Abstract
The cervix and/or fallopian tubes of pigtailed macaques were experimentally infected with Chlamydia trachomatis. Their sera were collected at varying time points and screened for identification of immunodominant antigens using a whole-genome protein microarray. The effect of doxycycline treatment on the antibody response generated in these macaques was also investigated. Twenty-five female macaques were infected with C. trachomatis serovars D or E in the cervix and/or fallopian tubes. Bloods were collected at baseline and at various intervals after challenge. Serum samples were tested for antibodies using a C. trachomatis serovar D protein microarray. Twenty chlamydial antigens reacted with sera from at least 68% (17/25) of the macaques. In addition to some well-known chlamydial antigens, nine different proteins, not previously recognized as immunodominant, including four hypothetical proteins (CT005, CT066, CT360 and CT578), were identified. Monkeys infected in the fallopian tubes developed a more robust antibody response than animals inoculated in the cervix. Treatment with doxycycline significantly decreased Chlamydia-specific antibody levels. In summary, using protein microarray serum samples from experimentally infected pigtailed macaques were screened for immunodominant chlamydial antigens. These antigens can now be tested in animal models for their ability to protect and as markers of disease progression.
Keywords: Chlamydia trachomatis, pigtailed macaques, Macaca nemestrina, antigen, protein microarrays, antibody response, serum, cervix, fallopian tubes, antibiotics
Introduction
In developed countries C. trachomatis is the most common bacterial sexually transmitted infection while in regions of the world with poor sanitary conditions this pathogen is the most common cause of preventable blindness [1, 2]. Attempts to control these infections using antibiotics have not been effective. For example, in regions where screening programs, followed by antibiotic treatment, have been implemented, an increase in the number of cases has been reported [3, 4]. Therefore, to eradicate Chlamydia, a vaccine is likely the most efficacious approach. A whole organism vaccine against trachoma resulted in a hypersensitivity reaction in some immunized individuals upon re-exposure to this pathogen [2, 5, 6]. The possibility that an antigenic component of Chlamydia mediated this hypersensitivity reaction prompted the abandonment of a whole organism vaccine and stimulated the search for a subunit formulation [7–9]. Due to the large number of proteins present in most pathogens it is difficult to identify those that are antigenic. However, recent advances generating whole proteome arrays have led to rapid screening methods to identify proteins that can generate an immune response [10–12].
Non-human primates are the only animal model naturally susceptible to infection with the C. trachomatis human serovars and therefore, are the ideal system for testing vaccines [13–15]. The genetic variability in the human population requires Chlamydia vaccines that include antigens that can be recognized by individuals with multiple immunogenetic backgrounds. In this study, we screened sera from 25 pigtailed macaques (Macaca nemestrina) previously infected in the lower and/or upper genital tract with C. trachomatis. We identified 20 immunodominant antigens that can now be tested for their ability to induce protective responses. The effects of antibiotic therapy on the immune response to chlamydial infection were also investigated.
Materials and Methods
Animals
Sexually mature female pigtailed macaques (Macaca nemestrina) were enrolled in these studies. All 25 macaques were housed at the Washington National Primate Research Center at the University of Washington, Seattle, WA [15]. Approval was obtained from the Institutional Animal Care and Use Committee at the University of Washington. Animals were handled humanely within the National Institutes of Health's animal use guidelines. To rule out previous or current chlamydial infection all animals were prescreened by cervical culture and serology.
Infection of pigtailed macaques with C. trachomatis
Animals underwent different C. trachomatis infection regimes using serovars D (P0124) or E (MTW477) (Table 1) [13, 14]. In one study, a single cervical inoculation with serovar E was delivered with a 1 ml tuberculin syringe into the vaginal fornix, thereby exposing the cervix to the organism. In the second study, five weekly cervical challenges were done to establish chronic chlamydial infection. To establish a chronic chlamydial infection of the upper reproductive tract, in the third study, the fallopian tubes were inoculated directly through the fimbrial os at 2-week intervals. Bloods were collected at baseline and at various intervals after challenge. Following infection macaques were treated with antibiotics or placebo as shown in Table 1. At the time the experiments were performed combination therapy with different agents was included.
Table 1.
