Summary
EDT1/HGD11 coordinately upregulates gene families of cell-wall-loosening proteins to alter cell-wall extensibility and promote primary root elongation.
Key words: Cell-wall-loosening protein genes, cellulase, edt1, expansin, HDG11, pectin-related enzymes, XTH.
Abstract
The gain-of-function mutant edt1 shows significantly enhanced drought tolerance and a well-developed root system including deeper primary roots and more lateral roots. To explore the molecular mechanisms underlying the improved root system of edt1, we performed transcriptome comparison between the wild-type and edt1 roots. One of the interesting findings from the analysis was that several gene families of cell-wall-loosening proteins were upregulated in the mutant roots, including expansins, extensins, xyloglucan endotransglucosylase/hydrolases (XTHs), pectin-related enzymes, and cellulases. Most of these genes contain HD-binding cis-elements in their promoters predominantly with the TTTAATTT sequence, which can be bound by HDG11 in vitro and in vivo. The coordinated expression of these gene families overlaps fast root elongation. Furthermore, overexpression of AtEXPA5, which was dramatically upregulated in edt1, resulted in longer primary roots because cells were more extended longitudinally. When combined by crossing the AtEXPA5-overexpression lines with one pectin methylesterase inhibitor family protein (PMEI) gene (At5g62360)- or one cellulase (CEL) gene (At2g32990)-overexpression lines, the primary roots of the progeny even exceeded both parents in length. Our results demonstrate that HDG11 directly upregulates cell-wall-loosening protein genes, which is correlated with altered root system architecture, and confirm that cell-wall-loosening proteins play important roles in coordinating cell-wall extensibility with root development. The results of transgene experiments showed that expansin works together with PMEI and CEL to generate synergistic effects on primary root elongation, suggesting that different cell-wall-loosening protein families may function in combination to generate optimal effects on root extensibility.
Introduction
A root system is crucial for plants to absorb water and nutrients from the soil, in addition to anchoring and supporting the plant. A better-shaped root-system architecture would benefit a plant with increased fitness (Aiken and Smucker, 1996; de Dorlodot et al., 2007; Smith and De Smet, 2012). Root development and growth are not only genetically programmed but also constantly influenced by environmental cues. Roots perceive various environmental signals such as water and nutrients, and process these signals to reprogramme their development to adapt to the environment. In this sense, roots have to be very flexible during their development and growth.
It is well established that plant hormones play essential roles in root development and growth (Brunoud et al., 2012; Dello Ioio et al., 2012; Durbak et al., 2012). Among those hormones, auxin seems to play the most important role in root development. It has been found that most hormones that affect the root development seem to act ultimately by modulating auxin activity (Moubayidin et al., 2010; Perilli et al., 2012). Hormones control root development in many ways, one of which is that hormones can change the expression of root cell-wall-loosening protein genes or the activity of cell-wall-loosening proteins (Jan et al., 2004; Liu et al., 2007; Zhao et al., 2012). Regulated cell expansion allows plants to adapt their morphogenesis to survive in different environmental conditions.
Turgor pressure works with cell-wall loosening proteins to regulate cell expansion (Cosgrove, 2005). Cell expansion is driven by turgor pressure created by water uptake and is circumscribed by the extensibility of the cell wall (Brux et al., 2008). However, much less is known about the change in cell walls that must be coordinately loosened during root development for root cell expansion and lateral root emergence.
Plant cells encase themselves within a complex polysaccharide wall, and thus cell expansion must depend on cell-wall loosening through a process of controlled polymer creep. Plant cells loosen their cell walls through molecular modifications of the wall network that result in relaxation of the wall stress. Wall stress relaxation results from scission of a stress-bearing crosslink or from sliding of such a crosslink along a scaffold. Cell-wall enlargement occurs secondarily because of cellular water uptake. Four molecular mechanisms for cell-wall loosening are generally accepted. The four wall-loosening agents are expansins (Cosgrove et al., 2002; Yennawar et al., 2006; Won et al., 2010; ZhiMing et al., 2011), xyloglucan endotransglucosylase/hydrolase (XTHs) (Vissenberg et al., 2003, 2005; Osato et al., 2006; Maris et al., 2009; Tominaga-Wada et al., 2009), endo-(1,4)-β-d-glucanase (Yoshida et al., 2006; Zhu et al., 2006; Yang et al., 2011) and hydroxyl radical (•OH) (Schopfer et al., 2002; Liszkay et al., 2003; Muller et al., 2009).
Expansins are pH-dependent wall-loosening proteins that are first identified from ‘acid growth’. They are involved in cell enlargement and in a variety of developmental processes requiring cell-wall modifications. Expansins are a superfamily that contains four families: α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA) and expansin-like B (EXLB).The polyphyletic group of non-plant expansins, such as those in Dictyostelium, can be referred to as expansin-like family X (EXLX). EXPA and EXPB proteins have been demonstrated experimentally to cause cell-wall loosening (Cho and Kende, 1997; Guo et al., 2011), whereas EXLAs and EXLBs proteins are known only from their gene sequences.
XTHs are enzymes involved in the modification of load-bearing cell-wall components. They are also encoded by large multigene families (Maris et al., 2009; Yokoyama et al., 2010). A comprehensive expression analysis of all 33 members of the Arabidopsis XTH gene family revealed their tissue specificity and distinct response to hormonal stimulation. Among them, at least 10 genes were expressed predominantly in roots (Yokoyama and Nishitani, 2001).
Cellulose is the major component of the plant cell wall. Besides XTH, plants have a family of secreted endo-(1,4)-β-d-glucanases (also called ‘cellulases’), which belong to glycoside hydrolase family 9 (Henrissat, 1991). In Arabidopsis, there are 25 members in this family, and three of them are membrane-bound endoglucanases that are involved in cellulose formation (Cosgrove, 2005). The remaining 22 members are secreted enzymes, mostly of unknown function. The role of endo-(1,4)-β-d-glucanases in wall loosening merits greater attention.
A hydroxyl radical (•OH) is a highly active form of reactive oxygen species and has important roles in signalling and cell death. It was reported that •OH produced in the cell wall can loosen it through destroying the hydrogen bonds by non-enzymatically removing a hydrogen atom from polysaccharides (Fry, 1998; Schopfer, 2001; Lionetti et al., 2007).
