Summary
Chemical and transgenic approaches can activate ABA signalling via crop PYR/PYL ABA receptors; quinabactin can selectively activate tomato ABA receptors, and overexpression of monomeric-type receptors confers enhanced plant drought resistance.
Key words: Abscisic acid (ABA), drought resistance, tomato ABA receptor, tomato clade A PP2C.
Abstract
Abscisic acid (ABA) plays a crucial role in the plant’s response to both biotic and abiotic stress. Sustainable production of food faces several key challenges, particularly the generation of new varieties with improved water use efficiency and drought tolerance. Different studies have shown the potential applications of Arabidopsis PYR/PYL/RCAR ABA receptors to enhance plant drought resistance. Consequently the functional characterization of orthologous genes in crops holds promise for agriculture. The full set of tomato (Solanum lycopersicum) PYR/PYL/RCAR ABA receptors have been identified here. From the 15 putative tomato ABA receptors, 14 of them could be grouped in three subfamilies that correlated well with corresponding Arabidopsis subfamilies. High levels of expression of PYR/PYL/RCAR genes was found in tomato root, and some genes showed predominant expression in leaf and fruit tissues. Functional characterization of tomato receptors was performed through interaction assays with Arabidopsis and tomato clade A protein phosphatase type 2Cs (PP2Cs) as well as phosphatase inhibition studies. Tomato receptors were able to inhibit the activity of clade A PP2Cs differentially in an ABA-dependent manner, and at least three receptors were sensitive to the ABA agonist quinabactin, which inhibited tomato seed germination. Indeed, the chemical activation of ABA signalling induced by quinabactin was able to activate stress-responsive genes. Both dimeric and monomeric tomato receptors were functional in Arabidopsis plant cells, but only overexpression of monomeric-type receptors conferred enhanced drought resistance. In summary, gene expression analyses, and chemical and transgenic approaches revealed distinct properties of tomato PYR/PYL/RCAR ABA receptors that might have biotechnological implications.
Introduction
Drought, high salinity, and cold have adverse effects on plant growth and seed production. Abscisic acid (ABA)-induced changes play a central role among the various biochemical and physiological processes required to acquire abiotic stress tolerance (Verslues et al., 2006). Thus, in order to maintain water, ABA promotes stomatal closure through the control of membrane transport systems (Osakabe et al., 2014). On the other hand, shoot growth is inhibited whereas the root growth rate is maintained to gain access to water (Sharp et al., 2004; Des Marais et al., 2012). Gene expression is widely regulated by ABA and, as a result, genes encoding proteins involved in protection and damage repair are up-regulated, such as late embryogenesis abunant (LEA)/dehydrins, reactive oxygen species (ROS) scavengers, or osmolyte biosynthetic enzymes (Verslues et al., 2006). Extensive knowledge of ABA perception and signal transduction has emerged in recent years in Arabidopsis thaliana (Cutler et al., 2010). PYR/PYL/RCAR (PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS) receptors perceive ABA intracellularly and, as a result, form ternary complexes with clade A protein phosphatase type 2Cs (PP2Cs), thereby inactivating them (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009a ; Umezawa et al., 2009; Vlad et al., 2009; Nishimura et al., 2010). This allows the activation of downstream targets of the PP2Cs, such as the sucrose non-fermenting 1-related subfamily 2 (SnRK2s) protein kinases, namely SnRK2.2/D, 2.3/I, and 2.6/OST1/E, which are key players in the regulation of the transcriptional response to ABA and stomatal aperture (Cutler et al., 2010; Finkelstein, 2013). PP2Cs also dephosphorylate other classes of kinases or kinase-regulated proteins (Finkelstein, 2013; Rodrigues et al., 2013). According to the oligomeric nature of the apo receptors, they can be classified in two major classes: dimeric (PYR1 and PYL1–PYL3) or monomeric (PYL4–PYL10, except the untested PYL7) (Dupeux et al., 2011a ; Hao et al., 2011). Upon ligand binding, dimeric receptors dissociate to make available the PP2C interaction surface (Dupeux et al., 2011a ). In particular, PYL3 suffers a severe cis- to trans-dimer transition by a protomer rotation of 135 º to facilitate subsequent dissociation (Zhang et al., 2012). Monomeric receptors are able to interact with PP2Cs in the absence of ABA; however, the presence of ABA is required for a major inhibition of PP2Cs when protection of phosphorylated protein substrates by ABA receptors is evaluated (Antoni et al., 2012; Pizzio et al., 2013).
Several biochemical and structural studies of apo/ABA-bound PYLs and ternary receptor–ABA–phosphatase complexes have provided the molecular details of the ABA perception and signalling mechanism (Melcher et al., 2009; Miyazono et al., 2009; Santiago et al., 2009b ; Nishimura et al., 2009; Yin et al., 2009). Two loops located between the β3–β4 and β5–β6 sheets control the access of the ABA molecule to the ABA-binding pocket. These loops generate an open conformation of the ligand-binding pocket in the apo receptors. In response to ABA, conformational changes occur in these loops that serve as a gate and latch to stabilize the closed conformation of the ABA-bound receptor. A conserved serine residue flips out of the β3–β4 loop and inserts into the phosphatase catalytic site, blocking access of potential substrates. The mechanism of PP2C inhibition is further explained by the structure of the ternary receptor–ABA–phosphatase complex. In particular, a conserved tryptophan residue located in a β-hairpin of PP2C establishes contact with the gate and latch loops and indirectly with ABA’s ketone group through a hydrogen bond mediated by a critical water molecule (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009; Dupeux et al., 2011b). As a result of this gate–latch–lock mechanism and PP2C interaction, the ternary complex shows high affinity for ABA binding (K d between 20nM and 40nM) (Ma et al., 2009; Santiago et al., 2009a).
Different works have shown the potential applications of Arabidopsis PYR/PYL receptors to enhance plant drought resistance, through either genetic engineering or chemical approaches (Santiago et al., 2009a; Saavedra et al., 2010; Mosquna et al., 2011; Cao et al., 2013; Okamoto et al., 2013; Pizzio et al., 2013). Thus, through overexpression of either wild-type, constitutively active receptors or mutated versions that enhance ABA-dependent inhibition of PP2Cs, enhanced drought resistance could be conferred to Arabidopsis plants (Santiago et al., 2009a; Saavedra et al., 2010; Mosquna et al., 2011; Pizzio et al., 2013). On the other hand, chemicals acting as ABA agonists have proved to be effective for similar purposes (Cao et al., 2013; Okamoto et al., 2013). Given the physiological and practical implications of the ABA signalling pathway in agriculture, it is expected that similar approaches could be implemented in crops. Identification of core components of the ABA signalling pathway taking advantage of the knowledge generated in Arabidopsis is now possible (Ben-Ari, 2012). Thus, since the discovery of the PYR/PYL/RCAR ABA receptor family in Arabidopsis, several reports have described orthologous genes in commercial crops, such as tomato (Sun et al., 2011), strawberry (Chai et al., 2011), rice (Kim et al., 2012), grape (Boneh et al., 2012), sweet orange (Romero et al., 2012), and soybean (Bai et al., 2013). Recently, overexpression of OsPYL5 in the monocot rice was shown to confer enhanced drought tolerance (Kim et al., 2014). In this work, a comprehensive identification of putative PYR/PYL/RCAR receptors was carried out in the dicot tomato and different studies were performed to validate their function and to show that they encode functional ABA receptors in plant cells. As a result, it was found that they inhibited tomato clade A PP2Cs in an ABA-dependent manner and some of them could be activated by the ABA agonist quinabactin (QB), which induced tomato stress-responsive genes. The overexpression in Arabidopsis of two tomato receptors from the monomeric subgroups AtPY4-6 and AtPYL7-10 conferred enhanced drought resistance, whereas overexpression of a tomato dimeric receptor from the subgroup AtPYL1 failed to confer this phenotype.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana and Solanum lycopersicum (cv. Moneymaker) plants were routinely grown under greenhouse conditions (40–50% relative humidity) in pots containing a 1:3 vermiculite–soil mixture. For Arabidopsis plants grown under growth chamber conditions, seeds were surface sterilized by treatment with 70% ethanol for 20min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10min, and, finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4 ºC for 3 d. Then, seeds were sown on Murashige and Skoog (MS) plates composed of MS basal salts, 0.1% 2-[N-morpholino]ethanesulphonic acid, 1% sucrose, and 1% agar. The pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber at 22 ºC under a 16h light, 8h dark photoperiod at 80–100 μE m–2 s–1. Tomato seeds were surface sterilized by treatment with commercial bleach (2.5% sodium hypochlorite) containing 0.05% Tween-20 for 30min and four washes with sterile distilled water. MS plates for tomato seeds contained 0.5× MS salts.