Pigtailed macaques entered in the experiment C. trachomatis serovar used to infect, site of inoculation, antibiotic treatment and number of samples collected
| # Pigtaile d macaqu es |
Inoculati on site |
# serum samples before inoculati on |
Chlamy dia serovar |
# of inoculatio ns |
# serum samples after inoculati on |
# Pigtaile d macaqu es |
Treatment | # serum sampl es after tx |
|---|---|---|---|---|---|---|---|---|
| 15 | Fallopian tubes |
6 | Doxycyclin e: 2.2mg/kg/d ay orally, 10 days Doxycyclin e: |
16 | ||||
| 9 | 6 × 106 Serovar D (PO124) |
3X | 13 | 1 | 2.2mg/kg/d ay orally, 10 days + Triamcinolo ne: 0.2mg/kg each, 3 days |
1 | ||
| 2 | Placebo x 10 days Doxycyclin |
9 | ||||||
| 2 | 5 × 106 Serovar E |
2X | 3 | 2 | e: 2.2mg/kg/d |
3 | ||
| (MTW47 7) | ay orally, 10 days Doxycyclin e: |
|||||||
| 1 | 2.2mg/kg/d ay orally, 10 days Doxycyclin |
1 | ||||||
| 4 | 6 × 106 Serovar E |
2X | 6 | e: 2.2mg/kg/d ay orally, |
||||
| (MTW47 7) | 2 | 10 days + Triamcinolo ne: 0.2mg/kg each, 3 days |
4 | |||||
| 1 | Placebo x 10 days |
0 | ||||||
| 5 × 104 Serovar |
4 | Placebo x 14 days |
7 | |||||
| 10 | Cervix | 7 | E (MTW47 7) |
5X | 10 | 3 | Placebo x 14 days |
0 |
| 3 | 5 × 104 Serovar E (MTW47 7) |
1X | 8 | 3 | Azithromyci n: 14mg/kg each, 5 days |
0 |
Production of C. trachomatis proteome microarray chips
The C. trachomatis protein microarray chips were prepared following a three steps process: 1) PCR amplification of the 894 open reading frames (ORF); 2) in vivo recombination cloning, and 3) in vitro transcription/translation followed by microarrays chip printing (Antigen Discovery, Inc., Irvine, CA). The C. trachomatis serovar D (UW-3/Cx; ATCC) genomic specific PCR primers were designed using 20 bp of the gene-specific sequence and 33 bp of adapter sequences [10, 11, 16, 17]. The adapter sequences were designed to be homologous to the cloning site of the linearized T7 expression vector pXT7 and allowed the PCR products to be cloned by homologous recombination in DH5α E. coli cells. At the 5' end of the fusion protein a polyhistidine (His) fragment was incorporated and at the 3' end, a hemagglutinin (HA) sequence and a T7 terminator were included. Plasmids with Chlamydia specific sequences were expressed using an in vitro transcription-translation system (RTS 100 kit; Roche). Microarrays were printed onto nitrocellulose coated glass slides (GraceBio) using an OmniGrid Accent microarrays printer (Digilab). Full-length protein expression was monitored in the microarrays by using anti-polyhistidine (clone His-1; Sigma) and anti-hemagglutinin antibodies (clone 3F10; Roche).
Microarray probing and data collection
A total of 106 serum samples were tested for the presence of antibodies using the C. trachomatis serovar D microarray. Briefly, serum samples were diluted 1:100 with 1X protein array blocking buffer (Whatman, Piscataway, NJ) containing 10% Escherichia coli lysate (McLab, San Francisco, CA) and incubated at room temperature for 30 minutes with constant agitation. The microarrays were rehydrated in 1X protein array blocking buffer and probed with serum samples [10]. The slides were washed and incubated with biotin-conjugated goat anti-human antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). The bound secondary antibodies were detected using streptavidin-conjugated Sensilight P3 (Columbia Biosciences, Columbia, MD). The slides were scanned in a ScanArray Express HT microarray scanner (Perkin Elmer, Waltham, MA), and the fluorescence signal was quantified (QuantArray software; Perkin Elmer, Waltham, MA). Proteins were spotted in triplicate on each array
Bioinformatics analysis
Data processing and normalization
Antigen-specific signal intensities were first corrected for background noise (QuantArray software; Perkin Elmer). Antigens were spotted in triplicate on each array and the average of the three background corrected spots was used as the signal intensity. The signal intensities were then normalized using the variance stabilization and calibration for microarray data (VSN) package implemented in R [18, 19]. The VSN model was build using only the 192 no DNA control spot intensities for each sample, and then the model was applied to the antigen signal intensities.
