Abstract
The computer-based design of protein-protein interactions is a challenging problem because large desolvation and entropic penalties must be overcome by the creation of favorable hydrophobic and polar contacts at the target interface. Indeed, many computationally designed interactions fail to form when tested in the laboratory. Here, we highlight strategies our laboratory has been pursuing to make interface design more tractable. Our general approach has been to make use of structural motifs found in native proteins that are predisposed to interact with a particular binding geometry, and then further bolster these anchor points with favorable hydrophobic contacts. We describe the use of three different anchor points – β-strand pairing, metal binding, and the docking of an α-helix into a groove – to successfully design new interfaces. In several cases high- resolution crystal structures show that the design models closely match the experimental structure. Additionally, we have tested the use of buried hydrogen bond networks as a source of affinity and specificity at interfaces. In these cases the designed complexes did not form, highlighting the challenges associated with designing buried polar interactions.
Keywords: Computational Protein Design, Rosetta, Protein-Protein Interfaces, De Novo Protein Design
Introduction
Protein-protein interactions (PPIs) mediate a wide array of signaling pathways in living organisms and the design of new PPIs promises the development of powerful therapeutics and research tools. Computational protein design is a relatively new method for engineering novel PPIs, and offers many benefits, such as fine-grained control over binding orientation. Most studies that have explored the computational design of new interactions have made use of high-resolution structures of protein monomers as the starting point for the design process. The proteins of interest are computationally docked against each other and then sequence optimization algorithms are used to search for mutations at the protein-protein interface that will stabilize the new complex (Fig. 1). Both proteins can be mutated to induce binding, or if the goal of the project is to target a naturally occurring protein then only one side of the interface is optimized. In the last few years this approach has been used to design a novel protein inhibitor, homodimers, nanocages and a crystal, all with atomic-level accuracy [1–9]. Additionally, these methods can be used to redesign naturally occurring interfaces for enhanced affinity and altered specificity [10,11]. Despite these successes, computational design of PPIs has proven to be a difficult challenge, and the vast majority of designs characterized in the laboratory fail to exhibit the desired behavior [12]. It is not uncommon to characterize on the order of 50 design predictions in order to discover a weak binder, although success rates are highly dependent on the specific design goal.
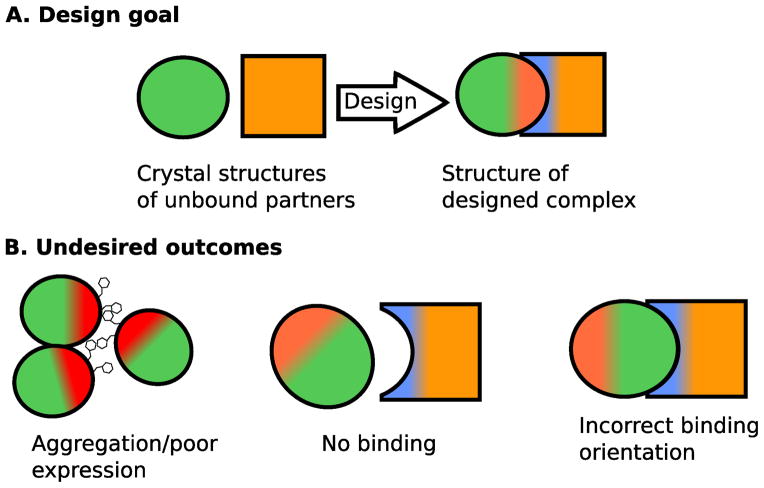
Figure 1.
Diagram illustrating the goal and potential outcomes of protein interface design. (A) The typical starting point and desired outcome of a protein design project. (B) Potential undesired outcomes common to protein interface design.