Besides cellulose, pectin is one of the main components of the plant cell wall. Pectin-related enzymes have important functions in cell-wall loosening such as pectate lyase, pectin esterase/pectin methylesterase (PME), pectin methylesterase inhibitor family protein (PMEI), and pectinase. Pectin is secreted in a highly methyl-esterified form and is dimethyl-esterified by PME, whose activity leads to stiffer pectin gels and reduced cell growth (Derbyshire et al., 2007; Siedlecka et al., 2008). This is probably one of multiple mechanisms for cell-wall stiffening. Regulation of PME activity by specific PMEIs can therefore play a positive role in cell growth. Overexpression of PMEIs results in root length increases (Lionetti et al., 2007). However, opposite results have also been reported (Peaucelle et al., 2011; Hongo et al., 2012). PME and PMEI may have diverse roles in regulating the ratio of methyl-pectin and demethyl-pectin to affect cell-wall extensibility.
The previously reported gain-of-function Arabidopsis mutant edt1 has enhanced drought tolerance with a more extensive root system than the wild-type plant (Yu et al., 2008). In edt1 mutant, an HD-ZIP IV transcription factor, HDG11, is activated in most tissues. When overexpressed in rice, AtEDT1/HDG11 also confers drought tolerance and an improved root system (Yu et al., 2013). To uncover the molecular mechanisms underlying the improved root system architecture of edt1, we compared the root transcriptome between the edt1 mutant and the Columbia (Col-0) wild type at different root development stages using microarray analysis. It was found that cell-wall-related genes, most of which function in loosening the cell wall to facilitate its growth, were significantly upregulated in edt1, including expansins, XTHs, pectin-related enzymes, and cellulases. Using yeast-one-hybrid (Y1H) and chromatin immunoprecipitation (ChIP) assays, we further demonstrated that most of the upregulated cell-wall-loosening protein genes were directly regulated by HDG11 at the transcriptional level. Our results suggest that the cell-wall-loosening proteins function coordinately to loosen the cell wall during root development. The regulated expression of cell-wall-loosening protein genes is an indispensable component of root development and growth.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana Col-0 ecotype was used through this study. The edt1 mutant was identified previously in the same laboratory (Yu et al., 2008). Seeds were surface sterilized in 10% bleach for 10min and then rinsed five times in sterile water. To overcome dormancy, we imbibed seeds at 4 °C for 2–4 d. The seeds were then germinated and grown on Murashige and Skoog (MS) medium at 22 °C under 16h light/8h dark cycles.
Measurement of cell length of the root elongation zone
Five-day old seedling roots of wild-type and EXPA5-overexpression lines were observed under an Olympus light microscope and photographed. We used the measurement software RULER to measure the cell length of the elongation zone, for three cell columns for each root.
Microarrays
The seeds were germinated on MS solid medium with or without 50mg l–1 of glufosinate ammonium horizontally and the seedlings were then transferred to new vertical MS solid medium. After growing for 3, 6, 10, 15, and 20 d, the roots of edt1 and the wild type were collected and RNA was extracted using TRIzol reagent (Invitrogen). An Affymetrix gene chip was performed and analysed by Capitolbio Corporation.
Real-time reverse transcription (RT)-PCR
Total RNA was extracted using TRIzol reagent. First-strand cDNA was synthesized from 1 μg of total RNA in a 20 μl reaction mixture using a Prime Script RT regent kit (Takara). The transcript levels of EXPA5 were examined using the specific primers 5′-ttagtaatctcgcttctcgtggttc-3′ and 5′-ccataaccttggctatacagattgc-3′. The transcript levels of EXPB3 were examined using the specific primers 5′-atgcagctttttccagtcatgttag-3′ and 5′-tcacatccaccaacgtaccgtaa-3′. Arabidopsis UBQ5 was used as an internal control using the specific primers 5′-agaagatcaagcacaagcat-3′ and 5′-cagatcaagcttcaactcct-3′. The PCR was performed on an ABI7000 using the following conditions: 95 °C for 3min, followed by 40 cycles of 95 °C for 20 s, 62 °C for 20 s, and 72 °C for 31 s.
Semi-quantitative RT-PCR
Total RNA was extracted using TRIzol reagent from 20-d-old roots of the wild type and edt1. First-strand cDNA was synthesized from 1 μg of total RNA in a 20 μl reaction mixture using a Prime Script RT regent kit (Takara). The transcript levels of AT1g48930, AT5g62360, AT2g36870, At2g32990, At5g57560, At1g67750, and At1g54970 were examined using the specific primers listed in Supplementary Table S3 at JXB online.
Y1H assay
A Y1H assay was performed as described previously with minor modifications (Wang et al., 2007). A cDNA fragment encoding HDG11 was amplified with the primer pair 5′-ccgctcgagatgagtttcgtcgtcggcgt-3′ and 5′-gctctagaagctgtagttgaagctgtag-3′, and inserted into pAD-GAL4-2.1 to generate the pAD/HDG11 plasmid, and this plasmid was used to produce protein (translationally fused to the GAL4 AD domain) for DNA binding in the Y1H assay. Three copies of the cis-DNA elements, containing SacI and MluI adaptors, were annealed and cloned into the SacI and MluI sites of the reporter plasmid, pHIS2, which also contained the nutritional reporter gene, HIS3. The constructs were confirmed by sequencing.
The pAD/HDG11 construct and the reporter pHIS2 containing the cis-DNA element were co-transfected into yeast cells Y187. For the negative control, the pAD/HDG11 vector and the pHIS2 empty plasmid were co-transfected into Y187 yeast cells. Yeast was grown in SD/–Trp–Leu medium and then spotted on SD/–Trp–Leu–His medium in the presence of 10mM 3-aminotriazole (Sigma) at different dilutions. The plates were incubated at 30 ℃ for 4 d and the extent of yeast growth was determined. Normal growth of the cells on the SD/–Trp–Leu–His medium in the presence of 10mM 3-aminotriazole indicated that binding of the transcription factor to the corresponding cis-DNA element had occurred.
Constructs and generation of transgenic plants
To get 35S:: EXPA5 transgenic lines, we isolated the EXPA5 cDNA from wild-type Col-0 by RT-PCR with forward primer 5′-gtacaaaaaagcaggctatgggagttttagtaatctcgct-3′ and reverse primer 5′-gtacaagaaagctgggtttaataccgaaactgccctcc-3′, cloned it into pDONR207, and subsequently shuttled it into the expression binary vector pCB2004 (Lei et al., 2007).