Reactive antigen selection
To define reactive antigens the following treatment of the data was applied:
Transform VSN normalized data back to the raw scale
Calculate the mean and standard deviation (SD) of no DNA spots for each of the 106 samples
Subtract the no DNA mean plus two standard deviations (no DNA mean + 2*SD) from each antigen intensity
Subtract the pre-inoculation sample intensity
Remaining intensities greater than 0 are considered reactive
These criteria for defining reactivity were developed in previous projects based on the same technology [10, 12, 20]. This form of the data was not used for statistical analysis, but only for a coarse determination of reactivity with respect to background and the initial pre-inoculation
Prediction of cellular role
Computational prediction of protein cellular role, enzyme class, and gene ontology were downloaded from Comprehensive Microbial Resource website (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi).
Statistical analysis
Only the VSN normalized data was used for statistical analysis. The statistical significance of increases in signal intensity following inoculation was calculated using Cyber-T differential analysis software [21, 22] to perform a Bayes-regularized t-test. The groups being compared were (1) the pre-inoculation samples of the 25 subjects and (2) the latest post-inoculation time point samples before any treatment was given to the 25 subjects. For each protein, the null hypothesis was that the mean of the intensities of the pre-inoculation samples was greater than or equal to the mean of the intensities of the 25 post-inoculation samples. The alternative hypothesis was that the mean of the post inoculation samples was greater. The results of these tests are presented in the Results subsection “Identification of C. trachomatis immunodominant antigens” and in Figure 1.
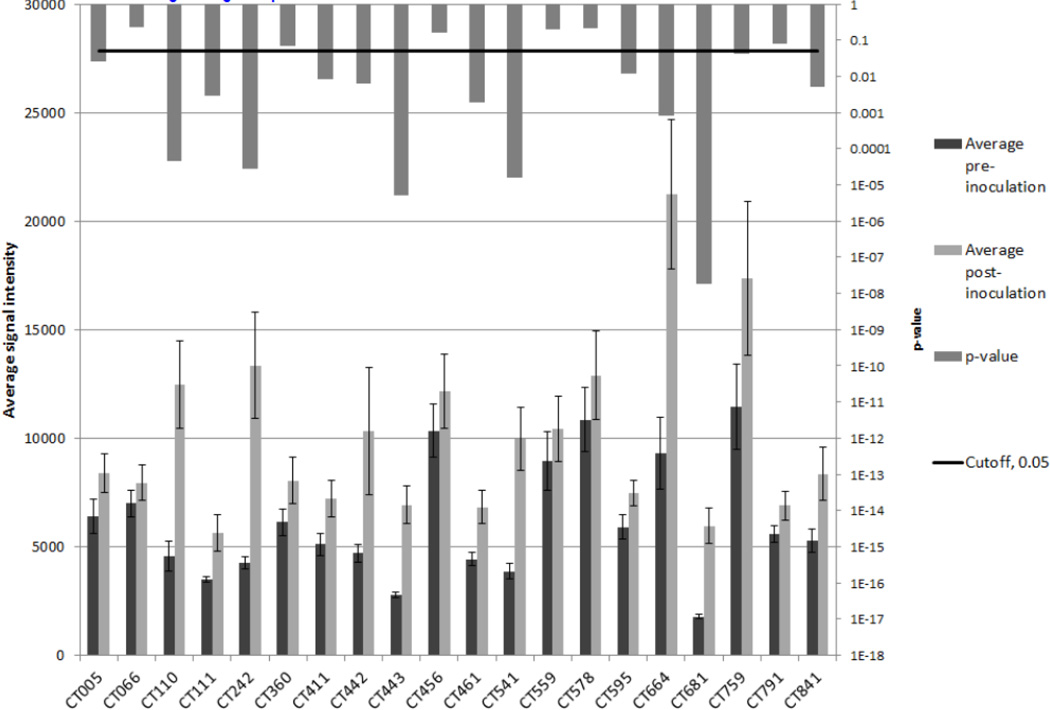
Figure 1. Comparison of pre-inoculation and post-inoculation average signal intensities for the immunodominant antigens.
The pre-inoculation samples (n=25) were compared to the set of the latest post-inoculation samples for the subjects (n=25). For each of the 20-immunodominant antigens the barplots compare the average signal intensities (left vertical axis) of the groups and the error bars show standard error. The vertical bars descending from the top of the figure indicate the p-values resulting from t-tests (right vertical axis).
