Abstract
Hepatitis C virus (HCV) and GB virus type C (GBV-C) are associated with impaired T cell function despite the fact that HCV replicates in hepatocytes and GBV-C in a small proportion of lymphocytes. Recently, we showed that HCV and GBV-C E2−envelope proteins reduce T cell activation via the T cell receptor (TCR) by competing for phosphorylation with a critical kinase in the TCR signaling cascade (Lck). E2 interfered with TCR signaling in E2 expressing cells and in bystander cells. The bystander effect was mediated by virus particles and extracellular microvesicular particles (exosomes). Multiple kinase substrate sites are predicted to reside on viral structural proteins and based on bioinformatic predictions, many RNA virus pathogens may interfere with TCR signaling via a similar mechanism. Identification of T cell inhibitory effects of virus structural proteins may provide novel approaches to enhance the immunogenicity and memory of viral vaccines.
INTRODUCTION
A large number of human viruses have RNA genomes, including viruses with double-stranded RNA (eg, rotovirus), single-strand RNA (eg, influenza, hepatitis C virus [HCV]), and RNA genomes that require transcription of a DNA intermediate for replication (retroviruses). Retroviruses integrate the DNA copy of their genome into host cell chromosomes resulting in persistent infection. In contrast, only two human RNA viruses that do not have a DNA intermediate, HCV and human Pegivirus (HPgV), cause persistent infection in humans who have otherwise normal immune systems. A variety of immune evasion mechanisms have been identified for RNA viruses, including the rapid selection of variants that escape recognition by antibodies and/or effector T cells, and interference with innate immune recognition. Here, we describe a novel mechanism by which HCV and HPgV reduce T cell activation, contributing to viral persistence.
HISTORY AND EPIDEMIOLOGY
HCV infection is transmitted primarily by exposure to infected blood, and is transmitted less efficiently by sexual intercourse and from mother to child. Approximately 70% of HCV-infected humans will develop persistent viremia, whereas the remainder spontaneously clears infection. Approximately 25% of persistently infected people will develop liver disease over 2 decades on average (reviewed in Hoofnagle [1]), whereas the majority of HCV-infected people do not develop clinical liver disease. Cofactors including alcohol ingestion and HIV enhance HCV pathogenesis, and due to the fact that there are an estimated 180 million people infected with HCV worldwide, the virus contributes significantly to global liver disease and hepatocellular carcinoma. HCV infection is also associated with extrahepatic diseases, including immune-complex−mediated renal disease, cryoglobulinemia, and with an increased risk of non-Hodgkin's lymphoma (reviewed in Hoofnagle [1]).
HPgV was discovered independently by two commercial virus discovery groups in 1995, and was named hepatitis G Virus (HGV) by Genelabs, and GB virus C (GBV-C) by Abbott Laboratories (reviewed in Mohr and Stapleton [2]). After several years of intense epidemiologic study of stored specimens obtained from people with liver disease and other illnesses, no disease association was convincingly identified with HPgV, thus the name hepatitis G was misleading. The other name of the virus (GBV-C) was also misleading, as this virus was named after a surgeon with the initials G.B. whose serum obtained during a case of acute hepatitis was inoculated into marmosets with resultant hepatitis. Following 12 passages in marmosets, two viruses were discovered and named GB virus A and GB virus B. Both turned out to be nonhuman primate viruses, and neither virus appears to replicate in humans (reviewed in Stapleton et al [3]). Using the sequences obtained from GBV-A and B, a human variant was identified in the serum of people with non-A, non-B, non-C hepatitis. This human virus was called GBV-C. However, no evidence exists that the surgeon GB had GBV-C, thus this name was also misleading. Based on genome sequence homology, similar viral genome organization, and considerable protein sequence homology, all three of the GB viruses (A, B, and C) were classified in the Flavivirus family. Subsequently two of these, GBV-A and GBV-C, were assigned to a new genus, the Pegiviruses, for persistent (Pe), “G” viruses, whereas GBV-B was assigned to the Hepacivirus genus, as it more closely resembles HCV. Because HGV and GBV-C are both inaccurate and misleading names, this virus (HGV/GBV-C) was renamed as the human pegivirus, or HPgV (3,4). Among human viruses, HPgV is the most closely related virus to HCV.