There are several ways that protein interface design can fail: no or weak binding, binding in an undesired conformation, aggregation between the unbound or bound proteins, and poor stability or expression of the binding partners (Fig. 1). For projects pursued in our laboratory, lack of binding has been the most common failure, but we have also observed low expression yields and aggregation in some projects. Achieving tight binding through a specific binding mode is challenging for a variety of reasons. In order for binding to be favorable, desolvation and entropic costs must be overcome by favorable interactions at the interface. This requires close packing between atoms at the interface and satisfying the hydrogen bonding potential of desolvated polar atoms. The desolvation penalty for binding can be minimized or perhaps eliminated by relying on hydrophobic interactions at the interface, but there are several potential pitfalls associated with this approach. Proteins with large hydrophobic patches on their surface are more likely to nonspecifically self-aggregate in the unbound state and hydrophobic interactions often lend little directional specificity to interactions as non-polar groups can interact favorably in a variety of geometries. In contrast, hydrogen bonds between polar groups have strong orientational preferences and can help specify binding geometry, but require greater accuracy in the design process as small deviations from ideality can result in unfavorable energies.
Because of the challenges inherent to de novo interface design, our laboratory has been testing specific design strategies that may make the problem more tractable. The general approach has been to use structural motifs found in native proteins that are predisposed to interact with a particular binding geometry, and then further bolster these anchor points with regions of hydrophobic packing. In this mini-review we describe three types of structural motifs that we have used to help successfully design new interfaces: β-strand pairing, metal binding, and the docking of an α-helix into a groove (Fig. 2). We conclude the review by discussing unsolved issues in interface design, including the use of side chain mediated hydrogen bond networks to create stability and specificity.
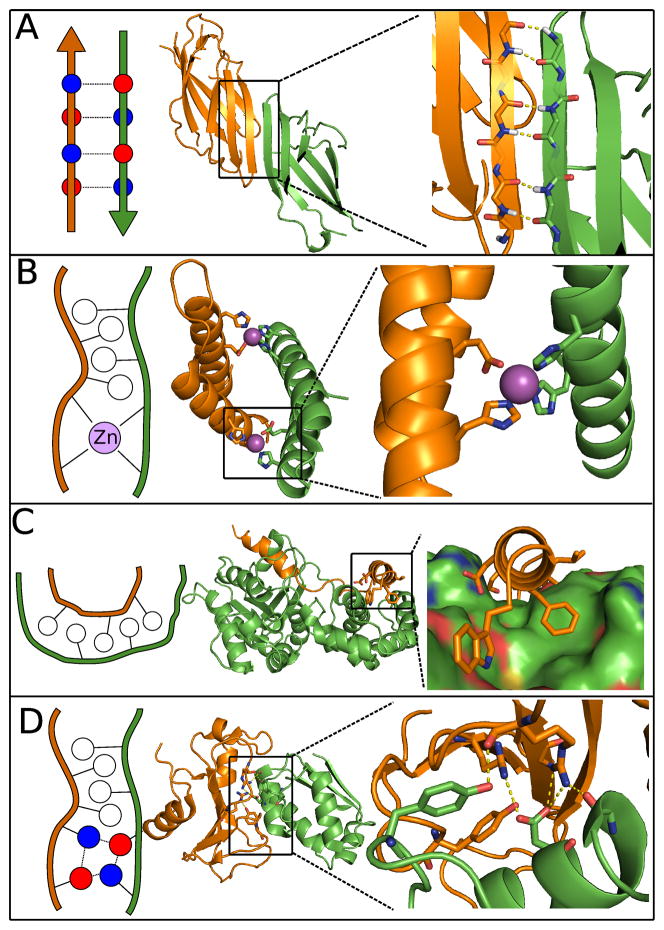
Figure 2.
Anchor motifs found in natural and designed protein interfaces. Polar residues illustrated in red and blue, hydrophobic residues in white. (A) β-strand pairing across a designed protein interface (PDB code 3ZY7). (B) Designed metal-mediated protein interface (PDB code 3V1E). (C) Designed helix-in-groove interface (PDB code 2XNS). (D) Naturally occurring interface mediated by hydrogen bonds (PDB code 1BGS).
Design of β-strand pairing across a protein-protein interface
β-strand pairing is a common naturally occurring binding motif found both in hetero- and homo-oligomers. Inter-protein β-sheets are often found in antibody-antigen interactions, and are prevalent in important signaling cascade interactions, such as those between G- proteins and their effectors [13]. β-strand pairing is an appealing interface design strategy for many reasons: The regular hydrogen-bonding pattern between strands provides favorable interaction energy and gives strong orientation preferences between the interacting partners. Furthermore, β-strands have predictable structures and sequence preferences. Finally, the predominantly backbone nature of β-sheet formation allows sidechains to remain free, lowering the entropic penalty of β-strand mediated binding compared to other strategies. The interface-forming tendencies of solvent exposed β-strands have led to the evolution of several promiscuity-protecting factors in β-proteins, such as strategically placed charges, and edge-strands with various irregularities [14]. Therefore, the “sticky” nature of solvent exposed β-strands must be careful considered during interface design to avoid aggregation.