The same approach was used to compare: (1) fallopian tube inoculation versus cervix inoculation – results in Figure 2a; (2) serovar D versus serovar E – results in Figure 2b; (3) pre-doxycycline treatment versus post-doxycycline treatment – results in Figure 3a; and (4) pre-placebo treatment versus post-placebo treatment – results in Figure 3b.
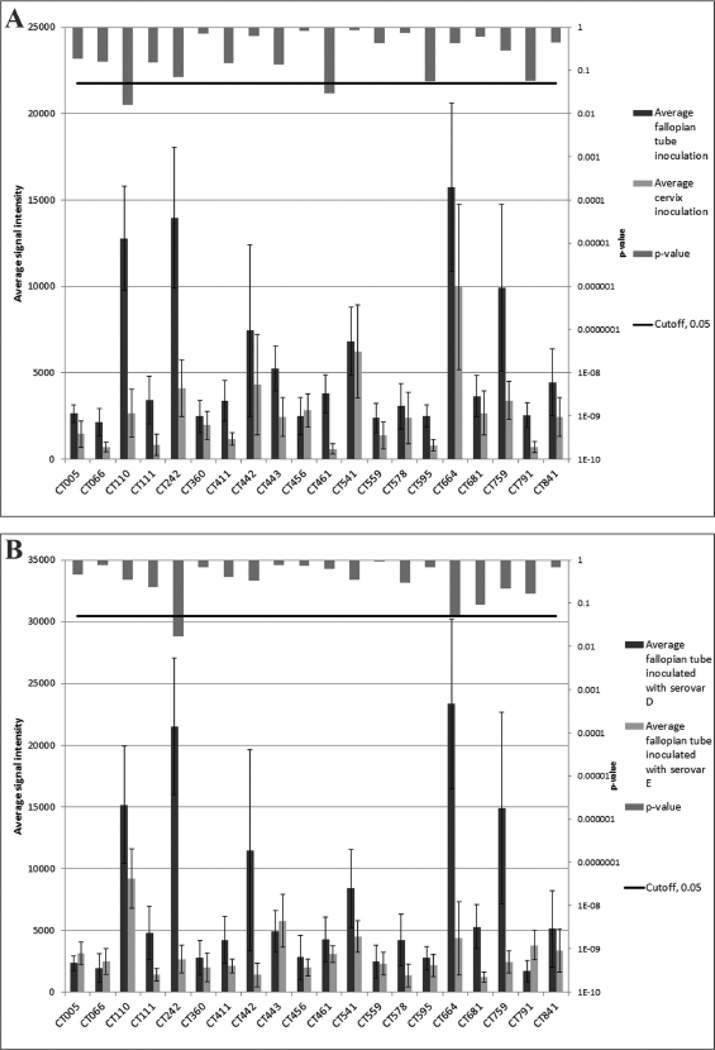
Figure 2. Comparison of inoculation site (a) and serovar type (b) average signal intensities to immunodominant antigens.
Panel (a) compares serum samples from pigtailed macaques inoculated in the cervix to those inoculated in the fallopian tubes. Panel (b) compares serum samples from pigtailed macaques inoculated with serovar D to those inoculated with serovar E. For each of the 20-immunodominant antigens the barplots compare the average signal intensities (left vertical axis) of the groups and the error bars show standard error. The vertical bars descending from the top of the figure indicate the p-values resulting from t-tests (right vertical axis).