HPgV appears to be lymphotropic, and similar to HCV is transmitted by blood and sexual exposure, and by maternal-to-fetal transmission. Sexual transmission appears to be more efficient than for HCV, probably due to lymphotropism and higher serum virus concentrations (3). However, HPgV is not as efficient at establishing persistent infection as HCV, with an estimated 25% of infections leading to persistence with the other 75% clearing infection within 2 years of infection (5). The global prevalence of HPgV infection is not well characterized, but the prevalence range in cross-sectional serum surveys of healthy blood donors detects 1% to 5% of individuals in developed countries with HPgV viremia, and up to 20% of people in developing countries have active infection (2). Based on these data, and the fact that the prevalence is increased among individuals with other blood-born and sexually transmitted infections (HIV, HCV, or frequent transfusion), an estimated 750 million people worldwide are actively infected (viremic), and another 1.5 to 2 billion have had and cleared HPgV infection. Thus HPgV is possibly the most common persistent RNA virus infection on earth and a major contributor to the microbiome.
CLINICAL EVIDENCE OF HPgV AND HCV IMMUNE MODULATION
The rationale for studying HPgV arose from clinical observations that HIV-infected people with HPgV coinfection survived longer than did those with HIV monoinfection (2,6–8). Approaches to understand potential mechanisms by which HPgV might improve survival initially focused on clinical and laboratory studies of HIV-infected cohorts, comparing those with and without HPgV coinfection. Although not entirely consistent, the majority of studies found higher CD4 counts, a slower rate of CD4 decline, and lower HIV viral loads (2,6–8). Because HIV pathogenesis is closely linked to the induction and maintenance of chronic immune activation, several studies were conducted to examine the effects of HPgV on the expression of surface T cell markers on CD4+ and CD8+ T cells. These found a lower rate of activation markers (CD38, CCR5, HLA-DR), proliferation markers (Ki67) on CD4+ and CD8+ T cells in those with HPgV-HIV coinfection when compared to those with HIV monoinfection (9–14). Furthermore, the proportion of naïve T cells was significantly higher among HPgV viremic subjects independent of HIV viral load, and in people with antiretroviral therapy-suppressed HIV RNA (13). Consistent with this, HPgV viremia was associated with a block in CD4+ T cell expansion in subjects receiving recombinant interleukin-2 (IL-2) therapy, suggesting an interaction between HPgV and IL-2 receptor−mediated signaling (15). Studies of HCV-infected cohorts have also described evidence supporting clinical immune suppression by HCV, including increased bacterial infections and reduced rates rejection after kidney transplantation (16,17).
Early studies found that the ex vivo proliferative and cytotoxic T cell responses to HCV antigens, and to a lesser extent to heterologous antigens, are reduced in HCV-infected individuals. The effects are most pronounced when intrahepatic T cells are studied; however, peripheral blood T (PBT) cell responses are also reduced in comparison to HCV-negative subjects (18) and reviewed in Pereira et al (16). Similarly, incubation of healthy PBT cells with serum from HCV-infected people reduced activation through the T cell receptor (TCR), suggesting that structural proteins present in HCV particles interact with T cells to modulate function.