Microarray analysis
Fruits at breaker stage were harvested from S. lycopersicum (‘Moneymaker’ and ‘Microtom’) and S. pimpinellifolium (‘TO-937’) plants. Pericarp and epidermis were excised manually with a sterile scalpel, frozen, and ground with liquid nitrogen to a fine powder. At least three biologically replicated samples for RNA isolation were prepared from each genotype and tissue from three or more pooled fruits. RNA was extracted from pericarp with the modified cetyltrimethylammonium bromide (CTAB) method (Powell et al., 2012) and from epidermis with the Trizol Reagent (Invitrogen). The RNA clean up protocol was done with the RNA Plant Mini Kit (Qiagen). The RNA pellet was resuspended in nuclease-free water. Samples of total RNA were checked for integrity and quality using an Agilent Bioanalyzer (Agilent Technologies). The three biologically replicated RNA samples were amplified, labelled, and hybridized to the 34K gene EUTOM3 Exon array (https://bioinformatics.psb.ugent.be/gdb/solanum) according to the manufacturer’s instructions (Affymetrix) at Unitat Central d’Investigació (Universitat de Valencia, Spain) as described in Powell et al. (2012). Data were pre-processed and analysed using Partek Genomic Suite software v6.6 (Partek Inc.) with the probes matching only once with the ITAG annotation 2.30. The configuration consisted of a pre-background adjustment for GC content, robust multiarray analysis for background correction, quantile normalization, and probe set summarization using median polishing (Irizarry et al., 2003). Library files were eutom3gene_v2_ucprobes.cdf and the annotation file version was eutom3-annotation-per-scaffold-modif.txt which represents 30 000 tomato genes.
Yeast two hybrid (Y2H) assays
The full-length coding sequences of Sl08g076960, Sl06g061180, Sl09g015380, Sl06g050500, Sl03g095780, Sl12g055990, and Sl03g007310 ABA receptors as well as Sl05g052980 PP2C were amplified by PCR from tomato leaf/fruit cDNA and cloned into the pCR8/GW/TOPO entry vector (Invitrogen). An N-terminal deleted version (ΔN) of Sl12g096020 PP2C was amplified from tomato leaf/fruit cDNA using primers that amplify the catalytic PP2C core (amino acid residues 178–509, ΔN Sl12g096020). All the primers used in this work are listed in Supplementary Table S1 available at JXB online. Appropriate restriction sites were introduced in some primers to allow the subsequent cloning steps, and all constructs were verified by DNA sequencing. Tomato ABA receptors were fused by Gateway recombination to the GAL4 DNA-binding domain (GBD) in pGBKT7GW. As preys, a set of Arabidopsis clade A PP2Cs fused to the GAL4 activation domain (GAD) in the pGADT7 vector was used (Lackman et al., 2011; Antoni et al., 2012). Tomato Sl05g052980 and ΔN Sl12g096020 PP2Cs were fused to the GAD in the pGADT7GW vector. Protocols for Y2H assays were similar to those described previously (Saez et al., 2008).
Purification of recombinant proteins
Sl06g050500, Sl03g095780, Sl12g055990, and Sl03g007310 coding sequences were cloned in pCR8/GW/TOPO, excised using NcoI/EcoRI double digestion, and subcloned into pETM11. Sl09g015380, Sl08g076960, and Sl06g061180 coding sequences have either EcoRI or NcoI internal restriction sites and were subcloned using a different strategy. Coding sequences of Sl08g076960 and Sl09g015380 were excised using NcoI/HindIII and NcoI/BamHI double digestion, respectively, and subcloned into pETM11, whereas Sl06g061180 was excised using EcoRI digestion and subcloned into pET28a. Escherichia coli BL21 (DE3) cells transformed with the corresponding pET28a/pETM11 construct were grown in 50ml of Luria–Bertani medium supplemented with 50 μg ml–1 kanamycin to an optical density at 600nm of 0.6–0.8. Then, 1mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cells were harvested 3h after induction and stored at –80 ºC before purification. The pellet was resuspended in 2ml of HIS buffer (50mM TRIS-HCl, pH 7.6, 250mM KCl, 10% glycerol, 0.1% Tween-20, and 10mM mercaptoethanol), and the cells were sonicated in a Branson sonifier. A cleared lysate was obtained after centrifugation at 14 000 g for 15min, and it was diluted with 2 vols of HIS buffer. The protein extract was applied to a 0.5ml nickel–nitrilotriacetic acid (Ni-NTA) agarose column, and the column was washed with 10ml of HIS buffer supplemented with 20% glycerol and 30mM imidazole. Bound protein was eluted with HIS buffer supplemented with 20% glycerol and 250mM imidazole.
In order to obtain enough protein for size exclusion chromatography (SEC) analysis, 8ml of an overnight culture were subcultured into 800ml of fresh 2TY broth (16g of Bacto tryptone, 10g of yeast extract, 5g of NaCl per litre of solution) plus kanamycin (50 μg ml–1). Protein expression was induced with 0.3mM IPTG, and the cells were harvested after overnight incubation at 16 ºC. Pellets were resuspended in 25mM TRIS-HCl pH 8.0, 200mM NaCl, 50mM imidazole, 5mM β-mercaptoethanol, and disrupted by sonication. After centrifugation for 40min at 40 000 g, the clear supernatant was filtered (pore diameter 0.45 μm; Millipore Corporation, Bedford, MA, USA). The 6His-tagged proteins were purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. Proteins were eluted with the following elution buffer: 25mM TRIS-HCl pH 8.0, 200mM NaCl, 500mM imidazole, 5mM β-mercaptoethanol, and cleaved with Tobacco etch virus (TEV) protease (1:100). Proteins 8g076960, 6g061180, and 6g050500 were concentrated to 10mg ml–1 and 12g055990 was concentrated to 0.7mg ml–1. Finally, each purified protein was subjected to gel filtration using a prep grade Superdex200 10/30 (Amersham Biosciences Limited, UK) previously equilibrated with 25mM TRIS-HCl pH 8.0, 200mM NaCl, 5mM β-mercaptoethanol. Approximately 1mg of 8g076960, 6g061180, or 6g050500 was loaded onto the column, whereas 12g055990 was difficult to solubilize and only 0.1mg was loaded.
PP2C activity assays
Phosphatase activity was measured using the RRA(phosphoT)VA peptide as substrate, which has a K m of 0.5–1 μM for eukaryotic PP2Cs (Donella et al., 1990). Assays were performed in a 100 μl reaction volume containing 25mM TRIS-HCl, pH 7.5, 10mM MgCl2, 1mM dithiothreitol (DTT), 25 μM peptide substrate, and 0.5 μM PP2C. When indicated, PYR/PYL/RCAR recombinant proteins and ABA or QB (Life Chemicals) were included in the PP2C activity assay. ABA and QB concentrations were 0.1, 0.5, 1, 5, 10, and 50 μM. After incubation for 60min at 30 ºC, the reaction was stopped by the addition of 30 μl of molybdate dye (Baykov et al., 1988), and the absorbance was read at 630nm with a 96-well plate reader. Appropriate controls including dimethylsulphoxide (DMSO) and buffer HIS were included.
ABA and QB treatment of tomato seedlings
Ten-day-old tomato seedlings (cv. Moneymaker) were mock treated or treated with 10 μM ABA or QB for 3h. Total RNA was extracted using a NucleoSpin RNA plant kit. Synthesis of cDNA and quantitaive real-time PCR (qRT-PCR) analyses were performed as described (Saez et al., 2006). Amplification of the ABA- and stress-responsive Sl02g084850, Sl06g067980, and Sl06g019170 genes was done using the primers described in Supplementary Table S1 at JXB online. Expression was normalized using the values obtained with Sl06g009970 (SlEF1a).