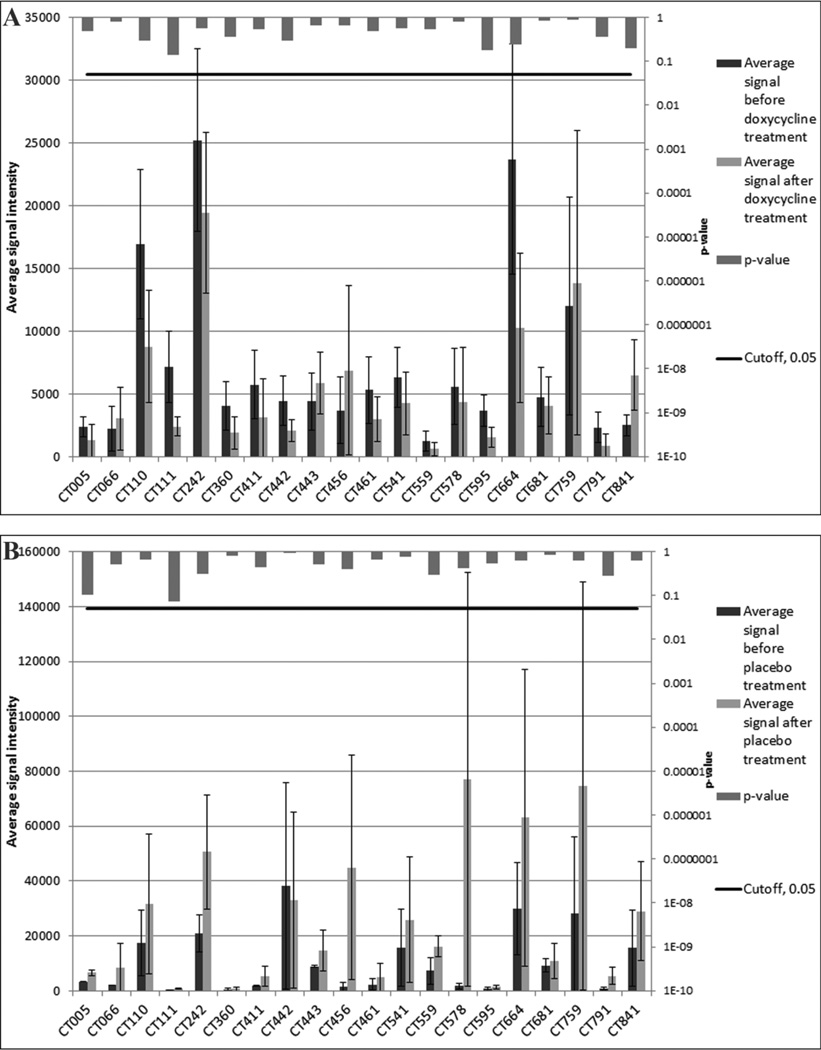
Figure 3. Comparison of pretreatment and post-treatment with doxycycline (a) and placebo (b) average signal intensities to immunodominant antigens.
Panel (a) compares serum samples from pigtailed macaques treated with doxycycline before and after treatment. Panel (b) compares serum samples from pigtailed macaques treated with placebo before and after treatment For each of the 20 immunodominant antigens the barplots compare the average signal intensities (left vertical axis) of the groups and the error bars show standard error. The vertical bars descending from the top of the figure indicate the p-values resulting from t-tests (right vertical axis).
The Chi-square and Fisher’s exact tests (http://www.graphpad.com/quickcalcs/contingency1.cfm) were used to calculate p-values for the inoculation site, serovar E versus F, and the cell function comparisons.
Results
Identification of C. trachomatis immunodominant antigens
The protein microarray included the expression products of 894 unique ORFs of C. trachomatis serovar D as well as positive and negative controls. Tag detection was used as a measure of protein expression. Of the 894 ORF arrayed, 864 (96.4%) were positive for both the N-terminal poly-His and the C-terminal HA tags. A summary of the intra- and inter- array coefficients of variation (CVs) is presented in Supplementary Materials.
Microarrays were used to profile the antibody response in 106 serum samples collected from 25 pigtailed macaques that included 25 baselines samples, 40 after infection with Chlamydia and 41 after antibiotic or placebo treatment (Table 1). A total of 789 antigens were reactive with at least one serum sample, 184 were reactive with at least 52% (13/25) of the pigtailed macaques, and 54 were reactive with at least 68% (17/25). Considering only sera from macaques infected in the fallopian tubes with serovar D, 674 antigens reacted with at least one sera. Considering only sera from animals inoculated with serovar E, 600 antigens were reactive with at least one sera sample (Tables 1 & 2; on-line). Considering only sera from animals infected in the cervix with serovar E, 684 antigens were reactive. A total of 517 antigens were reactive with at least one serum from the three subsets of subjects described above.
From the 517 common reactive antigens we selected 20 that gave a positive signal with at least one serum sample from 17 (68%) of the 25 macaques and with a signal greater than 2,000 in at least one of the inoculation routes (Table 2). Figure 1 (on line) provides a global view of the increased reactivity levels following inoculation with each dot representing one of the 894 antigens. In the figure the x- and y-axis represent the pre- and post-inoculation mean reactivity values and the 20 selected antigens are indicated by larger dots.
Table 2.