Recently, our laboratory used β-strand mediation to design a novel homodimer [9]. In this study, the PDB was searched for monomeric proteins with solvent exposed β-strands. Models of β-strand mediated dimers were generated by duplicating the monomer structures and docking the two units on their exposed β-strands. The sequences of the dimers were then optimized for binding using the molecular modeling program Rosetta [15]. Two separate strategies were employed during sequence optimization, one that designed mostly hydrophobic residues, and another that designed mostly polar amino acids. Complementary charges were designed on the interacting β-strands in an attempt to prevent aggregation. Using this method a homodimer with an equilibrium dissociation constant of 1uM was generated. The solved structure of this homodimer matched the design model with an all-backbone atom RMSD of 1.0 Angstroms, with a near perfect match of the interface β-strands. This strong success demonstrates the benefit of using well-defined secondary structural elements at protein interfaces, as well as the value of β-strand pairing in promoting predictable orientation between interacting pairs. Notably, the successful design from this project was generated from the mostly-hydrophobic sequence design strategy. The inability to design multiple polar contacts is a common theme in protein interface design, and emphasizes the difficulties of designing de novo hydrogen bond networks.
Design of metal-mediated protein-protein interfaces
Metal ions play a vital role in cellular function, and frequently coordinate tertiary and quaternary protein structure. Metal coordination produces very specific geometries, which make them ideally suited as the anchor point for a designed protein-protein interface. Furthermore, the hydrogen bonds involved in metal binding are stronger than protein-only hydrogen bonds and van der Waals contacts, imparting valuable affinity in designed PPIs. Beyond energetic benefits, metal mediated design is attractive due to the minimal number of required mutations. The sum of these advantages has led to a rich history of metal-binding in nearly all aspects of protein design [16–19]. One such project undertaken by our laboratory in collaboration with Akif Tezcan’s laboratory attempted to mimic a potential evolutionary pathway for a simple PPI [20]. In this study a weak homotetrameric protein interface, constituted almost entirely of a metal-binding site, was stabilized through the addition of hydrophobic contacts at the primitive interface. The redesigned interface stabilize the tetrameric state by nearly three orders of magnitude [20]. Crystal structures of the redesigned interfaces showed very little change to the overall tetramer fold, in agreement with the design models. To further explore the potential of using metal mediation to design protein interfaces, our laboratory aimed to design a de novo zinc-mediated symmetric homodimer [21]. In the design models, each side of the interface would coordinate zinc with two histidine residues. The remainder of the interface was designed for affinity using mainly hydrophobic residues. Here, a zinc-mediated homodimer with a kD of 30nM was generated, and the overall topology of the zinc-bound crystal structure matched well with the design model. Interestingly, a crystal structure of the dimer in its apo form, which associates with a weakened kD of 4.3uM, demonstrates a binding orientation that is very different from the design model, highlighting the difficulty of relying on hydrophobic interactions to promote specific binding orientations. Conversely, the crystal structure of the zinc-bound dimer revealed several unfavorable structural rearrangements, such as partial loss of helicity, which formed in order to accommodate metal binding. This result underlines the advantage of using an anchor motif with strict geometric requirements, as well as the affinity-producing power offered by metal-mediation, which can function to compensate for modeling errors in other parts of the design interface. In addition to this result, the benefits of designing metals into protein quaternary structure are further established through successes by other groups, including design of diiron coiled-coils [22].
One disadvantage of utilizing metal mediation in design of PPIs is the requirement of being able to design coordinating residues on both of the interacting partners. This requirement limits the ability of using metal mediated design for targeting natural proteins. One potential way to work around this limitation is by using single residues on a given target protein to complete the coordination sphere of the interface metals. However, a recent result from our group, in which a de novo ubiquitin-binding protein was designed, demonstrated that a single metal-coordinating residue on the target protein was not enough to generate strong orientational preference between the interacting proteins [23].