Generation of transgenic lines
Sl06g050500, Sl03g007310, and Sl08g076960 coding sequences in the pCR8/GW/TOPO entry clone were recombined by LR reaction into the Gateway-compatible ALLIGATOR2 vector (Bensmihen et al., 2004). The ALLIGATOR2 vector drives expression of the recombined gene under control of the Cauliflower mosaic virus (CaMV) 35S promoter and introduces a triple haemagglutinin (HA) epitope at the N-terminus of the encoded protein. Selection of transgenic lines is based on the visualization of green fluorescent protein (GFP) in seeds, whose expression is driven by the specific seed promoter At2S3. The ALLIGATOR2 constructs were transferred to Agrobacterium tumefaciens C58C1 (pGV2260) (Deblaere et al., 1985) by electroporation and used to transform Columbia wild type by the floral dip method. T1 transgenic seeds were selected based on GFP visualization and sown in soil to obtain the T2 generation. Homozygous T3 progeny was used for further studies, and expression of HA-tagged protein was verified by immunoblot analysis using anti-HA-peroxidase (Roche).
Seed germination assays
After surface sterilization of the tomato seeds, ~100 seeds were sown on 0.5× MS plates lacking (control plates) or supplemented with either 1 μM or 10 μM ABA or QB. Seeds were germinated in the dark at 23 ºC for 3 d. In order to score seed germination, radical emergence was analysed at 72h after sowing. Since QB was dissolved in DMSO, control MS plates for QB experiments were supplemented with 0.1% DMSO.
Root growth assays
Arabidopsis seedlings were grown on vertically oriented MS plates for 3 d. Afterwards, 20 plants were transferred to new MS plates lacking or supplemented with the indicated concentrations of ABA. The plates were scanned on a flatbed scanner after 10 d to produce image files suitable for quantitative analysis of root growth using the NIH Image software ImageJ v1.37.
Drought stress and water status measurement
Plants grown under greenhouse conditions (10 individuals per experiment, three independent experiments) were grown under normal watering conditions for 15 d and then subjected to drought stress by stopping irrigation for 20 d. Next, watering was resumed and the survival rate was calculated after 3 d by counting the percentage of plants that had more than four green leaves. Photographs were taken at the start of the experiment (day 0), after 16 d and 20 d of drought, and 3 d after re-watering. The relative water content (RWC) of the plants was measured in rosette leaves at 11, 14, and 17 d. Samples of 10 leaves from five plants were collected and their fresh weight (FW) was obtained. Leaves were then floated for 3h on demineralized water and weighed again in order to obtain their turgid weight (TW). Finally, leaves were dried for 16h at 70 °C and weighed to obtain the dry weight (DW). The RWC was calculated as (FW–DW/TW–DW)×100 and each measurement was made in triplicate.
Results
The tomato genome encodes 15 putative PYR/PYL/RCAR ABA receptors
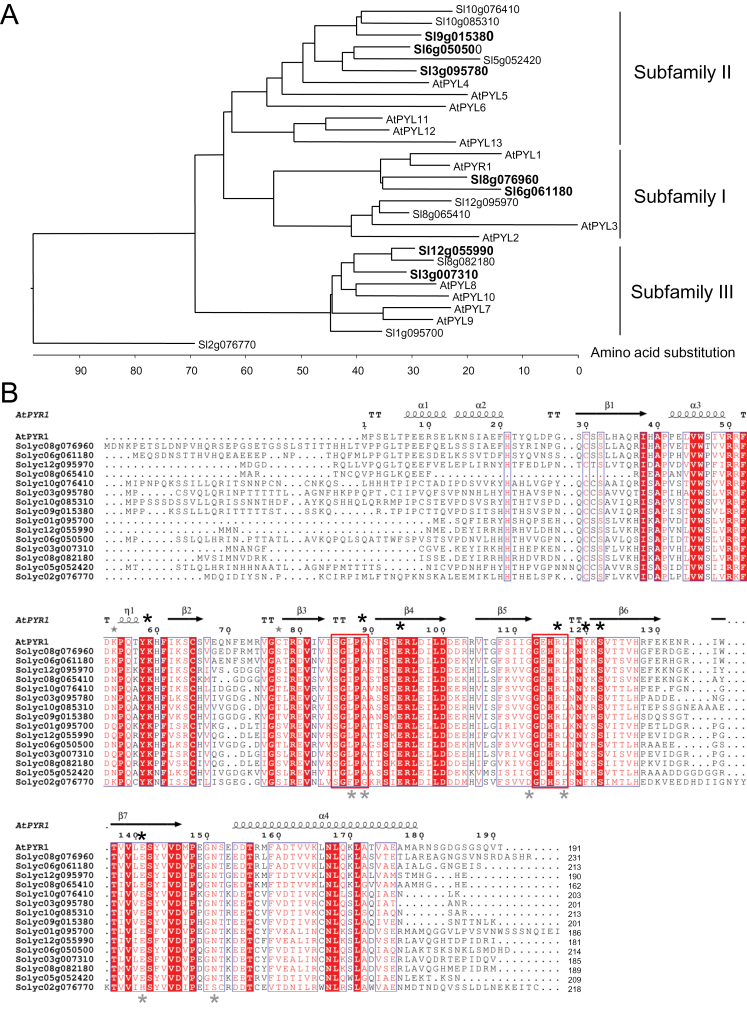
A partial analysis of the tomato PYR/PYL/RCAR family was published by Sun et al. (2011), leading to the discovery of eight receptors. The analysis has now been extended to the complete tomato genome (Tomato Genome Consortium, 2012) and, as a result, 15 receptors have been identified (Fig. 1A). With the exception of 2g076770, they were distributed in three subfamilies, which matched the corresponding groups from Arabidopsis PYR/PYL/RCAR receptors. Since it is possible that biochemical or physiological features already known in Arabidopsis receptors might be translated to crop receptors, an attempt was made to correlate tomato receptors with the corresponding groups in Arabidopsis (Fig. 1A). On this basis, a new nomenclature is proposed. Thus, in subfamily I, two tomato receptors, 8g076960 and 6g061180, closely related to Arabidopsis PYL1/PYR1 (Supplementary Fig. S1 at JXB online), and two other receptors, 12g095970 and 8g065410, more closely related to AtPYL2/PYL3, were found. In subfamily II, six tomato receptors related to AtPYL4/PYL5/PYL6 were found and in subfamily III four tomato receptors related to AtPYL7/PYL8/PYL9/PYL10 were found. No close relative for the Arabidopsis group AtPYL11/12/13 was found in tomato. Finally, the putative tomato receptor 2g076770 was ungrouped, probably because it lacked key conserved residues of the PYR/PYL/RCAR family, such as the conserved leucine of the β3–β4 and β5–β6 loops and the asparagine residue before the α4-helix (Fig. 1B). Additionally, 2g076770 gene expression could not be detected by RNA-Sequencing (RNA-Seq) analysis in vegetative or fruit tissue (see below Fig. 2); therefore, it remains to be established whether 2g076770 is a functional ABA receptor. In contrast, the key residues of both gate-like and latch-like loops were conserved in the remaining 14 tomato receptors (Fig. 1B).
Fig. 1.
Cladogram and amino acid sequence alignment of tomato PYR/PYL ABA receptors. (A) Cladogram of the multiple sequence alignment of tomato and Arabidopsis PYR/PYL receptors, indicating three major subfamilies and the ungrouped 2g076770. Those tomato receptors further described in the text are in bold face. (B) Sequence and secondary structure alignment of tomato PYR/PYL ABA receptors and Arabidopsis PYR1 protein. The predicted secondary structure of the tomato proteins was indicated, taking as a model the crystallographic structure of PYR1 (Protein DataBank Code 3K90) and using the Espript interface (http://espript.ibcp.fr/). Boxes indicate the position of the gate and latch loops. Black asterisks mark residues K59, A89, E94, R116, Y120, S122, and E141 of PYR1 involved in ABA binding. Grey asterisks mark conserved residues of the tomato receptor family that differ in tomato 2g076770 protein. (This figure is available in colour at JXB online.)