UNRAVELING THE MECHANISM OF T CELL INHIBITION
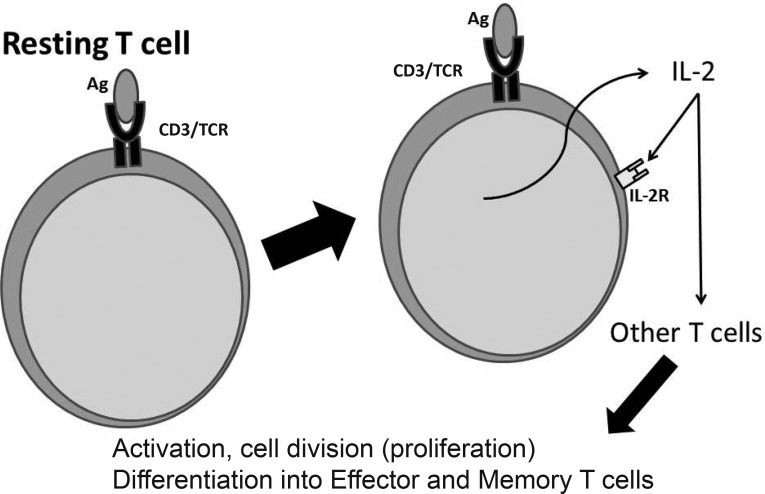
Based on the clinical associations between HPgV and HCV with immune modulation, studies were undertaken to examine viral particle interactions with T cells, specifically T cell activation initiated by engagement of the TCR. Figure 1 shows that, after TCR activation with antigen, resting T cells are induced to release IL-2, which interacts with the IL-2 receptor (IL-2R) on other T cells leading to T cell activation and proliferation. This is necessary before naïve T cells can differentiate into effector cells, some of which will regress to become memory T cells. We recently showed that PBT cells incubated with serum-derived HPgV RNA-containing particles precipitated from infected subjects inhibited TCR signaling. The particles were prepared using an exosome isolation kit and have properties consistent with exosomes (density of 1.11 – 1.19 g/mL); however, we cannot rule out the presence of HPgV virions in this preparation (19,20). Regardless, incubation of T cells with these particles before activation mediated through the TCR using anti-CD3/anti-CD28 antibodies inhibited activation as measured by both the release of IL-2 from cells and upregulation of CD69 and CD25 as compared to cells incubated in HPgV uninfected serum particles (19,20).
Fig. 1.
T cell receptor (TCR) engagement and interleukin 2 (IL-2). After engagement of the TCR with a foreign antigen, resting T cells are induced to produce IL-2 and the high affinity IL-2 receptor (IL-2R) alpha subunit. This leads to a positive feedback loop resulting in proliferation, activation and differentiation of T cells from resting cells into effector T cells. IL-2 is also necessary for the development of T cell memory, which depends on the expansion of the number and function of antigen-selected T cell clones.
The finding that virus particles inhibit TCR signaling suggests that viral structural proteins are involved in this interaction. Consequently, interactions between the HPgV and HCV major envelope glycoprotein (E2) and T cell activation were studied in vitro. First, cell lines expressing either the HPgV E2 protein or the E2 coding region with a frame-shift introduced to abrogate protein expression (frame-shift [FS]) were stimulated with anti-CD3/CD28 antibodies. Cells expressing E2 protein significantly inhibited IL-2 release and CD69 upregulation compared to the FS and parental cell controls (20). Furthermore, adding recombinant HPgV E2 to primary human peripheral blood mononuclear cells (PBMCs) inhibited both CD4+ and CD8+ T cell activation responses after TCR engagement (19;20). IL-2R signaling was also inhibited by HPgV E2 protein expression, contributing to the clinical finding that HPgV-HIV coinfected subjects did not respond to infusion of IL-2 whereas HIV-monoinfected subjects did (15).
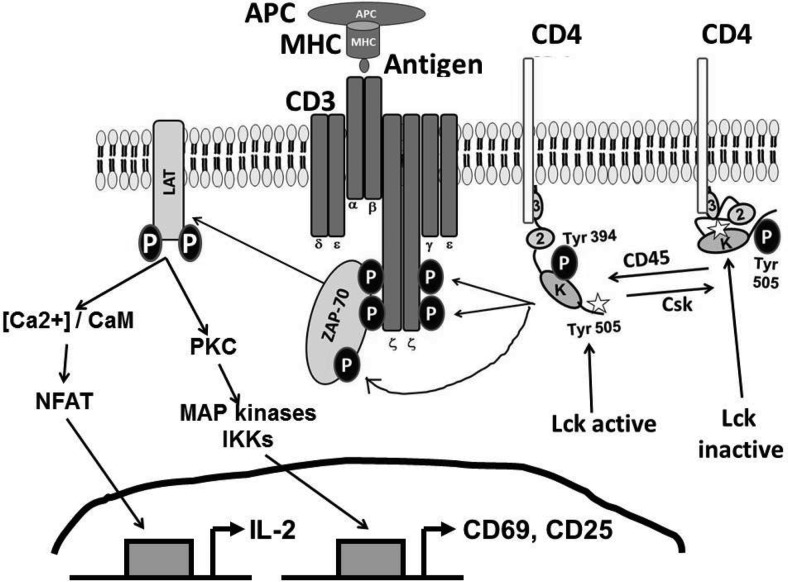
Sequential deletion mapping of the HPgV E2 protein identified a small peptide region that was sufficient to inhibit TCR signaling (20). This region includes a tyrosine (Y87) that is predicted to be a substrate for the most proximal molecule in the TCR signaling cascade (lymphocyte specific protein tyrosine kinase or Lck). After TCR engagement, Lck must be activated starting with dephosphorylation of Lck Y505 by the CD45 phosphatase, which leads to a change in conformation in Lck, opening up the tyrosine at 394 which is phosphorylated in trans by the kinase domain of Lck (Figure 2). Consistent with the TCR-mediated inhibition of IL-2 release and upregulation of CD69 (20), Lck phosphorylated HPgV E2 and an E2 