOI] [PubMed] [Google Scholar]
- Braham H, Mighri Z, Jannet H. B., Matthew S, Abreu PM. 2005. Antioxidant phenolic glycosides from Moricandia arvensis . Journal of Natural Products 68, 517–522 [DOI] [PubMed] [Google Scholar]
- Chang SJ, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116 [Google Scholar]
- Clauß K, von Roepenack-Lahaye E, Böttcher C, Roth MR, Welti R, Erban A, Kopka J, Scheel D, Milkowski C, Strack D. 2011. Overexpression of sinapine esterase BnSCE3 in oilseed rape seeds triggers global changes in seed metabolism. Plant Physiology 155, 1127–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell RL, Hill SJ, Ortori CA, Barrett DA. 2008. Quantitative profiling of nucleotides and related phosphate-containing metabolites in cultured mammalian cells by liquid chromatography tandem electrospray mass spectrometry. Journal of Chromatography B 871, 115–124 [DOI] [PubMed] [Google Scholar]
- Corner JJ, Swain T. 1965. Enzymatic synthesis of sugar esters of hydroxy-aromatic acids. Nature 207, 634–635 [DOI] [PubMed] [Google Scholar]
- D’Abrosca B, Fiorentino A, Ricci A, Scognamiglio M, Pacifico S, Piccolella S, Monaco P. 2010. Structural characterization and radical scavenging activity of monomeric and dimeric cinnamoyl glucose esters from Petrorhagia velutina leaves. Phytochemistry Letters 3, 38–44 [Google Scholar]
- D’Auria JC. 2006. Acyltransferases in plants: a good time to be BAHD. Current Opinion in Plant Biology 9, 331–340 [DOI] [PubMed] [Google Scholar]
- Dixon RA. 2005. Engineering of plant natural product pathways. Current Opinion in Plant Biology 8, 329–336 [DOI] [PubMed] [Google Scholar]
- Douglas CJ, Ehlting J, Harding SA. 2011. Phenylpropanoid and phenolic metabolism in Populus: gene family structure and comparative and functional genomics. In: Joshi CP, DiFazio SP, Kole C, eds. Genetics, genomics and breeding of poplars. Enfield, NH: Science Publishers; pp 304–326 [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, et al. 2006. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiology 140, 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläßgen W, Seitz H. 1992. Acylation of anthocyanins with hydroxycinnamic acids via 1-O-acylglucosides by protein preparations from cell cultures of Daucus carota L . Planta 186, 582–585 [DOI] [PubMed] [Google Scholar]
- Gou J-Y, Park S, Yu ×-H, Miller LM, Liu C-J. 2008. Compositional characterization and imaging of ‘wall-bound’ acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta 229, 15–24 [DOI] [PubMed] [Google Scholar]
- Grace SC, Logan BA, Adams WW. 1998. Seasonal differences in foliar content of chlorogenic acid, a phenylpropanoid antioxidant, in Mahonia repens . Plant, Cell and Environment 21, 513–521 [Google Scholar]
- Griesser M, Hoffmann T, Bellido ML, Rosati C, Fink B, Kurtzer R, Aharoni A, Muñoz-Blanco J, Schwab W. 2008. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiology 146, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SA, Jiang HY, Jeong ML, Casado FL, Lin HW, Tsai CJ. 2005. Functional genomics analysis of foliar condensed tannin and phenolic glycoside regulation in natural cottonwood hybrids. Tree Physiology 25, 1475– 1486 [DOI] [PubMed] [Google Scholar]
- Hüsken A, Baumert A, Strack D, Becker HC, Möllers C, Milkowski C. 2005. Reduction of sinapate ester content in transgenic oilseed rape (Brassica napus) by dsRNAi-based suppression of BnSGT1 gene expression. Molecular Breeding 16, 127–138 [Google Scholar]
- Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. 2001. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. Journal of Biological Chemistry 276, 4350–4356 [DOI] [PubMed] [Google Scholar]
- Kammerer B, Kahlich R, Biegert C, Gleiter CH, Heide L. 2005. HPLC-MS/MS analysis of willow bark extracts contained in pharmaceutical preparations. Phytochemical Analysis 16, 470–478 [DOI] [PubMed] [Google Scholar]