To get 35S::At5g62360 (PMEI) transgenic lines, we isolated the At5g62360 cDNA from wild-type Col-0 by RT-PCR with forward primer 5′-gtttgtacaaaaaagcaggctatgggtgaatcttttagattat-3′ and reverse primer 5′-ctttgtacaagaaagctgggtttagccatgaatagaagcaaag-3′, cloned it into pDONR207, and subsequently shuttled it into the expression binary vector pCB2004.
To get 35S::At2g32990 [cellulase (CEL)] transgenic lines, we isolated the At2g32990 cDNA from wild-type Col-0 by RT-PCR with forward primer 5′-gtttgtacaaaaaagcaggctatgactgtgatgaatcaccgac-3′ and reverse primer 5′-ctttgtacaagaaagctgggtctatctcttataagttgcaacc-3′, cloned it into pDONR207, and subsequently shuttled it into the expression binary vector pCB2004.
For ChIP assays, the 35S::HA-Tag-HDG11 construct was made by primers with a haemagglutinin (HA) tag: 5′-gtttgtacaaaaaagcaggctatgtacccatacgatgttccagattacgctatgagtttcgtcgtcggcgtcg-3′ and 5′-ctttgtacaagaaagctgggttcaagctgtagttgaagctgta-3′, cloned it into pDONR207, and subsequently shuttled it into the expression binary vector pCB2004.
The constructs were introduced into Agrobacterium tumefaciens C58C1, which was used to transform the Col-0 wild-type plants as described previously (Clough and Bent, 1998).
ChIP assay
A ChIP assay was performed as described previously (Lee et al., 2007) using an HA tag-specific monoclonal antibody (Cali-Bio) for immunoprecipitation. Approximately 0.5g of 10-d-old roots was used in each ChIP experiment. The immunoprecipitated chromatin was extracted with phenol/chloroform and the DNA was precipitated with ethanol. ChIP-PCR was then used to verify each promoter segment of related genes using the primers listed in Supplementary Table S4 at JXB online.
Results
A large number of cell-wall-loosening protein genes are upregulated in edt1 roots
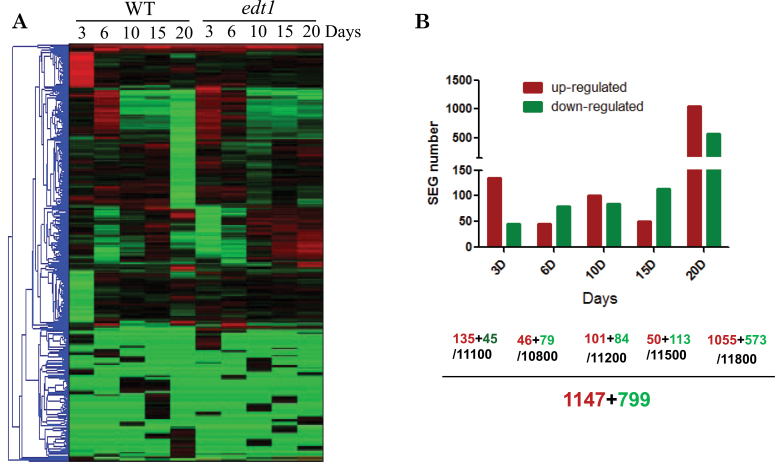
The edt1 mutant is a gain-of-function mutant with enhanced drought tolerance and a well-developed root system (Yu et al., 2008). To gain a view of global expression patterns in the root of the edt1 mutant, we compared the root transcriptome between edt1 and the wild type (Col-0). The transcription profiling results showed that there were dramatic changes in expression profiles in edt1 compared with the wild type (Fig. 1A). In our analysis, the genes were defined as significantly expressed genes (SEGs) if their change was greater than or equal to 2-fold that of the wild type. At different developmental stages, there were different numbers of SEGs. The stage of 20 d had the largest number of SEGs. In total, there were 1147 upregulated SEGs and 799 downregulated SEGs (Fig. 1B).
Fig. 1.
Microarray analysis of edt1 roots. RNA samples isolated from 3-, 6-, 10-, 15-, and 20-d-old roots of edt1 and wild-type plants were analysed by microarray. (A) Overview of gene clusters. (B) SEG numbers at different growth stages. Upregulated and downregulated SEG numbers are indicated, together with the total number of genes identified at each specific stage. (This figure is available in colour at JXB online.)
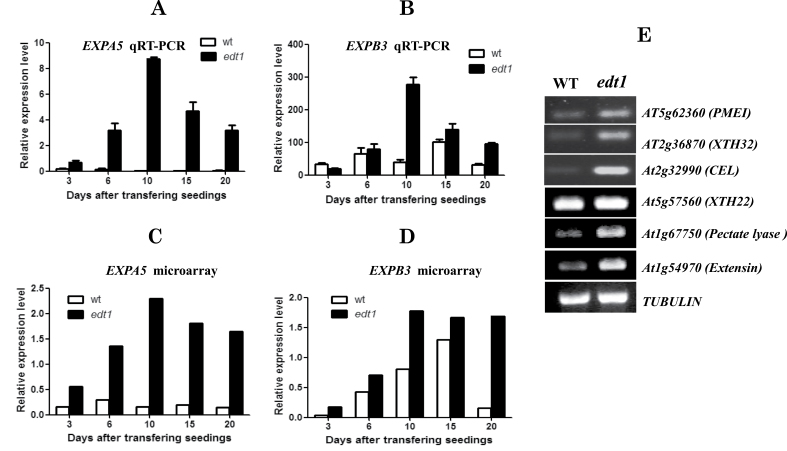
Gene Ontology (GO) term enrichment analysis of upregulated SEGs showed that cell-wall subcellular localization genes were significantly enriched (Table 1). Moreover, when we looked into the cell-wall-related genes, we found that most of the 72 cell-wall-related genes had functions in cell-wall loosening (Supplementary Table S1 at JXB online), such as expansins, XTHs, pectin-related enzymes, glycosyl hydrolases, and extensins. Considering that there are many gene products that do not localize on cell wall but that have functions in cell-wall loosening, we searched all 1147 upregulated genes and found 11 expansins, 13 XTHs, 20 pectin-related enzymes (six pectate lyase lyases, six pectin esterases, five PMEIs, and three pectinases), 29 glycosyl hydrolases, and 24 extensins (Supplementary Table S2 at JXB online). These kinds of gene all play key roles in cell-wall loosening, especially expansins and XTHs. EXPA5 was the most notably changed gene; it had almost no expression in wild-type roots but very high expression in edt1 roots (Fig. 2A, C, and Supplementary Fig. S1A at JXB online). EXPB3, expressed mainly in roots of the wild type with reference to the compiled microarray data (Supplementary Fig. S1B), was also significantly upregulated in edt1 roots (Fig. 2B, D). The upregulation of a representative member for other cell-wall-loosening protein families was also confirmed by semi-quantitative RT-PCR (Fig. 2E). Our data revealed that many families of cell-wall-loosening protein genes are upregulated significantly in edt1 roots, which may facilitate root growth and development.