synthetic peptide in vitro, and synthetic peptides including this motif inhibited TCR-mediated activation in primary CD4+ and CD8+ T cells. Substitution of an alanine for the tyrosine at amino acid 87 (Y87A) abolished TCR signaling inhibition, further showing the specificity of the interaction (20). HPgV has two conserved SH3 binding domains, and E2 directly interacts with Lck, as measured in coprecipitation experiments. Although the SH3 binding domain may facilitate the T cell inhibitory effect in vivo, it is not required for TCR inhibition in vitro based on sequential deletion mapping of the C- and N-termini (20).
Fig. 2.
Molecular signaling events after T cell receptor (TCR) engagement. Lck is comprised of three domains, an SH3 binding domain (3), an SH2 domain (2), and a kinase domain (K). The kinase associates with the cytoplasmic tails of CD4 and CD8 (CD4 is shown). When antigen presenting cells (APC) present a foreign antigen in the context of MHC, TCR engagement triggers the activation of Lck. This is initiated by the dephosphorylation of Y505 which leads to a conformation change making Lck Y394 accessible for autophosphorylation in trans. Activated Lck then phosphorylates the intracellular chains of CD3 and the ζ-chains of the TCR complex, allowing ZAP-70 to bind. Lck then phosphorylates ZAP-70, which then phosphorylates LAT (linker of activated T cells). LAT is a docking site for other several other proteins (not shown). The cascade initiated by Lck culminates in the intracellular mobilization of calcium ions and activation of other important signaling cascades (PKC, MAP, etc), and activation of transcription factors such as NFAT, NF-kB, and AP-1 which regulate numerous gene products, including IL-2, CD69, and CD25 (IL-2R).
The interference with TCR signaling was specific for Lck activation, as activation (IL-2 and CD69 upregulation) was not inhibited in T cells incubated with phorbol myristate acetate-ionomycin (PMA+I), which activates downstream of Lck. Furthermore, expression of the E2 protein from a chimpanzee Pegivirus variant, and deletion mutants not containing the Lck substrate site did not inhibit TCR signaling (20). In addition to measuring IL-2 release, cell activation was also measured by determining the proportion of cells with the cell surface expression of activation markers (CD69, CCR5, and CD25 [IL-2 receptor alpha]) after activation through the TCR.
Unfortunately, kinase prediction algorithms are not sufficient to determine if a protein amino acid sequence residing on the viral envelope is functional. HPgV has two predicted Lck sites, both completely conserved among all human isolates (20). In addition to the Y87, T cell inhibitory site, there is a conserved Y292 that is predicted to be phosphorylated by Lck, yet this site does not inhibit T cell activation. This may reflect the accessibility of the predicted substrate site within the context of the entire protein, and how the protein is folded during different parts of the viral replication cycle. Interestingly, although the Y292 does not inhibit TCR-signaling, it does inhibit HIV particle entry, whereas the Y87 region does not (21). In addition to Lck, Y292 is predicted to serve as a substrate for a different kinase, Abl, and previous studies implicate a role for Abl in facilitating HIV entry at the post-hemifusion stage (22). The HPgV Y292 region resides in a peptide region proposed to reside in the viral fusion domain, thus it is possible that this region is exposed during viral fusion (reviewed in Mohr and Stapleton [2]). Studies to assess interactions between E2 (Y292) and ABL are underway.