Helix-in-groove strategy
A helical peptide or protein fragment binding to a hydrophobic groove is a very common natural mechanism for imparting shape-specificity to a PPI. The simplicity and prevalence of this binding motif make it an attractive strategy for interface design. A particularly appealing aspect of this strategy is the fact that polar contacts, which have proven to be difficult to model accurately, can be largely omitted from designs. Instead, orientational specificity is dictated by complementary between secondary structure elements combined with tightly interlaced hydrophobic sidechains. A design project pursued by our laboratory aimed to use the helix-in-groove strategy to improve affinity of a weakly bound peptide by designing the sequence and structure of a 16-residue extension. Unlike previously described interface design projects that used a fixed backbone representation of the interacting partners, this protocol sampled the conformational space of the protein backbone through a fragment insertion protocol [24]. The resultant helical peptide increased affinity by approximately 25-fold, and the solved structure matched the design model closely. More recently, Fleishman et al. designed a de novo protein that binds to the conserved stem region of influenza hemagglutinin. In this case, hydrophobic “hotspot” residues were grafted onto a helix that was docked into a groove on the target protein [7]. The initial design models, which bound weakly to the target, underwent affinity maturation resulting in a final protein that bound the target protein in the low nanomolar range. Crystal structures of the matured protein revealed that incorporation of backbone sampling into the design protocol could have predicted the mutations produced by the experimental affinity maturation. Each of these projects highlights a disadvantage of designing helix-into-groove interfaces; the tightly packed residues required for specificity from hydrophobic residues often benefits greatly from sampling degrees of freedom in the protein backbone. Sampling backbone degrees of freedom greatly increases the conformational search space, which can create issues of under sampling for even the smallest protein interfaces. One particularly interesting advantage of designing hydrophobic helix-in-groove interactions is the fact that there are likely several sequences compatible with a given groove. The design of multiple sequences that bind a conserved target interface would constitute a natural first step towards the de novo design of multi-functional protein interface.
Design of inter-protein hydrogen bond networks
Hydrogen bonds are often found at protein-protein interfaces providing both affinity and specificity. The surface polar residues involved in these interactions serve the dual purpose of promoting solubility by breaking up patches of hydrophobic residues, while simultaneously conferring orientation specificity through strict geometric requirements. In many ways, the previously described interface design strategies, metal-site and β-strand mediation, can be thought of as simple methods to form predictable hydrogen bond networks. Despite the similarities, design of novel hydrogen bond networks at protein interfaces has proven to be a very difficult design problem. In a recent evaluation of failed and successful interface designs we found that all the successfully designed interfaces were formed largely by hydrophobic interactions [12]. In contrast, natural protein interfaces can be formed by much larger percentages of polar residues. Additionally, the successes in design of inter-protein hydrogen bond networks, which have all started from a pair of known binders, have reduced binding affinity by several orders of magnitude [25].
The difficulties in design of hydrogen bond networks are due to a variety of reasons including imperfect geometric and energetic scoring of the interacting atoms. Development of accurate scoring for hydrogen bond geometries is an active field of research, and improvement in this area will likely lead to more successful hydrogen bond designs. Another potential reason design of hydrogen bond networks has been unsuccessful is due to the fact that solvent exposed hydrogen bonds are relatively weak interactions, and thus formation of small numbers of hydrogen bonds isn’t strong enough to compensate for modeling inaccuracies in other parts of the protein interface. Finally, the conformational sampling required to identify complex hydrogen bond networks is enormous, and sampling algorithms are likely missing large percentages of favorable conformations. One could imagine addressing the issue of sampling by attempting to graft entire hydrogen bond motifs into protein interfaces. This strategy is similar in spirit to the metal and β-strand mediated designs outline above, as well as with residue hotspot-centric design strategies used by other groups.