Table 1.
GO term enrichment analysis of SEGs in edt1 rootsGO analysis was performed on the MIPS website (http://mips.helmholtz-muenchen.de/proj/funcatDB/). The lower the P value, the higher the degree of enrichment. The top four enrichment functional categories were chosen and are listed in this table. ‘Abs set’ means the gene number in this functional category out of input and ‘Rel set’ means its percentage of the input gene number (which in this table is 1147). ‘Abs genome’ means the gene number in this functional category out of the Arabidopsis genome and ‘Rel genome’ means its percentage of the Arabidopsis genome.
| Functional category | Abs set | Rel set | Abs genome | Rel genome | P value |
|---|---|---|---|---|---|
| Cell wall | 72 | 6.28 | 431 | 1.51 | 6.79E–25 |
| Oxidative stress response | 39 | 3.4 | 200 | 0.7 | 2.14E–16 |
| Oxygen and radical detoxification | 40 | 3.49 | 261 | 0.91 | 3.88E–13 |
| Osmosensing and response | 32 | 2.79 | 208 | 0.73 | 7.78E–11 |
Fig. 2.
Cell-wall loosening-related genes are coordinately upregulated in edt1. (A–D) RNA samples isolated from 3-, 6-, 10-, 15-, and 20-d-old roots of edt1 and wild-type plants were analysed by real-time RT-PCR using specific primers and a microarray. Results are shown as the relative expression level of EXPA5 (A) and EXPB3 (B) by real-time RT-PCR, and the relative expression level of EXPA5 (C) and EXPB3 (D) by microarray. (E) Expression levels of other cell-wall-loosening protein genes by RT-PCR. RNA samples isolated from 20-d-old roots of edt1 and wild-type plants were analysed by semi-quantitative RT-PCR using specific primers.
HD-binding cis-elements are highly enriched in the promoters of cell-wall-loosening protein genes
As AtHDG11 is a transcription factor belonging to the HD-ZIP IV subfamily, which can bind specific HD-binding cis-elements to regulate gene expression, we analysed the promoters of these upregulated SEGs and found that 523 out of the 1147 genes contained HD-binding cis-elements in their promoters. The result of GO term enrichment analysis of the 523 genes is shown in Table 2. The cell-wall-related genes were still the most enriched functional category. The ratios of genes with an HD-binding cis-element in different functional categories were all significantly higher than the average for the Arabidopsis genome (Table 3), which indicates that the upregulated SEGs could be directly regulated by HDG11. Moreover, among the upregulated cell-wall-loosening protein genes, eight of the 11 expansins, six of the 12 XTHs, 14 of the 20 pectin-related enzymes, 18 of the 29 glycosyl hydrolases, and 14 of the 24 extensins contained an HD-binding cis-element in their promoters (Supplementary Table S2). There were several types of HD-binding cis-element in the upregulated cell-wall loosening protein genes promoters; however, most of them contained AAATTAAA (Table 4). Thus, the AAATTAAA motif appeared to be the predominant HD-binding cis-element.
Table 2.
GO term enrichment analysis of SEGs containing HD-binding cis-element in edt1 rootsSee Table 1 legend for abbreviations.
| Functional category | Abs Set | Rel set | Abs genome | Rel genome | P value |
|---|---|---|---|---|---|
| Cell wall | 31 | 5.92 | 431 | 1.51 | 1.42E–10 |
| Oxidative stress response | 19 | 3.63 | 200 | 0.7 | 6.45E–09 |
| Osmosensing and response | 17 | 3.25 | 208 | 0.73 | 3.57E–07 |
| Oxygen and radical detoxification | 16 | 3.05 | 261 | 0.91 | 3.07E–05 |
Table 3.
The ratio of genes with HD-binding cis-element in different functional categories
| Functional category | Gene no. | Gene no. with HD-binding cis-element | Ratio |
|---|---|---|---|
| Cell wall | 72 | 31 | 43.1% |
| Oxidative stress response | 39 | 19 | 48.7% |
| Oxygen and radical detoxification | 40 | 17 | 42.5% |
| Osmosensing and response | 32 | 16 | 50.0% |
| Arabidopsis genome | 29110 | 10010 | 34.4% |
Table 4.
The HD-binding cis-element in the promoters of cell-wall loosening protein genes upregulated in edt1 roots (including the genes coding for proteins that do not localize in the wall)
| Gene family | Locus | HD-binding cis-element |
|---|---|---|
| Expansin | At3g29030 | aaattaaa |
| At4g28250 | aaattaaa | |
| At2g03090 | aaattaaa | |
| At1g20190 | aaattaaa ×2 | |
| At2g20750 | aaattaaa ×2 | |
| At2g39700 | aaattaaa ×3 | |
| At3g45970 | aaattaaa, aaattagt, actaattt | |
| At1g12560a | aaattagt ×2, actaattt ×2 | |
| XTH | At2g06850 | aaattaaa ×4 |
| At2g36870 | aaattaaa ×3 | |
| At4g03210 | aaattaaa | |
| At4g30270 | aaattaaa ×3 | |
| At4g37800 | aaattaaa ×3 | |
| At5g57560 | aaattaaa, aaattagt, actaattt | |
| Pectate lyase | At1g67750 | aaattaaa ×3 |
| At4g24780 | aaattaaa ×2 | |
| At1g04680 | aaattaaa, aaattagt, actaattt | |
| At3g24670a | aaattagt ×2, actaattt ×2 | |
| Pectinesterase | At2g45220 | aaattaaa ×2, aaattagt, actaattt |
| At1g02810 | aaattaaa | |
| At5g47500 | aaattaaa, aaattagt, actaattt | |
| At3g10720 | aaattaaa | |
| Pectinase | At1g65570a | actaattt ×2, aaattagt ×2 |
| At5g14650 | aaattaaa ×2, aaattagt, actaattt | |
| At3g07970 | aaattaaa, aaattagt, actaattt | |
| PMEI | At5g62350 | aaattaaa ×3, aaattagt, actaattt |
| At1g62770 | aaattaaa | |
| At5g62360 | aaattaaa ×2 | |
| Glycosyl hydrolase | At3g60130 | aaattaaa |
| At1g51470 | aaattaaa | |
| At1g47600 | aaattaaa | |
| At3g07320 | aaattaaa | |
| At4g18340 | aaattaaa | |
| At5g63800 | aaattaaa | |
| At5g34940 | aaattaaa | |
| At5g15870 | aaattaaa | |
| At4g02290 | aaattaaa | |
| At4g19810 | aaattaaa ×2 | |
| At2g32990 | aaattaaa ×2, aaattagt ×2, actaattt ×2 | |
| At3g28180 | aaattaaa ×3, aaattagt ×2, actaattt ×2 | |
| At1g60140 | aaattaaa ×3, aaattagt, actaattt | |
| At3g60140 | aaattaaa ×4, aaattagt ×2, actaattt ×2 | |
| At1g61820 | aaattaaa ×5 | |
| At1g02640 | aaattaaa ×5 | |
| At2g27500a | aaattagt | |
| At5g49360a | aaattagt, actaattt | |
| Extensin | At4g38770 | aaattaaa |
| At4g02270 | aaattaaa | |
| At5g05500 | aaattaaa, aaattagt, actaattt | |
| At3g54580 | aaattaaa ×2 | |
| At5g49080 | aaattaaa | |
| At4g08410 | aaattaaa, aaattagt, actaattt | |
| At1g21310 | aaattaaa | |
| At5g06630 | aaattaaa | |
| At3g28550 | aaattaaa ×2, aaattagt ×2, actaattt ×2 | |
| At3g06750 | aaattaaa | |
| At1g76930 | aaattaaa ×2 | |
| At5g49280 | aaattaaa | |
| At5g09480 | aaattaaa | |
| At2g43150 | aaattaaa |
a Genes whose promoter does not contain the ‘aaattaaa’ motif.