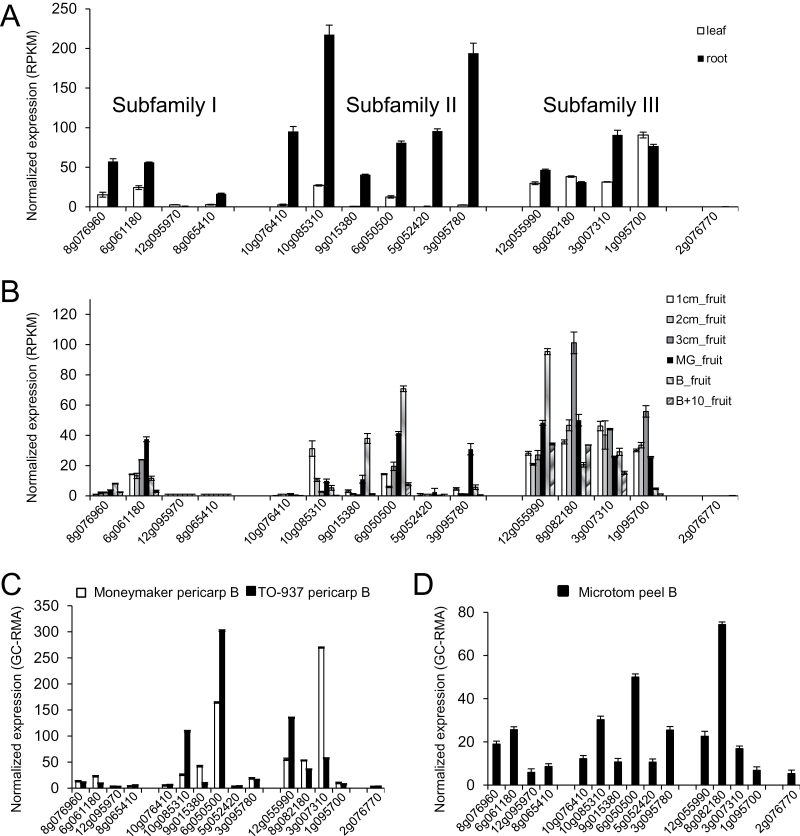
Fig. 2.
Relative gene expression of tomato ABA receptors in leaf, root, and fruit was determined by RNA-Seq and microarray analysis. (A, B) The transcriptome of the inbred tomato cultivar Heinz 1706 was analysed using Illumina RNA-Seq technology (Tomato genome Consortium, 2012). Data show gene transcription of tomato receptors grouped in three subfamilies and the ungrouped 2g076770 in leaf and root (A) and during fruit development and ripening (B). Significant expression of 2g076770 and 12g095970 was not detected in these tissues. RPKM, reads per kilobase of exon model per million mapped reads; MG, mature green stage; B, breaker stage. (C, D) Relative RNA abundance based on GC-RMA values (GC content–robust multiarray analysis) obtained from the Affymetrix exon tomato microarray (EUTOM3) hybridized with fruit pericarp RNA of Moneymaker and TO-937 (C) and fruit epidermis RNA of the Microtom background (D) at the breaker stage.
Differential gene expression of tomato ABA receptors in leaf, root, and fruit
Relative expression of all tomato genes has recently been reported using Illumina RNA-Seq technology, providing gene expression data for the transcriptome of the inbred tomato cultivar ‘Heinz 1706’ (Tomato Genome Consortium, 2012). Data mining was performed in the RNA-Seq transcriptome for the 15 tomato receptor genes in root, leaf, and six stages of fruit, and additionally a further microarray analysis was performed in other tomato accessions (Fig. 2). The expression pattern of ABA receptors in root and leaf indicates that some members clearly show a higher expression level compared with others (Fig. 2A). Two members from subfamily II, namely 10g085310 and 3g095780, showed the highest transcription level in root, whereas a member from subfamily III—1g095700—was the most transcribed receptor in leaf. In contrast, expression of some receptors was absent in these tissues. For instance, significant expression of 2g076770 and 12g095970 was not detected in either leaf or root (Fig. 2A). On the other hand, it was interesting to detect a high expression of several tomato ABA receptors in root tissue since ABA signalling is required at low water potentials to maintain primary root elongation, to increase root versus shoot biomass partitioning, to regulate root system architecture, and to promote root hydrotropism (Sharp et al., 2004; Des Marais et al., 2012; Duan et al., 2013). RNA-Seq data were also compiled from a tomato fruit development series composed of six fruit stages: 1cm, 2cm, 3cm, mature green, breaker (when colour becomes noticeable), and breaker +10 d. In general, members from subfamily III, for instance 12g055990 and 8g082180, showed high expression levels during fruit development, which suggests a role in this process (Fig. 2B). Additionally, recent studies suggest that ABA might be involved in regulating the onset of fruit ripening through triggering of ethylene biosynthesis and, consequently, ABA content peaks at breaker stages compared with the mature green stage (Zhang et al., 2009; Sun et al., 2011). Some tomato ABA receptors whose expression peaked during breaker stages were identified in subfamily II and III, such as 6g050500 and 12g055990; therefore they are candidate genes to regulate fruit ripening (Fig. 2B).
In order to obtain further data on fruit expression of tomato PYR/PYL genes, new studies were performed through microarray expression analysis in fruit pericarp at the breaker stage for cv. Moneymaker and S. pimpinellifolium accession TO-937 (Fig. 2C). Both 6g050500 and 12g055990 were among the top three most expressed genes at the breaker stage, although, depending on the cultivar considered, other tomato receptors also appeared to be highly expressed (for instance 3g007310 in Moneymaker and 10g085310 in TO-937). In any case, the comparison between Heinz, Moneymaker, and TO-937 indicates that 6g050500 was highly expressed in all three genetic backgrounds at the breaker stage, which suggests that it could be relevant in the regulation of tomato ripening. ABA receptors of subfamily III may also be important but in an accession-specific manner.
Fruit epidermis might be a putative target of ABA action to minimize fruit water loss, for instance through regulation of cuticle thickness or cuticle-dependent sensing of osmotic stress (Wang et al., 2011), and fruit peel also represents the point of entry for pathogen attack. Different genetic resources in the tomato Microtom background are available, including transgenic lines that modify fruit peel features (Shi et al., 2013). Therefore, it was of interest to obtain gene expression data for ABA receptors in fruit peel. Microarray expression data indicated different receptors from subfamily I, II, and III that were expressed at above average levels in fruit peel and, are, therefore, candidates to regulate ABA action at the fruit epidermis (e.g. 6g050500 and 8g082180; Fig. 2D).
Tomato ABA receptors interact with Arabidopsis and tomato clade A PP2Cs
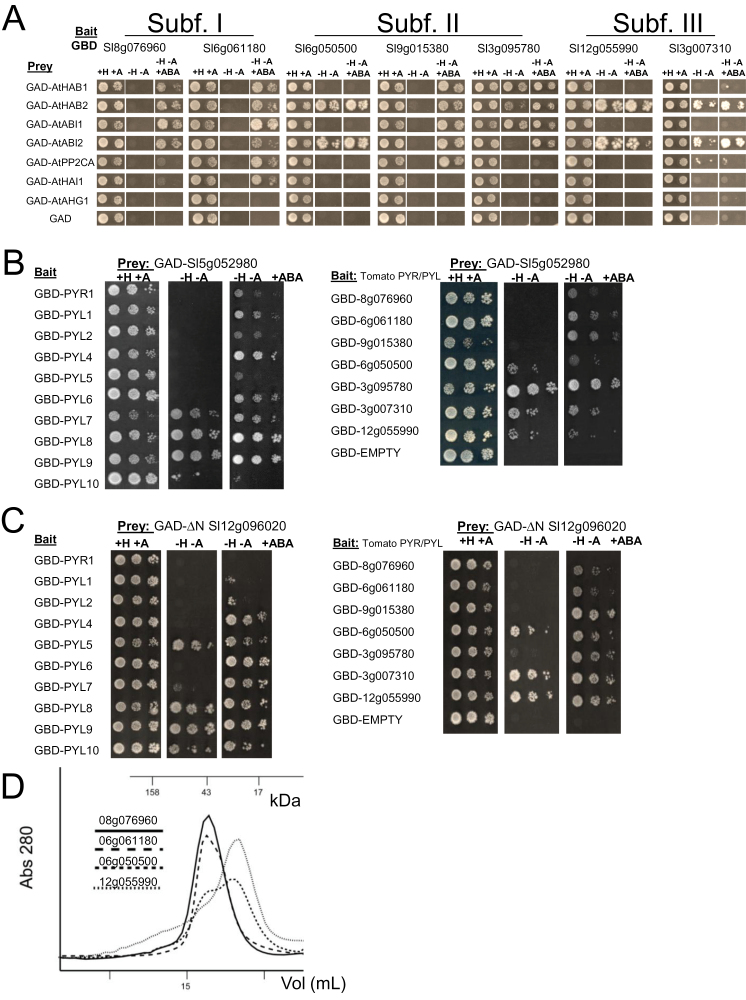
A key aspect of receptor function is its ability to interact and inhibit clade A PP2Cs. Both ABA-independent and ABA-dependent Y2H interactions among ABA receptors and PP2Cs have been reported in Arabidopsis, which probably reflect the monomeric/dimeric nature of the receptor as well as different K ds in yeast for particular receptor–phosphatase interactions (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009a ). ABA-independent interactions can be detected in Y2H assays; however, major inhibition of PP2C activity relies on the presence of ABA (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009a ). First of all, it was tested whether tomato PYR/PYLs were able to interact with Arabidopsis clade A PP2Cs. Tomato members were selected from the three subfamilies and both ABA-independent and ABA-dependent interactions with Arabidopsis PP2Cs were found (Fig. 3A). Interestingly, both 8g076960 and 6g061180, which are thought to be dimeric receptors according to their similarity to AtPYL1, showed ABA-dependent interactions with Arabidopsis PP2Cs (Fig. 3A). Dimeric receptors occlude their surface of interaction with PP2Cs and require ABA-induced dissociation to form ternary receptor monomer–ABA–phosphatase complexes (Dupeux et al., 2011a ). Three members from subfamily II were assayed and it was found that two of them showed ABA-independent interactions with some PP2Cs. The third member, 9g015380, is a close relative of AtPYL4 and it showed ABA-dependent interactions with PP2Cs, as was reported previously for AtPYL4 (Lackman et al., 2011). Finally, two members from subfamily III were assayed and ABA-independent interactions with AtHAB2 and AtABI2 were found. The Arabidopsis PP2C AHG1 is resistant to inhibition by ABA receptors because it lacks the conserved tryptophan residue required for formation of ternary complexes (Dupeux et al., 2011b ). Accordingly, this phosphatase did not interact with any tomato receptor, suggesting a similar tryptophan-dependent mechanism for formation of ternary complexes with tomato receptors. Finally, the PP2C HAI1, which shows a more restrictive pattern of interaction with Arabidopsis receptors, only interacted with one tomato receptor (Bhaskara et al., 2012).