Number of monkeys (% +) that gave positive signals with the 20 immunodominant antigens following infection with C. trachomatis
|
C. trachomatis geneID no. |
No. (%) of positive monkeys w/ fallopian tube inoc |
No. (%) of positive monkeys w/ cervix inoc |
No. (%) of positive monkeys w/ serovar D fallopian tube inoc |
No. (%) of positive monkeys w/ serovar E fallopian tube inoc |
Total number of positive monkeys (%) |
Common name for protein | Nucleic acid length |
|---|---|---|---|---|---|---|---|
| CT005 | 13 (87) | 7 (70) | 7 (78) | 6 (100) | 20 (80) | Hypothetical protein CT005 | 1092 |
| CT066 | 13 (87) | 6 (60) | 8 (89) | 5 (83) | 19 (76) | Hypothetical protein CT066 | 477 |
| CT110 | 14 (93) | 6 (60) | 8 (89) | 6 (100) | 20 (80) | Chaperonin GroEL; 60-kDa hsp | 1635 |
| CT111 | 13 (87) | 4 (40) | 8 (89) | 5 (83) | 17 (68) | Co-chaperonin GroES; 10-kDa hsp | 309 |
| CT242 | 13 (87) | 7 (70) | 9 (100) | 4 (67) | 20 (80) | OmpH-like outer membrane protein | 522 |
| CT360 | 11 (73) | 9 (90) | 7 (78) | 4 (67) | 20 (80) | Hypothetical protein CT360 | 627 |
| CT411 | 14 (93) | 6 (60) | 8 (89) | 6 (100) | 20 (80) | Lipid-A-disaccharide synthase | 1824 |
| CT442 | 11 (73) | 9 (90) | 7 (78) | 4 (67) | 20 (80) | 15 kDa cysteine-rich protein | 453 |
| CT443 | 14 (93) | 5 (50) | 8 (89) | 6 (100) | 19 (76) | 60kD cysteine-rich outer membrane protein |
1662 |
| CT456 | 11 (73) | 9 (90) | 6 (67) | 5 (83) | 20 (80) | Hypothetical protein CT456 | 3018 |
| CT461 | 14 (93) | 4 (40) | 9 (100) | 5 (83) | 18 (72) | Phosphohydrolase | 990 |
| CT541 | 14 (93) | 7 (70) | 8 (89) | 6 (100) | 21 (84) | Peptidyl-prolyl cis-trans isomerase | 732 |
| CT559 | 12 (80) | 7 (70) | 7 (78) | 5 (83) | 19 (76) | Yop proteins translocation lipoprotein J | 981 |
| CT578 | 12 (80) | 7 (70) | 9 (100) | 3 (50) | 19 (76) | Hypothetical protein CT578 | 1464 |
| CT595 | 13 (87) | 6 (60) | 8 (89) | 5 (83) | 19 (76) | Thio:disulfide interchange protein | 2079 |
| CT664 | 15 (100) | 7 (70) | 9 (100) | 6 (100) | 22 (88) | FHA domain-containing protein | 2490 |
| CT681 | 13 (87) | 6 (60) | 8 (89) | 5 (83) | 19 (76) | Major outer membrane protein | 1182 |
| CT759 | 10 (68) | 9 (90) | 6 (67) | 4 (67) | 19 (76) | Muramidase | 738 |
| CT791 | 13 (87) | 4 (40) | 8 (89) | 5 (83) | 17 (68) | Excinuclease ABC subunit C | 1797 |
| CT841 | 13 (87) | 7 (70) | 8 (89) | 5 (83) | 20 (80) | ATP-dependent zinc protease | 2742 |
The selected immunodominant antigens included five proteins known to give positive signals with sera from humans infected with Chlamydia and with sera from experimental animals, mainly mice and guinea pigs: CT110 (chaperonin GroEL; 60k–Da heat-shock protein; hsp60), CT111 (chaperonin GroES; 10-kDa heat-shock protein; hsp10), CT442 (15-kDa-cysteine-rich protein; 15-kDa crp), CT443 (60-kDa-cysteine-rich protein; 60-kDa crp) and CT681 (major outer membrane protein; MOMP). Two additional proteins identified here, CT841 and CT456, have also being recognized as immunodominant in humans and another four, CT541, CT559, CT664 and CT759, were previously detected using mouse sera. The other nine proteins, including four hypothetical proteins (CT005, CT066, CT360 and CT578), have not previously being identified as immunodominant antigens.
The average signal intensity of pre-inoculation samples and post-inoculation samples and the associated significance of the difference for each of the immunodominant antigens are presented in Fig. 1. The unadjusted p-values of 14 out of the 20 immunodominant antigens were less than 0.05. Five of these antigens were significant after Benjamini and Hochberg (BH) multiple test correction [23]: CT681, CT443, CT541, CT242, and CT110. Of the remaining 874 antigens 36 had unadjusted p-values less than 0.05 and none were significant after BH correction.