HDG11 can directly regulate cell-wall-loosening protein gene expression
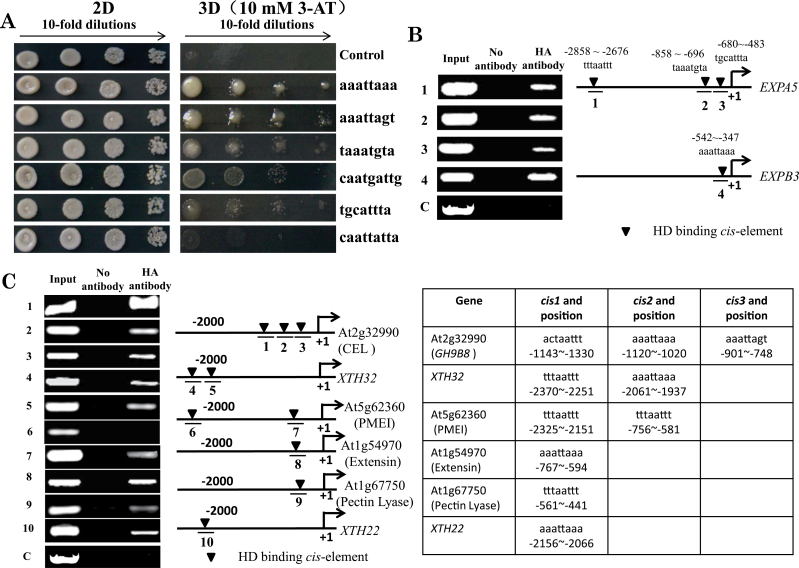
HD-ZIP IV transcription factors show a binding preference for variant HD-binding sequences. HDG7, HDG9, ATML1, and PDF2 recombinant proteins bind to the GCATT(A/T)AATGC consensus sequence, which overlaps with the L1 box sequence TAAATG(C/T)A recognized in vitro by ATML1 and GL2 (Ariel et al., 2007; Tominaga-Wada et al., 2009). According to the Arabidopsis Gene Regulatory Information Server (AGRIS), we chose six sequences, listed in Fig. 3A, as potential HDG11-binding sites in a Y1H assay. HDG11 showed different binding affinities to these sequences. The sequence aaattaaa was the favourite sequence, following by aaattagt, taaatgta (the L1 box), caatgattg, and tgcattta. HDG11 failed to bind caattatta (Fig. 3A). Most of the upregulated cell-wall-loosening protein genes contained aaattaaa sequence in their promoters (Table 4), indicating the possibility of HDG11 transcriptionally regulating these cell-wall-loosening protein genes.
Fig. 3.
HDG11 binds to the promoters in Y1H and ChIP assays. (A) Y1H assay in yeast strain Y187. The two vectors pGADT7/HDG11 and pHIS2/cis-element were transformed into Y187, and pGADT7/HDG11 and empty pHIS2 were used as negative controls. (B) ChIP-PCR for EXPA5 and EXPB3 promoters. The predicted HD-binding elements are indicated with inverted triangles, above which their sequence and position are shown. 1, EXPA5 promoter –2858 to –2676bp containing tttaattt; 2, EXPA5 promoter –858 to –696bp containing taaatgta; 3, EXPA5 promoter –680 to –483bp containing tgcattta; 4, EXPB3 promoter –542 to –347bp containing aaattaaa; C, negative control using TUB8 promoter. (C) ChIP-PCR for the GH9B8, XTH32, PMEI, extensin, pectin lyase, and XTH22 promoters. The predicted HD-binding elements and their sequence and position are listed in the table on the right. 1, GH9B8 promoter –1143 to –1330bp containing actaattt; 2, GH9B8 promoter –1120 to –1020bp containing aaattaaa; 3, GH9B8 promoter –901 to –748bp containing aaattagt; 4, XTH32 promoter –2370 to –2251 containing bp tttaattt; 5, XTH32 promoter –2061 to –1937bp containing aaattaaa; 6, PMEI promoter –2325 to –2151bp containing tttaattt; 7, PMEI promoter –756 to –581bp containing tttaattt and aaattaaa; 8, extensin promoter –767 to –594bp containing aaattaaa; 9, pectin lyase promoter –561 to –441bp containing tttaattt; 10, XTH22 promoter –2156 to –2066bp containing aaattaaa; C, negative control using the TUB8 promoter.