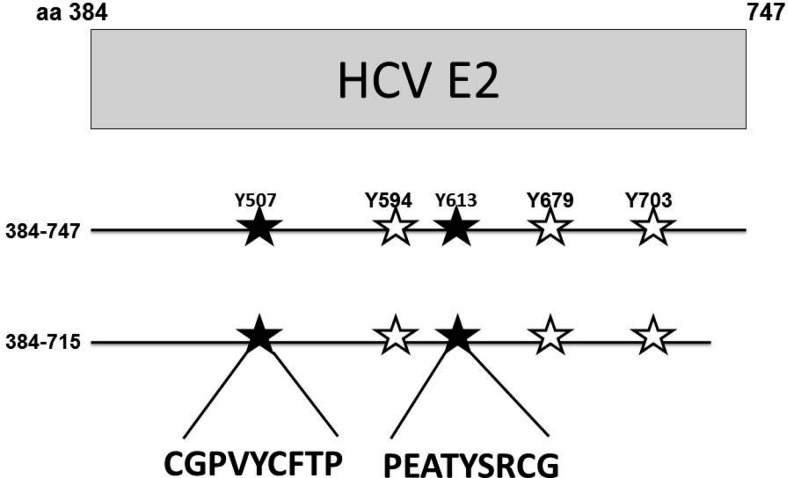
As noted above, serum-derived HCV virus particles interfere with T cell activation (17,23,24), suggesting that the T cell activation inhibitory effect might be conserved between these two closely related human viruses. Because HCV and GBV-C share similar genome organization and considerable amino acid homology, we examined the possibility that HCV E2 protein contains Lck substrate motifs. HCV E2 sequences from 687 HCV isolates including those used to update the HCV nomenclature scheme (25) and those published in a Los Alamos online database (26) were aligned. These sequences represent isolates of all seven genotypes (gt) and 66 characterized subtypes. Conserved tyrosines were examined using a Bayesian decision theory−based online program that predicts PK-specific phosphorylation sites (PPSP) (27,28). Although individual isolates have several conserved tyrosines within the deduced E2 amino acid sequences, many of these are not predicted to be Lck substrate motifs. Among sites on specific isolates, many predicted Lck substrate sites are not conserved. For example, there are five predicted Lck substrate sites in a widely used infectious clone E2 sequence (Figure 3), yet only two of these five sites are conserved among all of the isolates (Y507 and Y613). The Y613 of HCV E2 aligns with HPgV Y87, the tyrosine shown to be involved in TCR-signaling inhibition (20,29), and studies are underway to further characterize the region of HCV E2 involved in inhibition of TCR-signaling.
Fig. 3.
Predicted substrate motifs for the lymphocyte specific protein tyrosine kinase Lck. Five tyrosines on a genotype 1b E2 sequence are predicted to be substrate sites (507, 594, 613, 697, 703). Analysis of 687 HCV E2 sequences representing all seven genotypes and 67 subtypes found that two of these tyrosines (507 and 613) are completely conserved suggesting positive selection (solid stars). CD4+ T cell lines expressing the full-length HCV E2 protein (384–747) and an E2 protein truncated to remove the C-terminal transmembrane domain (384–715) have been generated, and studies are underway to assess these proteins effects on T cell activation. The two conserved Lck substrate motifs are shown (CGPVYCFTP and PEATYSRCG). By alignments, the Y613 site aligns with the HPgV Y87 T cell inhibitory motif.
SUMMARY
HPgV and HCV E2 proteins contain a TCR-inhibitory motif that reduces T cell activation and proliferation. The T cell inhibition is not complete, otherwise HCV- and HPgV-infected people would have life-threatening immunosuppression. Nevertheless, clinical data support a mild reduction in immune function in both HCV and GBV-C, and both viruses are associated with the development of non-Hodgkin's lymphoma, suggesting impaired immune surveillance (16,17,23,24).
Because envelope proteins are the first viral proteins to interact with immune cells and T adaptive immune responses rely on T cell activation, interference with T cell activation will blunt a variety of immune responses to foreign antigens. The T cell inhibitory effect contributes to the development of virus persistence and may explain in part the slow and inefficient HCV antibody development and poor T cell responses observed in HCV infection. Furthermore, the finding that virus structural proteins interfere with T cell responses explains why intrahepatic T cell responses are more impaired than are peripheral T cell responses because most if not all HCV E2 protein is expressed by hepatocytes and local concentrations of E2 will be greatest in the liver.