The selection of the 20 immunodominant antigens was performed as described above (i.e. clear increases following inoculation in at least 68% of the macaques) because in order to be of interest as a potential vaccine candidate, or to be useful as a marker for disease progression or treatment, an antigen must generate a strong immune response in a significant majority of the macaques. Alternative approaches for selecting antigens (e.g. statistical significance, mean increase) do not guarantee that these criteria would be met; however, the average intensity increases following inoculation were significant for 5 of 894 proteins after BH correction and all 5 were among the set of 20 immunodominant antigens.
The immunodominant antigens were also utilized for the additional comparisons in this study (i.e. inoculation site, serovar, and treatment). When all 894 proteins are tested for each of the additional comparisons none of the results are significant after BH correction; thus, no significant result is excluded by presenting results only for the immunodominant antigens.
Distribution of positive immunodominant antigens based on site of infection and C. trachomatis serovar used to infect
To determine if we could identify antigens preferentially associated with upper or lower genital tract infections the results were evaluated based on inoculation site (Table 2). More macaques inoculated in the fallopian tubes gave positive signals with the immunodominant antigens than animals infected in the cervix. Four of the 20 antigens CT111, CT443, CT461 and CT791, were positive at clearly higher frequency in animals infected in the fallopian tubes when compared with those infected in the cervix (Table 3). These four proteins were positive in 87 to 93% of the monkeys after tubal challenge but were only positive in 40 to 50% of the monkeys after cervical challenge. For the remaining 16 proteins, 15 showed similar percentage of positive monkeys in fallopian tubes and cervix infection groups. One protein, CT759, had a higher percentage of positive monkeys in the cervix inoculation group.
Table 3.
Antibody responses in monkeys to the 20 immunodominant C. trachomatisantigens following antibiotic/placebo treatment
| ID | Fallopian tube Serovar D, Doxy tx |
Fallopian tube Serovar D, Placebo tx |
p-value | ||
|---|---|---|---|---|---|
| Increase | Decrease | Increase | Decrease | ||
| CT005 | 1 | 4 | 2 | 0 | 0.1429 |
| CT066 | 5 | 1 | 1 | 1 | 0.4643 |
| CT110 | 1 | 5 | 2 | 0 | 0.1071 |
| CT111 | 0 | 6 | 2 | 0 | 0.0357 |
| CT242 | 2 | 4 | 2 | 0 | 0.4286 |
| CT360 | 1 | 5 | 1 | 0 | 0.2857 |
| CT411 | 0 | 6 | 2 | 0 | 0.0357 |
| CT442 | 2 | 3 | 1 | 1 | 1.0000 |
| CT443 | 4 | 1 | 1 | 1 | 1.0000 |
| CT456 | 1 | 4 | 2 | 0 | 0.1429 |
| CT461 | 1 | 5 | 1 | 1 | 0.4643 |
| CT541 | 2 | 4 | 2 | 0 | 0.4286 |
| CT559 | 1 | 4 | 2 | 0 | 0.1429 |
| CT578 | 1 | 5 | 2 | 0 | 0.1071 |
| CT595 | 0 | 6 | 1 | 1 | 0.2500 |
| CT664 | 0 | 6 | 1 | 1 | 0.2500 |
| CT681 | 1 | 4 | 1 | 1 | 1.0000 |
| CT759 | 2 | 2 | 2 | 0 | 0.4667 |
| CT791 | 0 | 6 | 2 | 0 | 0.0357 |
| CT841 | 5 | 1 | 2 | 0 | 1.0000 |
The signal intensity for the 20-immunodominant antigens was also stronger in sera from macaques inoculated in the fallopian tubes than in those infected in the cervix (Fig. 2a). Of the 20 antigens, 19 (95%) gave stronger average signals with sera from macaques infected in the fallopian tubes when compared with those inoculated in the cervix. Out of these 19 antigens with higher average signal for fallopian tubes CT110 and CT461 resulted in unadjusted p-values < 0.05; however, neither result is significant after BH correction [23]. Only CT456 had a stronger signal in macaques inoculated in the cervix when compared with those infected in the fallopian tubes.