In the EXPA5 promoter, there are three potential HDG11-binding sites, all of which are predicted to be bound by HDG11. In the EXPB3 promoter, there is one potential HDG11-binding site. To confirm this, we generated HA-tagged HDG11-overexpressing lines for a ChIP assay to test whether HDG11 could bind to the promoters of EXPA5 and EXPB3 in vivo. The ChIP-PCR results showed that HDG11 was able to bind all three sites in the EXPA5 promoter and the one site in the EXPB3 promoter (Fig. 3B), consistent with the Y1H results. We also tested the binding of HDG11 to the promoters of other cell-wall-loosening protein genes in vivo. The results in Fig. 3C showed that HDG11 could bind to most of the predicted HD-binding cis-elements in the promoter of the upregulated cell-wall-loosening genes. These results demonstrated that HDG11 can directly bind to the promoters of the cell-wall-loosening protein genes and potentially activate their expression to coordinate cell-wall extensibility with root development.
Overexpression of EXPA5 stimulates root cell elongation
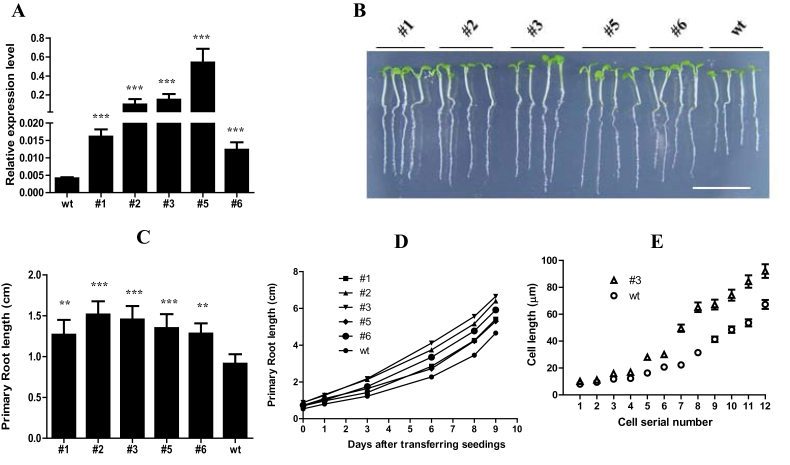
Among the upregulated cell-wall-loosening protein genes, EXPA5 was the most notable because its expression level was barely detectable in wild-type roots but was very high in edt1 roots (Fig. 2A and Supplementary Fig. S1A). We hypothesized that constitutively overexpressing this gene driven by the 35S promoter in the wild-type plant might affect the root growth. Five transgenic lines were obtained and their EXPA5 expression levels in roots are shown in Fig. 4A. The primary roots of these lines were significantly longer than that of the wild type (Fig. 4B, C). The root growth curves showed that primary root elongation of the transgenic lines was faster than that of the wild type from d 1 to d 9 in this experiment (Fig. 4D). The increased elongation was caused by increased cell length rather than cell number (Fig. 4E and Supplementary Fig. S2 at JXB online). Thus, altering the expression of a single expansin gene brought about a marked change in root length, which suggests that expansin proteins have a significant impact on cell-wall loosening and elasticity that are important during root development.
Fig. 4.
Root architecture of EXPA5-overexpression transgenic plants. (A) Relative expression level of EXPA5 in five transgenic lines. RNA samples isolated from 10-d-old roots of mutants and wild type were analysed by real-time RT-PCR using specific primers. Values are means±standard deviation (SD) (n=3). Statistically significant differences are indicated (***P<0.001). (B) The primary root of 5-d-old mutant seedlings (35S::EXPA5) was longer than that of the wild-type seedlings of the same age on MS medium. Bar, 1cm. (C) The primary root length of plants shown in (B). Presented are means±SD (n=3, >50 seeds per replicate experiment). Statistically significant differences are indicated (**P<0.05; ***P<0.001). (D) Primary root elongation curve of the wild-type and mutant seedlings grown on MS medium. (E) Length of cells from the elongation zone of the wild type and one of the transgenic lines. (This figure is available in colour at JXB online.)
Combined overexpression of EXPA5 with either PMEI or CEL generates a synergistic effect on primary root elongation, while overexpression of a PMEI or CEL gene alone does not
Having demonstrated that overexpression of EXPA5 could make primary roots elongate faster, we wondered if overexpression of other cell-wall-loosening protein genes could also have similar effects. Therefore, we tested one CEL gene (At2g32990) and one PMEI gene (At5g62360) and found that neither transgenic line had a primary root longer than the wild-type control (Supplementary Fig. S3 at JXB online). These results suggested that some of the cell-wall-loosening proteins may not function alone in root development and that some coordination among those proteins is probably required for their optimal function.
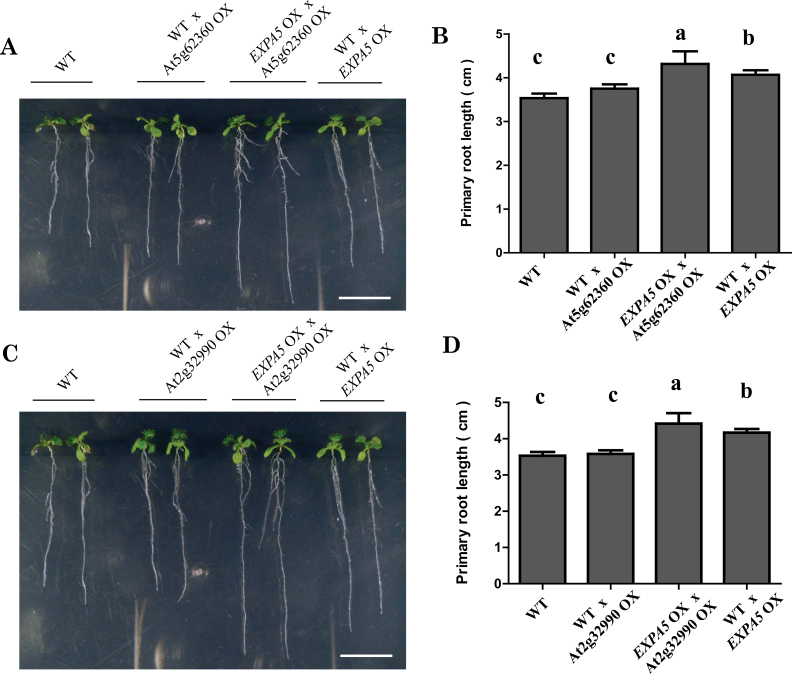
Cell-wall loosening is a strictly controlled progress that needs many proteins to coordinate properly. To test this idea, we crossed the EXPA5-overexpressing line with a PMEI- or CLE-overexpressing line. Interestingly, F1 plants overexpressing EXPA5 plus either PMEI or CLE produced primary roots that were significantly longer than those of either parents (Fig. 5). These results demonstrated that when either PMEI or CLE combined with EXPA5, the primary root elongation was promoted and the primary root length exceeded the EXPA5-overexpressing line, suggesting that cooperation may exist among the cell-wall-loosening proteins in regulating cell-wall elasticity at least between EXPA5 and PMEI or CLE proteins.