Fig. 3.
Interactions between PYR/PYL ABA receptors and clade A PP2Cs. Interaction was determined by growth assay on media lacking histidine and adenine (–H, –A), which were supplemented or not with 50 μM ABA (+ABA). Dilutions (10–1, 10–2, and 10–3) of saturated cultures were spotted onto the plates. (A) Interaction of tomato receptors with Arabidopsis PP2Cs. (B) Interaction of a tomato PP2CA-like phosphatase (5g052980) and Arabidopsis (left) or tomato (right) PYR/PYL ABA receptors. (C) Interaction of a tomato ΔN HAB1-like phosphatase (12g096020) and Arabidopsis (left) or tomato (right) PYR/PYL ABA receptors. (D) Elution profiles after size-exclusion chromatography of four tomato ABA receptors in the absence of ABA. The lines show the absorbance recorded at 280nm. Molecular mass markers are indicated in kDa. (This figure is available in colour at JXB online.)
Next the cDNA of two tomato clade A PP2Cs, 5g052980 and 12g096020, which are close relatives of Arabidopsis PP2CA and HAB1, respectively, were cloned (Supplementary Fig. S2 at JXB online). The N-terminal deleted version (ΔN) of 12g096020 was used for both Y2H and activity assays, since the N-terminus is dispensable for interaction with ABA receptors (Santiago et al., 2009a ). 5g052980 and ΔN 12g096020 showed both ABA-dependent and ABA-independent interactions with Arabidopsis as well as tomato receptors (Fig. 3B, C). Therefore, a similar mechanism to that described in Arabidopsis for receptor–phosphatase interaction seems to operate in tomato. Finally, according to Y2H analyses and sequence similarity, it was predicted that tomato relatives of AtPYL1, such as 8g076960 and 6g061180, might be dimeric receptors whereas members of subfamilies II and III might be monomeric receptors. To test this prediction, SEC analysis was performed for four tomato receptors in the absence of ABA (Fig. 3D). As a result, it was found that 8g076960 and 6g061180 migrated with an estimated molecular mass of 48kDa, which corresponds to a dimeric receptor, and 12g055990 migrated with an estimated molecular mass of 27kDa, which corresponds to a monomeric receptor (predicted molecular masses are 25.6, 23.7, and 25.8kDa, respectively). However, 6g050500 displays a two-peak profile that corresponds to a distribution between monomeric and dimeric species (predicted molecular mass is 24.5kDa). Recent results with the monomeric receptor AtPYL9 indicate that it can form dimers during crystal packing and a minor dimeric form also appears after SEC (Zhang et al., 2013).
Tomato ABA receptors inhibit both Arabidopsis and tomato clade A PP2Cs in an ABA-dependent manner
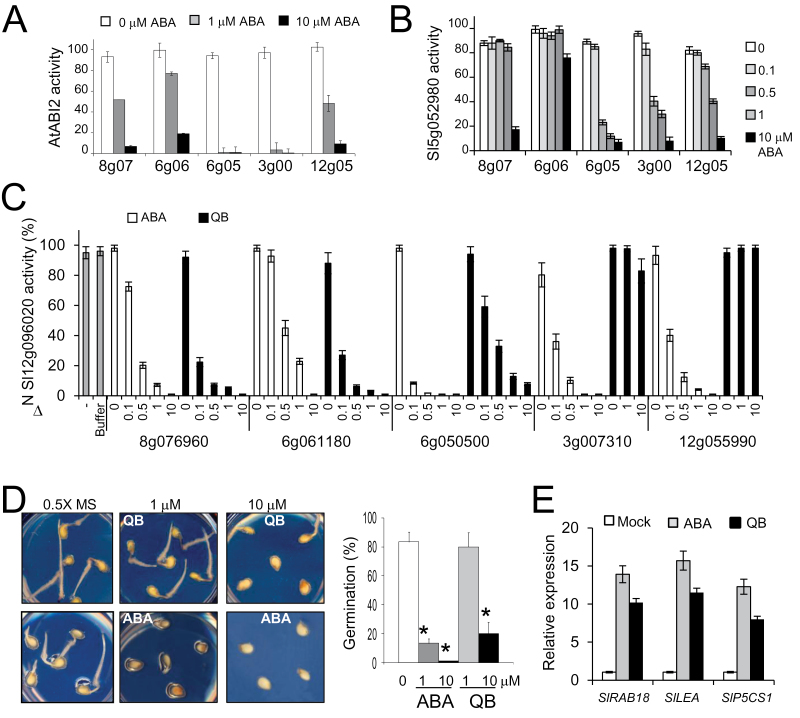
To investigate whether tomato PYR/PYL proteins are actually functional receptors, able to perceive ABA and to inhibit PP2Cs, phosphatase activity was measured using the RRA(phosphoT)VA phosphopeptide as substrate (Fig. 4A–C). To this end, the Arabidopsis PP2C ABI2, which showed interaction with all tomato receptors assayed in Y2H assays, and two tomato PP2Cs, 5g052980 and ΔN 12g096020, which are putative orthologous proteins from AtPP2CA and AtHAB1, respectively, were used. It was possible to purify recombinant soluble proteins for five tomato receptors representing the three subfamilies. All of them were able to inhibit phosphatase activity in an ABA-dependent manner, although to different extents. ABI2 and ΔN 12g096020 activity was sensitive to all receptors, whereas 5g052980 activity was hardly affected by 6g061180 and only at 10 μM ABA by 8g076960. Since tomato phosphatases belong to different sub-branches of the clade A PP2C family, these results are in agreement with the differential inhibition by ABA receptors described previously for AtPP2CA and AtHAB1 (Hao et al., 2011; Antoni et al., 2012; Pizzio et al., 2013).
Fig. 4.