Several important questions remain related to these observations. First, how does E2 come into contact with Lck in infected cells? Both HCV and HPgV have infectious viral RNA, and the viral polyprotein is translated immediately on release of RNA into the cell cytoplasm. In this setting, does some E2 protein escape ER localization and interact with Lck in the cell cytoplasm? Alternatively, do the viruses enhance E2 protein release from the endoplasmic reticulum via the endosomal sorting complexes required for transport (ESCRT) pathway and, before degradation, interacts with Lck via the SH3 binding domain with resultant competition for Lck phosphorylation? In the setting where virus particles are incubated with T cells, do virions or viral proteins in exosomes become exposed to Lck near the plasma membrane, and during internalization, serve as pseudosubstrates competitively reducing Lck activation? Clearly, understanding the localization of E2 in both particle-mediated and E2 expression−mediated T cell inhibition is needed. Second, do HCV and HPgV envelope proteins interact with other cell kinases, modulating immune function or other cellular functions in additional ways? If so, are there ways to identify which predicted kinase substrate site among several on envelope proteins is/are functional? Third, are these T cell inhibitory motifs present on other viruses? Use of bioinformatic sequence analyses identifies conserved Lck substrate sites on many, though not all viral envelope proteins, and the phenomenon of viral structural proteins competing for phosphorylation after interactions with immune cells raises the possibility of a novel and complex envelope-mediated regulation of immunity during viral infection. Finally, do the immunomodulatory motifs on viral proteins contribute to poor immunogenicity of recombinant viral envelope-based vaccines? If so, mutation of the key tyrosine(s) may enhance immunogenicity, although it may also alter protein structure such that key neutralization antigenic sites are lost. Nevertheless, characterizing how virus particles and envelope proteins interfere with host T cell responses may lead to novel approaches of treating and preventing RNA virus infections.
ACKNOWLEDGMENTS
We thank Dr Jon Houtman (University of Iowa) for helpful discussions related to T cell receptor signaling. This work was supported in part by a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R01-AI58740 [JTS]) and by Merit Review grants from the Department of Veterans Affairs (BX 000207 [JTS], CX000821 [JTS], and BX001241 [JX]).
Footnotes
Potential Conflicts of Interest: Drs Stapleton, Xiang, McLinden, and Bhattarai have a patent pending related to the work described in this manuscript.
DISCUSSION
Boyer, New Haven: Given the very high prevalence of the GB virus, is there any evidence in the population that are positive, that they have less autoimmune disease or less severe autoimmune disease?
Stapleton, Iowa City: This is certainly possible. I did some studies with Gary Gilchrest, who was a resident with me and is here at MUSC, South Carolina. And epidemiologically, in a small look at lupus patients, there was a hint that there might be reduced prevalence. Although the numbers were small, none of the lupus patients had persistent viremia. If you look at healthy blood donors, it's between 1% and 2%. So every day in the US about 1000 units of blood are transfused that have this virus in them. Further studies on the question of modulating or protecting against autoimmunity would certainly be interesting.
Bodenheimer, New York: Not everybody who is exposed to hepatitis C goes on to chronic infection, and so how much variability is there on the human side in terms of the susceptibility of suppression of T cell activity from your hepatitis?
Stapleton, Iowa City: Well, so there are certainly genetic polymorphisms that are associated with hep C clearance; the IL28B gene, for example. But there are probably many others as well. One of the hallmarks of clearance is having a brisk and diverse cytotoxic T lymphocyte response to hepatitis C, so T cell responses to hepatitis C are very important. But if you look in the liver-derived lymphocytes, they don't react to hepatitis C antigens. And so one of the questions we have, and we are trying to ask is, “Do people who clear hepatitis C have mutations in this region?” That would be lovely if it does. So far, based on published sequences of people who cleared, it is not obvious if that's the case; but it is clearly complicated and it's not just the virus in the clearance.
Quesenberry, Providence: This is kind of an exploding field now in extracellular vesicles, especially with regard to cell fate change, restoration of renal damage, and liver damage. I am wondering particularly what the size of your particles was? Was it in the exosome range?