- Kolb CA, Kaser MA, Kopecky j, Zotz G, Riederer M, Pfundel EE. 2001. Effects of natural intensities of visible and ultraviolet radiation on epidermal ultraviolet screening and photosynthesis in grape leaves. Plant Physiology 127, 863–875 [PMC free article] [PubMed] [Google Scholar]
- Kosonen M, Keski-Saari S, Ruuhola T, Constabel CP, Julkunen-Tiitto R. 2012. Effects of overproduction of condensed tannins and elevated temperature on chemical and ecological traits of genetically modified hybrid aspens (Populus tremula × P. tremuloides). Journal of Chemical Ecology 38, 1235–1246 [DOI] [PubMed] [Google Scholar]
- Kylli P, Nousiainen P, Biely P, Sipilä J, Tenkanen M, Heinonen M. 2008. Antioxidant potential of hydroxycinnamic acid glycoside esters. Journal of Agricultural and Food Chemistry 56, 4797–4805 [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL. 1995. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiology 109, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot A, Hodge D, Lim E-K, Vaistij F, Bowles D. 2008. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 228, 609–616 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C. 2000. Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. The Plant Cell 12, 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ. 2001. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana . Journal of Biological Chemistry 276, 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Higgins GS, Ll Y, Bowles DJ. 2003. Regioselectivity of glucosylation of caffeic acid by a UDP-glucose:glucosyltransferase is maintained in planta. The Biochemical Journal 373, 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-K, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ. 2001. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis . Journal of Biological Chemistry 276, 4344–4349 [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Bellido M, Aharoni A, Salentijn EMJ, Kaldenhoff R, Coiner HA, Muñoz-Blanco J, Schwab W. 2006. Cinnamate metabolism in ripening fruit. Characterization of a UDP-glucose:cinnamate glucosyltransferase from strawberry. Plant Physiology 140, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilan R, Ma C. 2006. Poplar (Populus spp.). In: Wang K, ed. Methods in molecular biology, vol. 34: Agrobacterium protocols . Totowa, NJ: Humana Press; pp 143–151 [DOI] [PubMed] [Google Scholar]
- Meißner D, Albert A, Böttcher C, Strack D, Milkowski C. 2008. The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana . Planta 228, 663–674 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Baumert A, Strack D. 2000a. Cloning and heterologous expression of a rape cDNA encoding UDP-glucose:sinapate glucosyltransferase. Planta 211, 883–886 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Baumert A, Strack D. 2000b. Identification of four Arabidopsis genes encoding hydroxycinnamate glucosyltransferases. FEBS Letters 486, 183–184 [DOI] [PubMed] [Google Scholar]
- Mittasch J, Böttcher C, Frolov A, Strack D, Milkowski C. 2013. Reprogramming the phenylpropanoid metabolism in seeds of oilseed rape by suppressing the orthologs of REDUCED EPIDERMAL FLUORESCENCE1. Plant Physiology 161, 1656–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittasch J, Strack D, Milkowski C. 2007. Secondary product glycosyltransferases in seeds of Brassica napus . Planta 225, 515–522 [DOI] [PubMed] [Google Scholar]
- Mock H-P, Strack D. 1993. Energetics of the uridine 5′-diphosphoglucose: hydroxycinnamic acid acyl-glucosyltransferase reaction. Phytochemistry 32, 575–579 [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CHR. 2006. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiology 141, 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula R, Babst B, Nelsen M, Harding S, Tsai C-J. 2009. Glycosylation-mediated phenylpropanoid partitioning in Populus tremuloides cell cultures. BMC Plant Biology 9, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula RS, Tay KHC, Tsai C-J, Harding SA. 2011. The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. The Plant Journal 65, 757–770 [DOI] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG. 1986. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 25, 223–230 [Google Scholar]