ABA-dependent PP2C inhibition mediated by tomato ABA receptors. PP2C activity was measured in vitro using a phosphopeptide substrate in the absence or presence of ABA at a 1:4 ratio of phosphatase:receptor (0.5:2 μM stoichiometry). Data are averages ±SD for three independent experiments. (A) ABA-dependent inhibition of AtABI2 by tomato receptors. Values represent percentage activity compared with 100% in the absence of receptor and ABA. (B) Phosphatase activity of tomato 5g052980 in the presence of tomato receptors. (C) Phosphatase activity of tomato ΔN 12g096020 PP2C in the presence of tomato receptors. PP2C activity was measured in the absence or presence of 0, 0.1, 0.5, 1, or 10 μM ABA or QB. The column labelled as buffer contained an equivalent volume of HIS elution buffer and 0.5% DMSO. (D) Inhibition of tomato seed germination is more sensitive to ABA than QB. Seed germination was scored 72h after sowing. * indicates P<0.05 (Student’s t-test) when comparing data of plates supplemented with ABA or QB with plates lacking these chemicals. (E) QB treatment induces expression of ABA- and stress-responsive genes. Ten-day-old tomato seedlings were either mock treated or treated with 10 μM ABA or QB for 3h. The histograms indicate the relative induction by ABA or QB treatment of the indicated tomato genes with respect to mock conditions (value 1). (This figure is available in colour at JXB online.)
The ABA agonist quinabactin is selectively perceived by tomato ABA receptors and induces abiotic stress-responsive genes
QB is an ABA-mimicking ligand able to discriminate among the different Arabidopsis PYR/PYL receptors, showing preferential activation of dimeric receptors and certain activation of monomeric PYL5 and PYL7 (Cao et al., 2013; Okamoto et al., 2013). QB application in crop plants (soybean, barley, and maize) had ABA-like effects (Okamoto et al., 2013); however, its mechanism of action has not been investigated previously using crop ABA receptors and PP2Cs. Since QB represents a synthetic ABA agonist eliciting both seed and vegetative ABA responses, having the potential to enable plant protection against water stress, its effect was tested on tomato PYR/PYLs. Interestingly, QB inhibited (compared with ABA) ΔN12g096020 phosphatase activity efficiently through the dimeric tomato receptors 8g076960 and 6g061180, and also through 6g050500, although in this latter case less efficiently than ABA (Fig. 4C). 6g050500 belongs to the AtPYL4–AtPYL6 subfamily and therefore shows similarity to the QB-sensitive AtPYL5, which can explain its capacity to perceive QB. In contrast, two tomato receptors that belong to the AtPYL7–AtPYL10 family, namely 3g007310 and 12g055990, were not activated even by 10 μM QB. These results lend support to the selective effect of QB on ABA receptors and provide biochemical evidence that QB can be perceived by dicot crop receptors and inhibit the activity of a crop PP2C. In order to test whether QB has in vivo effects on tomato, tomato seed germination was analysed in the presence of the compound (Fig. 4D). QB was able to inhibit germination of tomato seeds, although at a higher concentration compared with ABA. These results suggest that tomato receptors not sensitive to QB are required for full regulation of seed germination or that QB-sensitive receptors are not expressed at high levels during this stage. Alternatively, since QB is less water soluble than ABA, the bioavailability of QB could be lower than that of ABA to inhibit seed germination or follow a less efficient transport system.
Finally, in order to assess the biological activity of QB in tomato seedlings, 10-day-old plants were treated with 10 μM QB or ABA for 3h. The transcriptional levels of ABA- and drought-responsive tomato genes were analysed using qRT-PCR (Fig. 4E). To this end, three tomato genes were selected that showed strong sequence similarity with either Arabidopsis RESPONSIVE TO ABA 18 (RAB18), LATE EMBRYOGENESIS ABUNDANT (LEA) family, or the DELTA 1-PYRROLINE-5-CARBOXYLATE SYNTHASE (P5CS1) genes, which were represented by the tomato loci 2g084850, 6g067980, and 6g019170, respectively. The Arabidopsis genes have been shown to be induced in response to drought, cold, salinity, and ABA, and therefore are good markers of plant response to these forms of abiotic stress (Saez et al., 2006). The three tomato genes were activated by both ABA and QB treatment, which indicated that ABA signalling was efficiently triggered by the ABA agonist QB in tomato (Fig. 4E).
Overexpression of tomato monomeric-type ABA receptors in Arabidopsis confers enhanced response to ABA and plant drought resistance
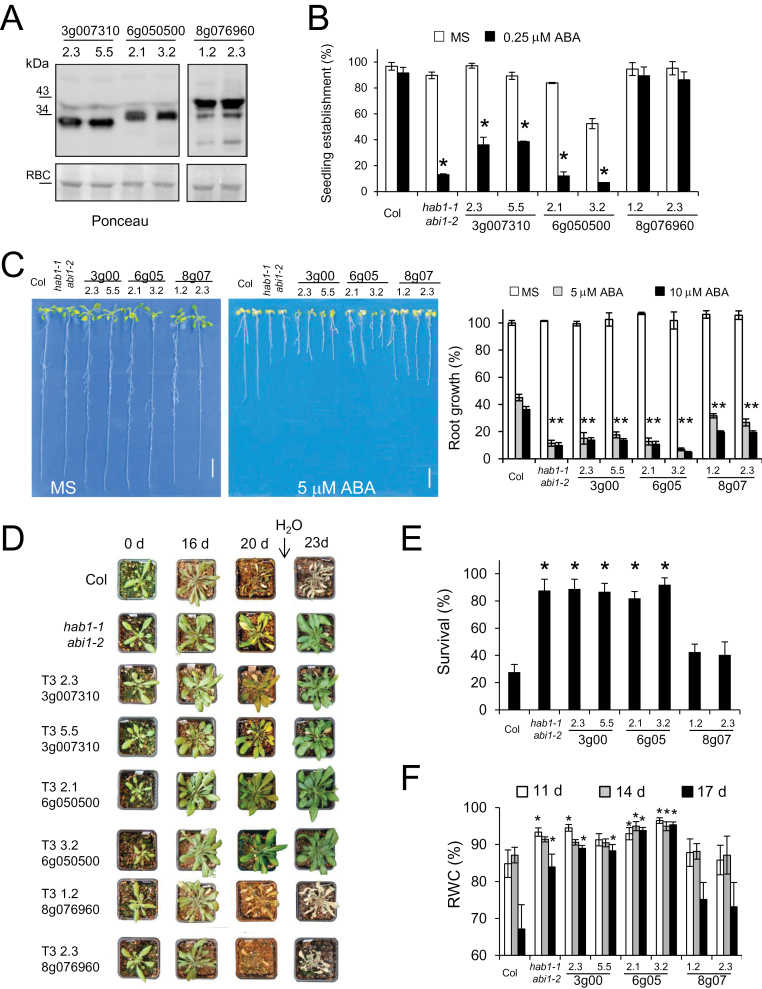
Overexpression of some monomeric Arabidopsis PYR/PYL receptors is known to enhance ABA response and plant drought resistance (Santiago et al., 2009a ; Saavedra et al., 2010; Pizzio et al., 2013). In order to investigate whether tomato PYR/PYLs are functional receptors in plant cells, transgenic plants that overexpress HA-tagged versions of either monomeric-type receptors, 6g050500 or 3g007310, or a dimeric receptor, 8g076960, were generated. Expression of HA-tagged tomato PYR/PYLs was verified by immunoblot analysis, and two independent transgenic lines were selected for further analysis (Fig. 5A). Overexpression of tomato monomeric-type receptors in Arabidopsis enhanced ABA-mediated inhibition of seedling establishment and root growth compared with non-transformed plants, a phenotype similar to that obtained by a double inactivation of the ABI1 and HAB1 PP2Cs (Fig. 5B, C) (Saez et al., 2006). Interestingly, overexpression of the tomato dimeric receptor did not enhance ABA-mediated inhibition of seedling establishment but enhanced root growth sensitivity to ABA and it also generated partial complementation of the ABA-insensitive phenotype of the 112458 pyr/pyl mutant (Fig. 5B, C; Supplementary Fig. S3 at JXB online).
Fig. 5.