Fig. 5.
Root architecture of PMEI- and EXPA5- or CEL- and EXPA5-overexpression transgenic plants. (A)The primary root of 8-d-old wild-type seedlings and F1 progeny of the cross between the wild type (♀)×PMEI-overexpressing line (OX) (♂), EXPA5 OX (♀)×PMEI OX, and the wild type (♀)×EXPA5 OX (♂) on MS medium. (B) The primary root length of plants shown in (A). Presented are means±SD (n=3, >50 seeds per replicate experiment). Different letters indicate significant differences using SPSS analysis. (C)The primary root of 8-d-old wild-type seedlings and F1 progeny of the cross between the wild type (♀)×CEL OX (♂), EXPA5 OX (♀)×CEL OX, and wild type (♀)×EXPA5 OX (♂) on MS medium. (D) The primary root length of the plants shown in (C). Presented are means±SD (n=3, >50 seeds per replicate experiment). Different letter indicate significant differences using SPSS analysis. (This figure is available in colour at JXB online.)
Discussion
To uncover the molecular mechanisms underlying the well-developed root system of the edt1 mutant, we compared the root transcript profiles of edt1 with that of the wild type and found that a series of cell-wall-loosening protein genes were upregulated in edt1. A promoter scan of these genes for cis-elements revealed that most of the cell-wall-loosening protein genes contained at least one HD-binding cis-element in their promoter. The aaattaaa sequence was the dominant cis-element (Table 4), and showed high affinity to the HDG11 protein in a Y1H assay (Fig. 3A). By Y1H and ChIP-PCR assays, we showed that HDG11 could bind to the promoter segments with an HD-binding cis-element of these cell-wall-loosening protein genes. Based on these results, it is clear that HDG11 can directly regulate cell-wall-loosening protein genes and hence alter cell-wall extensibility of roots, which at least partially contributes to the well-developed root system of edt1 mutant.
Plant growth and development need to break through the limit of the plant cell wall. The regulation of cell-wall extensibility is a necessary process accompanying cell expansion and division. In fast-growing tissues such as roots, loosening of the cell wall is a well-regulated process and is coordinated with development. Enhanced cell-wall loosening is also a response of roots to water deficit (Hsiao and Xu, 2000; Sharp et al., 2004). Our results with the edt1 mutant further demonstrate that cell-wall loosening is important to plant development and growth.
The cell-wall-loosening process is regulated by many factors including hormones and environmental cues and needs a set of proteins to cooperate in an orderly and closely regulated (Cosgrove, 2005). The results of our microarray analysis showed that a series of cell-wall-loosening protein genes are upregulated in edt1 roots, including expansins, XTHs, pectin lyases, cellulases, PMEIs, and extensins, which cover almost all the cell-wall-loosening protein gene families. Interestingly, the cell-wall-loosening protein genes were coordinately upregulated in edt1 roots, which was made possible by the HD-binding cis-elements in their promoters and activated expression of EDT1/HDG11. Moreover, the positive role of brassinosteroids in plant growth is performed partly by activating cell-wall-loosening protein genes (Nemhauser et al., 2004; Bai et al., 2012). Such coordinated upregulation may be a general mechanism in root development.
Among the cell-wall loosening protein genes, the expansin gene family has important roles in cell-wall loosening (Cosgrove et al., 2002). For instance, cell-wall modification is required for root-hair growth. The root-hair-specific expansin A family is required for root-hair elongation in rice. Loss of EXPA17 (OsEXPA17) produces short root hairs (ZhiMing et al., 2011). Lateral root formation is a major determinant of the root-system architecture and defines the capability of a plant to acquire water and nutrients from soil. During Arabidopsis lateral root emergence, EXPANSIN14 (EXP14) is directly activated by the LATERAL ORGAN BOUNDARIES DOMAIN 18 (LBD18) protein, a transcriptional activator playing important roles in lateral organ development (Lee et al., 2013). In the edt1 mutant, EXPA5 was dramatically upregulated in roots. When overexpressed, it improved root elongation (Fig. 4). Nevertheless, when there was overexpression of only the PMEI or CEL gene, root elongation was not significantly affected (Supplementary Fig. S3). However, when EXAP5 was co-overexpressed with these two cell-wall-loosening genes, the roots grew faster (Fig. 5). Although we did not test all of the cell-wall-loosening genes, from the microarray analysis and our experiment data, we speculate that the process of cell-wall loosening needs many proteins to cooperate closely. It has been reported that overexpression of one PMEI gene can increase root length, such as PMEI-1 (At1g48020) or PMEI-2 (At3g17220) (Lionetti et al., 2007). The PMEI (At5g62360) that we overexpressed has relatively low protein identity to these two PMEIs, which may be why overexpression of At5g62360 did not stimulate root elongation. Perhaps this PMEI needs to cooperate with other cell-wall-loosening proteins to play its role in root growth. If different combinations of cell-wall-loosening proteins generate different cell-wall modifications, this would increase the flexibility of the cell wall to meet the various demands of development in plants.
In conclusion, our study with the edt1 mutant demonstrates that the homeodomain transcription factor EDT1/HDG11 is able to directly and coordinately upregulate several gene families of cell-wall-loosening proteins, leading to the altered cell-wall extensibility required in root development. In addition, our results suggest that expansin may work in combination with PMEI or CEL to generate synergistic effects on cell-wall extension, which provides a mechanism for plants to adjust their cell-wall physical properties.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. EXPA5 (A) and EXPB3 (B) expression pattern by AtGenExpress Visualization Tool (AVT).
Supplementary Fig. S2. The cells in the elongation zone of the EXPA5-overexpressing line are longer than that of the wild type.
Supplementary Fig. S3. The root architecture of PMEI (At5g62360)- or CEL (At2g32990)-overexpressing transgenic plants.
Supplementary Table S1. The 72 cell-wall related genes upregulated in edt1 mutant roots.
Supplementary Table S2. The upregulated cell-wall-loosening protein genes with HD-binding cis-elements in their promoter and the ratio in each category.
Supplementary Table S3. Primer sequences for semi-quantitative PCR.
Supplementary Table S4. Primer sequences for ChIP-PCR.