Discussion
De novo design of PPIs is an exciting new frontier for computational modeling. The successes and failures in this field have taught us that small errors in sampling and scoring algorithms make precise interactions difficult to properly design, while design of less precise interactions frequently leads to protein aggregation and misfolding. Despite these challenges, there have been a handful of successful designs that give promise for the future. An analysis of several of these successes has demonstrated the usefulness of combining specificity determining interactions to dictate relative binding orientation, and hydrophobic interactions to increase affinity. This general strategy has been used successfully by many groups and should serve as a useful template for future design projects.
Acknowledgments
Funding
This work was supported by NIH grant GM073960.
References
- 1.London N, Ambroggio X. Journal of structural biology. Elsevier Inc; 2013. An accurate binding interaction model in de novo computational protein design of interactions: If you build it, they will bind. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead T, Baker D, Fleishman S. Methods in enzymology. Vol. 523. Elsevier Inc; 2013. Computational design of novel protein binders and experimental affinity maturation; pp. 1–19. [DOI] [PubMed] [Google Scholar]
- 3.Khare SD, Fleishman SJ. Emerging themes in the computational design of novel enzymes and protein-protein interfaces. FEBS letters, Federation of European Biochemical Societies. 2013;587:1147–54. doi: 10.1016/j.febslet.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Karanicolas J, Kuhlman B. Computational design of affinity and specificity at protein-protein interfaces. Current opinion in structural biology. 2009;19:458–63. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippow SM, Tidor B. Progress in computational protein design. Current opinion in biotechnology. 2007;18:305–11. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, André I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science (New York, NY) 2012;336:1171–4. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleishman S, Whitehead T, Ekiert D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science, American Association for the Advancement of Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanci CJ, MacDermaid CM, Kang S, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Computational design of a protein crystal. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7304–9. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stranges PB, Machius M, Miley MJ, Tripathy A, Kuhlman B. Computational design of a symmetric homodimer using β-strand assembly. Proceedings of the National Academy of Sciences of the United States of America, National Academy of Sciences. 2011;108:1–6. doi: 10.1073/pnas.1115124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nature chemical biology, Nature Publishing Group. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 11.Chen TS, Keating AE. Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods. Protein science : a publication of the Protein Society. 2012;21:949–63. doi: 10.1002/pro.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranges PB, Kuhlman B. A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein science : a publication of the Protein Society. 2013;22:74–82. doi: 10.1002/pro.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sprang SR. G protein mechanisms: insights from structural analysis. Annual review of biochemistry. 1997;66:639–78. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 14.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2754–9. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leaver-Fay A, Tyka M, Lewis SSM, Lange OF, Thompson J, Jacak R, Kaufman KW, Renfrew PD, Smith CA, Sheffler W, et al. Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. In: MLJ, LBBT-M, editors. Methods in enzymology. Vol. 487. Academic Press; 2011. pp. 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desjarlais JR, Clarke ND. Computer search algorithms in protein modification and design. Current opinion in structural biology. 1998;8:471–5. doi: 10.1016/s0959-440x(98)80125-5. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460:855–62. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemba M, Gardner KHK, Marino S, Clarke ND, Regan L. Nature structural biology. Vol. 2. New York, NY: Nature Pub. Co; 1995. Novel metal-binding proteins by design; pp. 368–373. c1994–c2003. [DOI] [PubMed] [Google Scholar]
- 19.Salgado E. Metal-directed protein self-assembly. Accounts of Chemical Research, American Chemical Society. 2010;43:661–672. doi: 10.1021/ar900273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado EN, Ambroggio XI, Brodin JD, Lewis Ra, Kuhlman B, Tezcan FA. Metal templated design of protein interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1827–32. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Der BS, Machius M, Miley MJ, Mills JL, Szyperski T, Kuhlman B. Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer. Journal of the American Chemical Society. 2012;134:375–85. doi: 10.1021/ja208015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. Computational de novo Design, and Characterization of an A2B2 Diiron Protein. Journal of Molecular Biology. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 23.Der B, Jha R. Combined computational design of a zinc binding site and a protein-protein interaction: One open zinc coordination site was not a robust hotspot for de novo ubiquitin. Proteins: Structure, Function, and Bioinformatics. 2013 doi: 10.1002/prot.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohl C, Strauss C, Misura K, Baker D. Protein structure prediction using Rosetta. Methods in enzymology. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 25.Joachimiak La, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. Journal of molecular biology. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]