Overexpression of monomeric-type tomato receptors in Arabidopsis confers enhanced response to ABA and drought resistance. (A) Immunoblot analysis using antibody against the haemagglutinin (HA) tag shows expression of tomato ABA receptors in 21-day-old seedlings (two independent Arabidopsis T3 transgenic lines for each tomato receptor). Ponceau staining is shown below. RBC indicates ribulose-1,5-bisphosphate carboxylase. (B) ABA-mediated inhibition of seedling establishment in transgenic lines compared with non-transformed Col plants and the hab1-1abi1-2 double mutant. * indicates P<0.05 (Student’s t-test) when comparing data of transgenic lines and the hab1-1abi1-2 mutant with non-transformed Col plants in the same assay conditions. Approximately 100 seeds of each genotype (three independent experiments) were sown on MS plates lacking or supplemented with 0.25 μM ABA. Seedlings were scored for the presence of both green cotyledons and the first pair of true leaves after 8 d. Values are averages ±SE. (C) Enhanced sensitivity to ABA-mediated inhibition of root growth of transgenic lines and the hab1-1abi1-2 mutant compared with non-transformed Col plants. Photographs show representative seedlings 10 d after the transfer of 4-day-old seedlings to MS plates lacking or supplemented with 5 μM ABA. Right panel: quantification of ABA-mediated root growth inhibition (values are means ±SE; growth of Col wild type on MS medium was taken as 100%). (D–F) Transgenic lines overexpressing monomeric-type receptors show enhanced drought resistance and survival, and higher RWC compared with non-transformed plants. (D) Two-week-old plants were deprived of water for 20 d and then re-watered. Photographs were taken at the start of the experiment (0-d), after 16 d and 20 d of drought, and 3 d after re-watering. (E) Percentage survival of non-transformed Col, hab1-1abi1-2, and transgenic lines 3 d after re-watering. (F) RWC of non-transformed Col, hab1-1abi1-2, and transgenic lines after 11, 14, and 17 d of water withdrawal. (This figure is available in colour at JXB online.)
Next, drought resistance experiments were performed under greenhouse conditions. Plants were grown under normal watering conditions for 2 weeks and then irrigation was stopped for 20 d (Fig. 5D). After 20 d without watering, non-transformed plants wilted and many rosette leaves yellowed, in contrast to transgenic lines that express tomato monomeric-type PYR/PYLs (Fig. 5D). Interestingly, transgenic lines expressing the tomato dimeric receptor showed a phenotype similar to the wild type (Fig. 5D). Watering was then resumed and survival of the plants was measured after 3 d. A remarkable enhanced survival (40–50%) was found in the drought-resistant hab1-1abi1-2 double mutant (Saez et al., 2006) and transgenic plants expressing tomato monomeric-type PYR/PYLs compared with non-transformed plants or transgenic lines expressing the dimeric receptor (Fig. 5E). Thus, a clear distinction regarding drought resistance was found between overexpressing tomato monomeric-type or dimeric receptors. During the drought stress experiment, the RWC of the rosette leaves was measured at 11, 14, and 17 d after water withdrawal. Both the hab1-1abi1-2 double mutant and transgenic lines expressing monomeric-type PYR/PYLs showed higher RWC compared with non-transformed plants or transgenic lines expressing the dimeric receptor (Fig. 5F). Thus, either knocking out clade A PP2Cs or overexpressing tomato monomeric-type receptors leads to plants that experience lower water loss compared with non-transformed plants.
Discussion
In this work distinct properties of tomato PYR/PYL ABA receptors according to gene expression and biochemical analyses, sensitivity to the ABA agonist QB, and capability to enhance plant drought resistance are revealed. It is demonstrated that both chemical and transgenic approaches can trigger activity of tomato PYR/PYL ABA receptors, leading to inhibition of crop PP2Cs. Thus, the results indicate that chemical treatment with an ABA agonist is effective to activate the ABA signalling pathway in a dicot crop plant, inducing key genes for drought stress response. Inhibition of PP2C activity by either overexpression of ABA receptors or combined insertional mutagenesis has proved to be an efficient approach to enhance plant drought resistance in Arabidopsis (Saez et al., 2006; Santiago et al., 2009a ). The results open the way for similar approaches in tomato given the feasibility of transgenic approaches and the availability of tomato mutant libraries and TILLING platforms (Okabe et al., 2012). According to results obtained here when tomato receptors were introduced into Arabidopsis and their effect on AtABI2 inhibition, it seems that ABA receptors can be functionally exchanged among different plants. Therefore, the generation of constitutively active receptors or mutated versions that enhance ABA-dependent inhibition of PP2Cs might be used as a transversal approach to enhance drought resistance in different plants (Mosquna et al., 2011; Pizzio et al., 2013). Since overexpression of monomeric-type tomato receptors in Arabidopsis conferred enhanced survival and higher RWC upon drought stress, it will be interesting to check it in tomato plants, through either constitutive or stress-induced expression. Interestingly, overexpression of a dimeric tomato receptor was not effective to enhance Arabidopsis drought resistance, which suggests that monomeric crop PYR/PYL ABA receptors might perform better to achieve such a goal. Indeed, in contrast to monomeric Arabidopsis PYR/PYL receptors, overexpression of dimeric receptor has not proved to be effective to enhance plant drought resistance, which might reflect structural constraints of dimeric receptors to interact with PP2Cs in the absence of ABA and therefore lack of basal activation of the pathway (Dupeux et al., 2011a ). In this case, chemical treatment with ABA agonists is an alternative and efficient approach to activate dimeric receptors (Cao et al., 2013; Okamoto et al., 2013).
Tomato is mainly produced in Mediterranean countries, where fresh water availability is a major problem that could be made worse in the event of climate change. When produced in soil-less greenhouses, solution recycling is mandatory and thus any reduction in the water used is relevant for producers. Gene transcription data suggest that a high number of tomato receptors operate in the root and their transcript levels are higher than those in leaves, which seems to support a relevant role for ABA perception and signalling in the root to cope with drought stress and promote a hydrotropic growth response (Sharp and LeNoble, 2002; Antoni et al., 2013). These data are in agreement with a recent transcriptional analysis of Arabidopsis genes involved in ABA synthesis and perception, which revealed that whereas genes involved in synthesis show higher levels in shoots than in roots, genes involved in perception show an opposite pattern (Boursiac et al., 2013). On the other hand, some reports have revealed a role for ABA in tomato fruit ripening and cell wall catabolism via regulation of ethylene biosynthesis and major catabolic genes (Zhang et al., 2009; Sun et al., 2012). Therefore, ABA signalling in tomato fruit might also be relevant to regulate fruit texture, shelf life, and water loss through the fruit epidermis (Sun et al., 2012; this work). In summary, both RNA-Seq and microarray expression analyses in tomato have indicated ABA receptors that showed a preferential expression (Sun et al., 2011; Wang et al., 2013; this work). These data together with their functional characterization open the door to a future biotechnological use in order to enhance tomato drought resistance or modify fruit properties regulated through ABA signalling.
In silico identification of the ABA signalling core components has been performed in several crops. For instance, the soybean and rice genome encode 23 and 13 putative ABA receptors, respectively (Bai et al., 2012; Kim et al., 2012). In general, the nomenclature of ABA receptors in crops has followed a numerical order that lacks correlation with Arabidopsis receptors. As a result, current nomenclature in crops makes it difficult to correlate biochemical and physiological properties of crop ABA receptors tentatively with Arabidopsis receptors. Since the ABA signalling pathway is universally conserved in land plants (Hauser et al., 2011), it seems sensible to take advantage of knowledge of PYR/PYL receptors in Arabidopsis. In this work, a nomenclature is proposed based on ascribing crop receptors to Arabidopsis subfamilies of PYR/PYL ABA receptors. This approach is supported by phylogenetic studies showing that PYR/PYL receptors can be grouped in three major clades and it allows tentative prediction of some receptor properties (Hauser et al., 2011). Indeed, it was possible to take advantage of this knowledge to infer some properties exhibited by tomato receptors regarding their oligomeric nature, inhibition of PP2C activity, and sensitivity to QB, for instance. Subfamily I corresponded to Arabidopsis dimeric receptors and, indeed, SEC analysis of two tomato members (closely related to AtPYL1) confirmed their dimeric nature. In agreement, Y2H interaction assays showed ABA-dependent interactions for dimeric receptors and PP2Cs, which presumably reflects the requirement for ABA-induced dissociation prior to interaction with the phosphatase. Subfamily II and III corresponded to AtPYL4–AtPYL 6 and AtPYL7–AtPYL 10 groups, respectively. SEC analysis of one tomato representative member of the AtPYL7– AtPYL10 group revealed a similar monomeric nature, whereas the tomato members of the AtPYL4–AtPYL6 group showed an elution profile that might be explained as a monomeric–dimeric mixture. It is possible that the high protein concentration present in the injected sample for SEC analysis might have promoted such equilibrium, as was described recently for the monomeric receptor AtPYL9 (Zhang et al., 2013).