Stapleton, Iowa City: Yes.
Quesenberry, Providence: So it's less than 100 nanometers?
Stapleton, Iowa City: They range from 40 to 100. They are heterogeneous. They are pulled down by anti-CD63, which is one of the findings that led to propose that some of these particles are exosomes. We were able to show CD81 and CD63 in the particles. So we know that some of them have the traits of exosomes, but there are many types of microvesicles. So I hesitate to say for sure that they are exosomes.
Quesenberry, Providence: And they are very heterogeneous, and I guess the other thing that is fascinating is a lot of other things can be going on with those vesicles.
Stapleton, Iowa City: Oh, absolutely. And there are two reports showing that hepatitis C virus envelope proteins are contained in exosomes released from infected cells in vitro. We think that interactions between exosomes containing viral envelope proteins may well be the mechanism by which these viruses reduce T cell activation and proliferation. If we are correct, this will represent a novel and innate mechanism of viral escape from T cell immunity.
Harrison, Nashville: You began by talking about the relationship of your virus with HIV survival that you discovered. Does this interaction with (Lck) lymphocyte-specific protein tyrosine kinase prevent expression of entry sites for HIV, like CCR5?
Stapleton, Iowa City: It does. It does interact and, in fact, we published a paper the year before last showing that if you look at lymphocytes from people who are HIV suppressed by drugs and compare their likelihood of a reactivation of latent HIV in the central memory T cells, that activation markers including CCR5 on T cells are significantly lower if you have GBVC coinfection, and the T cells don't proliferate as well. The interference in activation and proliferation may actually be a detriment to the cure strategies of trying to activate latent HIV to get it to replicate, and in one study we demonstrated that reactivation of latent HIV is reduced in people with GBV-C coinfection.
Mackowiak, Baltimore: We are going to have to make this question the last one. I'm sorry.
Reddy, Ann Arbor: A quick one. So given that these patients seem to have less T cell antibody responses, any association actually with modest or even severe immunodeficiency in kids, who get GB virus infections?
Stapleton, Iowa City: So kids don't get it very often. They usually get it from their mothers when they do. The prevalence goes up tremendously as children become sexually active, consistent with this being primarily a sexually transmitted infection, although it is also transmitted by blood products and maternal-fetal. One point to make is that there is not a large inhibition of T cell function. In fact, I don't think we would have found it if it were not in the setting of an immune activation disease, like HIV, where chronically ill patients have a high basal immune activation states, and the reduction of that actually is beneficial for people with HIV. But it's not an all or none or else these patients wouldn't survive. So this is a modest regulator of T cell function in response. So it's an anti-inflammatory, if you will.
REFERENCES
- 1.Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 2.Mohr EL, Stapleton JT. GB virus type C interactions with HIV: the role of enveope glycoproteins. J Viral Hepat. 2009;16:757–68. doi: 10.1111/j.1365-2893.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C(HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2012;92:233–46. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams MJ, King AMQ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2013;158:2023–230. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 5.Bhattarai N, Stapleton JT. GB virus C: the good boy virus? Trends Microbiol. 2012;20:123–30. doi: 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang J, Wuenschmann S, Diekema D, Klinzman DJ, Patrick KD, George SL, Stapleton JT. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med. 2001;345:707–14. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GBV-C viremia on survival of HIV infected individuals: a meta-analysis. HIV Med. 2006;7:173–80. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 8.Stapleton JT, Williams CF, Xiang J. GB Virus C: a beneficial infection? J Clin Micro. 2004;42:3915–9. doi: 10.1128/JCM.42.9.3915-3919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nattermann J, Nischalke HD, Kupfer B, et al. Regulation of CC chemokine receptor 5 in hepatitis G virus infection. AIDS. 2003;17:1457–62. doi: 10.1097/00002030-200307040-00006. [DOI] [PubMed] [Google Scholar]