Acknowledgements
This study was supported by grants from the National Nature Science Foundation of China (30830075 and 90917004), the Chinese Academy of Science (KSCX3-YW-N-007), and the Ministry of Science and Technology of China (2012CB114304 and 2011ZX08005-004).
Glossary
Abbreviations:
- CEL
cellulase
- ChIP
chromatin immunoprecipitation
- EXP
expansin
- GO
Gene Ontology
- MS
Murashige and Skoog
- PMEI
pectin methylesterase inhibitor
- RT-PCR
reverse transcription-PCR
- SD
standard deviation
- SEG
significantly expressed gene
- XTH
xyloglucan endotransglucosylase/hydrolase
- Y1H
yeast one-hybrid.
References
- Aiken RM, Smucker AJ. 1996. Root system regulation of whole plant growth. Annual Review of Phytopathology 34, 325–346 [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. 2007. The true story of the HD-Zip family. Trends in Plant Science 12, 419–426 [DOI] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY. 2012. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24, 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, et al. 2012. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 [DOI] [PubMed] [Google Scholar]
- Brux A, Liu TY, Krebs M, Stierhof YD, Lohmann JU, Miersch O, Wasternack C, Schumacher K. 2008. Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth Inhibition in Arabidopsis. Plant Cell 20, 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. 1997. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9, 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. 2002. The growing world of expansins. Plant and Cell Physiology 43, 1436–1444 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6, 850–861 [DOI] [PubMed] [Google Scholar]
- de Dorlodot S, Forster B, Pages L, Price A, Tuberosa R, Draye X. 2007. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends in Plant Science 12, 474–481 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, et al. 2012. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Current Biology 22, 1699–1704 [DOI] [PubMed] [Google Scholar]
- Derbyshire P, McCann MC, Roberts K. 2007. Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biology 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbak A, Yao H, McSteen P. 2012. Hormone signaling in plant development. Current Opinion in Plant Biology 15, 92–96 [DOI] [PubMed] [Google Scholar]
- Fry SC. 1998. Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochemical Journal 332, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhao J, Li X, Qin L, Yan X, Liao H. 2011. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. The Plant Journal 66, 541–552 [DOI] [PubMed] [Google Scholar]
- Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo S, Sato K, Yokoyama R, Nishitani K. 2012. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 24, 2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao TC, Xu LK. 2000. Sensitivity of growth of roots versus leaves to water stress: biophysical analysis and relation to water transport. Journal of Experimental Botany 51, 1595–1616 [DOI] [PubMed] [Google Scholar]
- Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, Matsumoto H, Komatsu S. 2004. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiology 136, 3670–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. 2013. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. The Plant Journal 73, 212–224 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z-Y, Zhao P, Cao M-J, Cui R, Chen X, Xiong L-Z, Zhang Q-F, Oliver DJ, Xiang C-B. 2007. High-throughput binary vectors for plant gene function analysis. Journal of Integrative Plant Biology 49, 556–567 [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. 2007. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea . Plant Physiology 143, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszkay A, Kenk B, Schopfer P. 2003. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 217, 658–667 [DOI] [PubMed] [Google Scholar]
- Liu YB, Lu SM, Zhang JF, Liu S, Lu YT. 2007. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226, 1547–1560 [DOI] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K. 2009. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. Journal of Experimental Botany 60, 3959–3972 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20, 1138–1143 [DOI] [PubMed] [Google Scholar]
- Muller K, Linkies A, Vreeburg RA, Fry SC, Krieger-Liszkay A, Leubner-Metzger G. 2009. In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiology 150, 1855–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology 2, E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. 2006. A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research 119, 153–162 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Hofte H. 2011. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Current Biology 21, 1720–1726 [DOI] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S. 2012. Growth and development of the root apical meristem. Current Opinion in Plant Biology 15, 17–23 [DOI] [PubMed] [Google Scholar]
- Schopfer P. 2001. Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. The Plant Journal 28, 679–688 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A, Bechtold M, Frahry G, Wagner A. 2002. Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta 214, 821–828 [DOI] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. 2004. Root growth maintenance during water deficits: physiology to functional genomics. Journal of Experimental Botany 55, 2343–2351 [DOI] [PubMed] [Google Scholar]
- Siedlecka A, Wiklund S, Peronne MA, Micheli F, Lesniewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ. 2008. Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiology 146, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. 2009. The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. The Plant Journal 60, 564–574 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K. 2005. Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant and Cell Physiology 46, 192–200 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen JP. 2003. Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays . Journal of Experimental Botany 54, 335–344 [DOI] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. 2007. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. The Plant Journal 52, 716–729 [DOI] [PubMed] [Google Scholar]
- Won SK, Choi SB, Kumari S, Cho M, Lee SH, Cho HT. 2010. Root hair-specific EXPANSIN B genes have been selected for Graminaceae root hairs. Molecules and Cells 30, 369–376 [DOI] [PubMed] [Google Scholar]
- Yang Z, Peng Z, Wei S, Yu Y, Cai P. 2011. Identification of differentially expressed genes in three-pistil mutation in wheat using annealing control primer system. Gene 485, 81–84 [DOI] [PubMed] [Google Scholar]
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ. 2006. Crystal structure and activities of EXPB1 (Zea m 1), a β-expansin and group-1 pollen allergen from maize. Proceedings of the National Academy of Sciences, USA 103, 14664–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. 2001. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology 42, 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Uwagaki Y, Sasaki H, Harada T, Hiwatashi Y, Hasebe M, Nishitani K. 2010. Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens . The Plant Journal 64, 645–656 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Imaizumi N, Kaneko S, Kawagoe Y, Tagiri A, Tanaka H, Nishitani K, Komae K. 2006. Carbohydrate-binding module of a rice endo-β-1,4-glycanase, OsCel9A, expressed in auxin-induced lateral root primordia, is post-translationally truncated. Plant and Cell Physiology 47, 1555–1571 [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. 2008. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. 2013. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiology 162, 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MR, Han YY, Feng YN, Li F, Wang W. 2012. Expansins are involved in cell growth mediated by abscisic acid and indole-3-acetic acid under drought stress in wheat. Plant Cell Reports 31, 671–685 [DOI] [PubMed] [Google Scholar]
- ZhiMing Y, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, Ming C, Hyung-Taeg C, Ping W. 2011. Root hair-specific expansins modulate root hair elongation in rice. The Plant Journal 66, 725–734 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen S, Alvarez S, Asirvatham VS, Schachtman DP, Wu Y, Sharp RE. 2006. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiology 140, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.