Y2H analyses performed among tomato ABA receptors and either Arabidopsis or tomato PP2Cs revealed both ABA-independent and ABA-dependent interactions (Fig. 3). However, major inhibition of PP2C activity by tomato ABA receptors was ABA dependent (Fig. 4). Therefore, these results suggest that the formation of stable ternary receptor–ABA–phosphatase complexes is required to achieve a major effect on the activation of tomato PP2C downstream targets. Clade A PP2Cs constitute a hub for regulation of different environmental responses, allowing the integration of stress signalling pathways into a coordinated response (Rodrigues et al., 2013). A fine tuning of their activity can be achieved by the selective or differential inhibition of PP2C activity carried out by ABA receptors, which was confirmed in the two tomato PP2Cs analysed in this work. Finally, it was also found that the ABA agonist QB was selective for some tomato receptors and promoted both in vitro inhibition of a tomato clade A PP2C and in vivo inhibition of tomato seed germination. These results, taken together with previous data from Okamoto et al. (2013), indicate that the ABA signalling pathway can be activated in crops by chemicals mimicking ABA action. Thus, chemical treatment of tomato seedlings with QB promoted expression of ABA- and stress-responsive genes. For instance, P5CS1, which encodes a key enzyme for proline biosynthesis and osmotic adjustment under drought stress, was efficiently induced by QB treatment (Fig. 4E). In summary, both chemical and transgenic approaches based on PYR/PYL ABA receptors might be effective to cope with water stress in tomato.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Amino acid sequence alignment of AtPYL1 and putative tomato orthologous ABA receptors.
Figure S2. Amino acid sequence alignment of AtHAB1, AtPP2CA, and putative tomato orthologous PP2Cs.
Figure S3. Complementation of the 112458 pyr/pyl mutant by tomato PYR/PYL receptors.
Table S1. List of oligonucleotides used in this work.
Acknowledgements
This work was supported by the Ministerio de Ciencia e Innovacion, Fondo Europeo de Desarrollo Regional and Consejo Superior de Investigaciones Cientificas (grants BIO2011-23446 to PLR, BFU2011-25384 to AA, and Fontagro and COST1106 for networking activities to AG); fellowships to LR and MP; and a Juan de la Cierva contract to MGG. Cristina Martinez-Andujar and the Bioinformatics Core Service of the IBMCP are acknowledged for the SlEF1a marker and help in bioinformatics analyses, respectively.
Glossary
Abbreviations:
- ABA
abscisic acid
- PP2C
protein phosphatase type 2C
- PYR/PYL/RCAR
PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS
- QB
quinabactin.
References
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL. 2012. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiology 158, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, et al. 2013. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signalling in root. Plant Physiology 161, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Yang DH, Zhao Y, Ha S, Yang F, Ma J, Gao XS, Wang ZM, Zhu JK. 2013. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Molecular Biology 83, 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykov AA, Evtushenko OA, Avaeva SM. 1988. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Analytical Biochemistry 171, 266–270 [DOI] [PubMed] [Google Scholar]
- Ben-Ari G. 2012. The ABA signal transduction mechanism in commercial crops: learning from Arabidopsis . Plant Cell Reports 31, 1357–1369 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. 2004. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Letters 561, 127–131 [DOI] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE. 2012. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiology 160, 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneh U, Biton I, Zheng C, Schwartz A, Ben-Ari G. 2012. Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Reports 31, 311–321 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Leran S, Corratge-Faillie C, Gojon A, Krouk G, Lacombe B. 2013. ABA transport and transporters. Trends in Plant Science 18, 325–333 [DOI] [PubMed] [Google Scholar]
- Cao M, Liu X, Zhang Y, et al. 2013. An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Research 23, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. 2011. FaPYR1 is involved in strawberry fruit ripening. Journal of Experimental Botany 62, 5079–5089 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signalling network. Annual Review of Plant Biology 61, 651–679 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. 1985. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Research 13, 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, McKay JK, Richards JH, Sen S, Wayne T, Juenger TE. 2012. Physiological genomics of response to soil drying in diverse Arabidopsis accessions. The Plant Cell 24, 893–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donella DA, Mac Gowan CH, Cohen P, Marchiori F, Meyer HE, Pinna LA. 1990. An investigation of the substrate specificity of protein phosphatase 2C using synthetic peptide substrates; comparison with protein phosphatase 2A. Biochimica et Biophysica Acta 1051, 199–202 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. 2013. Endodermal ABA signalling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. The Plant Cell 25, 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, et al. 2011. b Modulation of abscisic acid signalling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiology 156, 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, et al. 2011. a A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO Journal 30, 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. 2013. Abscisic acid synthesis and response. Arabidopsis Book 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N. 2011. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Molecular Cell 42, 662–672 [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. 2011. Evolution of abscisic acid synthesis and signalling mechanisms. Current Biology 21, R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG. 2012. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. Journal of Experimental Botany 63, 1013–1024 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, Byun MO, Kim ST, Jung KH, Kim BG. 2014. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. Journal of Experimental Botany 65, 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackman P, Gonzalez-Guzman M, Tilleman S, et al. 2011. Jasmonate signalling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proceedings of the National Academy of Sciences, USA 108, 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, et al. 2009. Structural basis of abscisic acid signalling. Nature 462, 609–614 [DOI] [PubMed] [Google Scholar]
- Mosquna A, Peterson FC, Park SY, Lozano-Juste J, Volkman BF, Cutler SR. 2011. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proceedings of the National Academy of Sciences, USA 108, 20838–20843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signalling by dimeric PYR1. Science 326, 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, et al. 2010. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis . The Plant Journal 61, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Asamizu E, Ariizumi T, Shirasawa K, Tabata S, Ezura H. 2012. Availability of Micro-Tom mutant library combined with TILLING in molecular breeding of tomato fruit shelf-life. Breeding Science 62, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR. 2013. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proceedings of the National Academy of Sciences, USA 110, 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. 2014. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytologist 202, 35–49 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio GA, Rodriguez L, Antoni R, Gonzalez-Guzman M, Yunta C, Merilo E, Kollist H, Albert A, Rodriguez PL. 2013. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signalling and plant drought resistance. Plant Physiology 163, 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AL, Nguyen CV, Hill T, et al. 2012. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715 [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, et al. 2013. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signalling in Arabidopsis. The Plant Cell 25, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Lafuente MT, Rodrigo MJ. 2012. The Citrus ABA signalosome: identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. Journal of Experimental Botany 63, 4931–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra X, Modrego A, Rodriguez D, Gonzalez-Garcia MP, Sanz L, Nicolas G, Lorenzo O. 2010. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signalling in seeds and stress. Plant Physiology 152, 133–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. 2006. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiology 141, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL. 2008. HAB1–SWI3B interaction reveals a link between abscisic acid signalling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis . The Plant Cell 20, 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. 2009. b The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. 2009. a Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. The Plant Journal 60, 575–588 [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME. 2002. ABA, ethylene, and the control of shoot and root growth under water stress. Journal of Experimental Botany 53, 33–37 [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. 2004. Root growth maintenance during water deficits: physiology to functional genomics. Journal of Experimental Botany 55, 2343–2351 [DOI] [PubMed] [Google Scholar]
- Shi JX, Adato A, Alkan N, et al. 2013. The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytologist 197, 468–480 [DOI] [PubMed] [Google Scholar]
- Sun L, Sun Y, Zhang M, et al. 2012. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiology 158, 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. 2011. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany 62, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. 2006. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. The Plant Journal 45, 523–539 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis . The Plant Cell 21, 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Xiong L, Li W, Zhu JK, Zhu J. 2011. The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. The Plant Cell 23, 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tao X, Tang XM, Xiao L, Sun JL, Yan XF, Li D, Deng HY, Ma XR. 2013. Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genomics 14, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. 2009. Structural insights into the mechanism of abscisic acid signalling by PYL proteins. Nature Structural and Molecular Biology 16, 1230–1236 [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. 2009. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. Journal of Experimental Botany 60, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jiang L, Wang G, Yu L, Zhang Q, Xin Q, Wu W, Gong Z, Chen Z. 2013. Structural insights into the abscisic acid stereospecificity by the ABA receptors PYR/PYL/RCAR. PLoS One 8, e67477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang Q, Xin Q, et al. 2012. Complex structures of the abscisic acid receptor PYL3/RCAR13 reveal a unique regulatory mechanism. Structure 20, 780–790 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.