- 10.Maidana Giret MT, Silva TM, Sauer MM, et al. GBV-C infection modulates T cell activation in recently HIV-infected subjects and is independent of HIV-1 viral load. AIDS. 2009;23:2277–87. doi: 10.1097/QAD.0b013e32832d7a11. [DOI] [PubMed] [Google Scholar]
- 11.Stapleton JT, Chaloner K, Martenson JA, et al. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PLoS One. 2012;7:e50563. doi: 10.1371/journal.pone.0050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stapleton JT, Martinson JA, Klinzman D, Xiang J, Desai SN, Landay A. GB virus C infection and B-cell, natural killer cell, and moncyte activation markers in HIV-infected individuals. AIDS. 2013;27:1829–32. doi: 10.1097/QAD.0b013e328363089f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarze-Zander C, Neibecker M, Othman S, et al. GB virus C coinfection in advanced HIV type-1 disease is associated with low CCR5 and CXCR4 surface expression on CD4(+) T-cells. Antivir Ther. 2010;15:745–52. doi: 10.3851/IMP1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattarai N, Rydze RT, Chivero ET, Stapleton JT. GBV-C viremia is associated with higher levels of double negative T cells and lower T cell activation in HIV-infected individuals on antiretroviral therapy. J Infect Dis. 2012;206:1469–72. doi: 10.1093/infdis/jis515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stapleton JT, Chaloner K, Zhang J, et al. GB virus C viremia is associated with reduced CD4 expansion following interleukin 2 therapy in HIV-infected people receiving HAART. AIDS. 2009;23:605–10. doi: 10.1097/QAD.0b013e32831f1b00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn YS. Subversion of immune responses by hepatitis C virus: immunomodulatory strategies beyond evasion? Curr Opin Immunol. 2003;15:443–9. doi: 10.1016/s0952-7915(03)00076-1. [DOI] [PubMed] [Google Scholar]
- 17.Pereira BJ, Natov SN, Bouthot BA, et al. Effects of hepatitis C infection and renal transplantation on survival in end-stage renal disease. The New England Organ Bank Hepatitis C Study Group. Kidney Int. 1998;53:1374–81. doi: 10.1046/j.1523-1755.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 18.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–52. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 19.Bhattarai N, McLinden JH, Xiang J, Kaufman TM, Stapleton JT. GB virus C envelope protein E2 inhibits T cell receptor induced IL-2 production and alters IL-2 signaling pathways. J Immunol. 2012;189:2211–6. doi: 10.4049/jimmunol.1201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattarai N, McLinden JH, Xiang J, Landay A, Chivero ET, Stapleton JT. GB virus C particles inhibit T cell activation via envelope E2 protein-mediated inhibition of T cell receptor signaling. J Immunol. 2013;190:6351–9. doi: 10.4049/jimmunol.1300589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang J, McLinden JH, Kaufman TM, et al. Characterization of a peptide domain within the GB virus C envelope glycoprotein (E2) that inhibits HIV replication. Virology. 2012;430:53–62. doi: 10.1016/j.virol.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmon B, Campbell N, Ratner L. Role of Abl kinase and the Wave2 signalng complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 2010;6:e1000956. doi: 10.1371/journal.ppat.1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner F, Greuner NH, Urbani J, et al. CD8-T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000;30:2479–87. doi: 10.1002/1521-4141(200009)30:9<2479::AID-IMMU2479>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Isaguliants MG, Petrakova NV, Kashuba EV, et al. Immunization with hepatitis C virus core gene triggers potent T-cell response, but affects CD4+ T-cells. Vaccine. 2004;22:1656–65. doi: 10.1016/j.vaccine.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 25.Smith DB, Bukh J, Kuiken C, et al. Expanded classification of hepatitis C virus into 7 genotypes and 66 subtypes, updated criteria and assignment web resource. Hepatology. 2013;59:318–27. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Los Alamos HIV Databases 2013. HCV Sequence Database. [Accessed February 26, 2014]. Available at: http://hcv.lanl.gov/content/index.
- 27.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarch. Mol Cell Proteomics. 2008;7:1598–608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Want X, Wang Y, et al. Prediction of posttranslational modification sites from amino acid sequences with kernel methods. J Theor Biol. 2014;344:78–87. doi: 10.1016/j.jtbi.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Chandriani S, Skewes-Cox P, Zhong W, et al. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc Natl Acad Sci U S A. 2013;110:1407–15. doi: 10.1073/pnas.1219217110. [DOI] [PMC free article] [PubMed] [Google Scholar]