- Ralph J, Bunzel M, Marita J, Hatfield R, Lu F, Kim H, Schatz P, Grabber J, Steinhart H. 2004. Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochemistry Reviews 3, 79–96 [Google Scholar]
- Ranjan P, Kao Y-Y, Jiang H, Joshi CP, Harding SA, Tsai C-J. 2004. Suppression subtractive hybridization-mediated transcriptome analysis from multiple tissues of aspen (Populus tremuloides) altered in phenylpropanoid metabolism. Planta 219, 694–704 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506 [DOI] [PubMed] [Google Scholar]
- Sheahan JJ. 1996. Sinapate esters provide greater UV-B attenuation than flavonoids in Arabidopsis thaliana (Brassicaceae). American Journal of Botany 83, 679–686 [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C. 2007. The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid:UDPG glucosyltransferase. The Plant Journal 49, 655–668 [DOI] [PubMed] [Google Scholar]
- Strack D. 2001. Enzymes involved in hydroxycinnamate metabolism. Methods in Enzymology 335, 70–81 [DOI] [PubMed] [Google Scholar]
- Sykes R, Yung M, Novaes E, Kirst M, Peter G, Davis M. 2009. High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. Methods in Molecular Biology 581, 169–183 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattini M, Galardi C, Pinelli P, Massai R, Remorini D, Agati G. 2004. Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytologist 163, 547–561 [DOI] [PubMed] [Google Scholar]
- Teusch M, Forkmann G, Seyffert W. 1987. Genetic control of hydroxycinnamoyl-coenzyme a: anthocyanidin 3-glycoside-hydroxycinnamoyltransferase from petals of Matthiola incana . Phytochemistry 26, 991–994 [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y. 2006. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus . New Phytologist 172, 47–62 [DOI] [PubMed] [Google Scholar]
- Tuominen L, Johnson V, Tsai C-J. 2011. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio SP, Jansson S, Bohlmann J, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N. 2007. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: chemical diversity, impacts on plant biology and human health. Biotechnology Journal 2, 1214–1234 [DOI] [PubMed] [Google Scholar]
- Vogt T. 2010. Phenylpropanoid biosynthesis. Molecular Plant 3, 2–20 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P. 2000. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends in Plant Science 5, 359–403 [DOI] [PubMed] [Google Scholar]
- Wang HQ, Tuominen LK, Tsai C-J. 2011. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics 27, 225–231 [DOI] [PubMed] [Google Scholar]
- Weiss M, Schmidt J, Neumann D, Wray V, Christ R, Strack D. 1999. Phenylpropanoids in mycorrhizas of the Pinaceae. Planta 208, 491–502 [Google Scholar]
- Wolfram K, Schmidt J, Wray V, Milkowski C, Schliemann W, Strack D. 2010. Profiling of phenylpropanoids in transgenic low-sinapine oilseed rape (Brassica napus). Phytochemistry 71, 1076–1084 [DOI] [PubMed] [Google Scholar]
- Xie D-Y, Sharma SB, Paiva NL, Ferreira D, Dixon RA. 2003. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399 [DOI] [PubMed] [Google Scholar]
- Xue L, Guo W, Yuan Y, et al. 2013. Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state, but not growth in transgenic Populus . The Plant Cell 25, 2714–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, et al. 2012. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana . The Plant Journal 69, 154–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Mori M, Kondo T. 2009. Blue flower color development by anthocyanins: from chemical structure to cell physiology. Natural Product Reports 26, 884–915 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chung J-D, Fu ×, Johnson VE, Ranjan P, Booth SL, Harding SA, Tsai C-J. 2009. Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 22020–22025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.