When comparing the percentage of pigtailed macaques infected with C. trachomatis serovars D or E in the fallopian tubes that gave a positive signal, no significant differences were observed for any of the 20-immunodominant antigens (Table 2). When comparing the signal intensities between macaques infected with serovar D versus serovar E in the fallopian tubes, CT242 was the only immunodominant antigen that resulted in an unadjusted p-value < 0.05; however, the result is not significant after BH correction (Fig. 2b). In addition, none of the other 894 antigens result in a significant difference after BH correction.
Effect of antibiotic treatment on antibody responses
To determine if antibiotic treatment affects antibody responses elicited by C. trachomatis infection we evaluated sera from monkeys infected with serovar D in the allopian tubes. As shown in Table 3, most macaques treated with doxycycline showed a decrease in signal intensity for 16 (80%) of the 20-immunodominant antigens. For three (15%) proteins, CT066, CT443, CT841, there was an increase in signal intensity and for (CT759), equal number of monkeys had an increase or a decrease in signal following antibiotic treatment. In animals treated with placebo the opposite trend occurred. In more macaques there was an increase in signal intensity for 13 (65%) of the antigens, while for the other seven proteins no change was observed.
The average signal intensity for the immunodominant antigens before and after doxycycline treatment of monkeys infected in the fallopian tubes with serovar D is shown in Fig. 3a. For 15 of the 20 antigens the average signals were higher before than after doxycycline treatment. For the other five antigens CT066, CT443, CT456, CT759 and CT841, the average signal intensities increased following treatment. In contrast, for the placebo treated monkeys the signal intensity increased for all, but one (CT442), of the antigens. Using an odds ratio confidence interval test, the observed decreased in signal in monkeys treated with antibiotic has an odds ratio of 10.1729, which falls within the 95% confidence interval (between 4.1288 to 25.0652). None of the doxycycline or placebo comparisons using the 20-immunodominant antigens resulted in an unadjusted p-value < 0.05 and none of the other 894 antigens was significant after BH correction.
Assignment of cell function
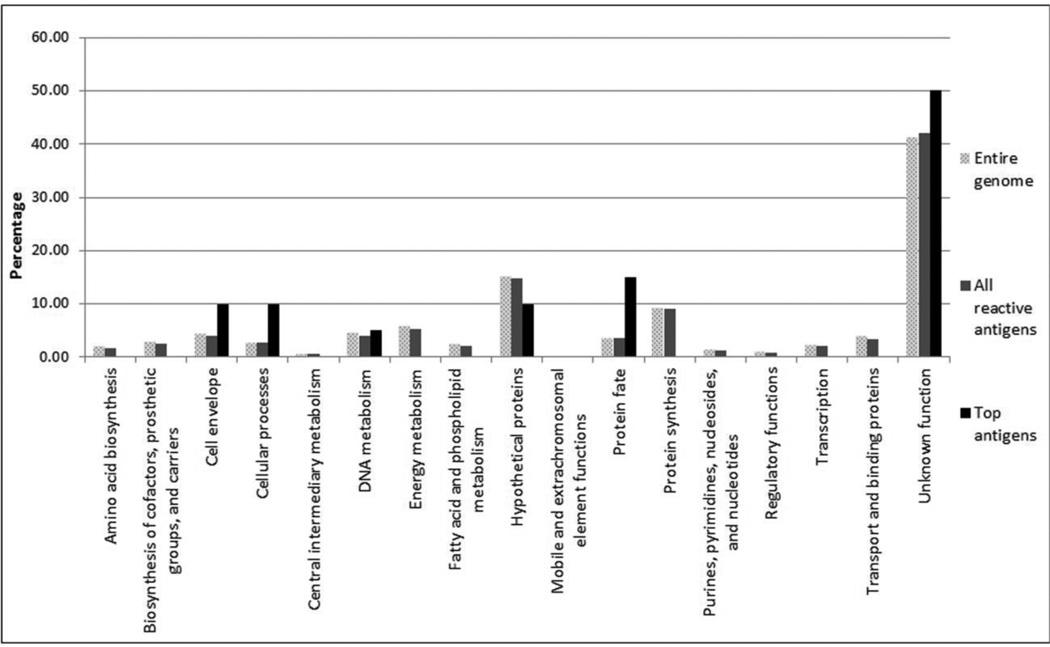
Using the Comprehensive Microbial Resource (CMR) from the J. Craig Venter Institute (JCVI; http://www.jcvi.org), we assigned the predicted cellular roles to the 894 Chlamydia ORFs, the 789 proteins that gave a positive signal with at least one serum and the 20-immunodominant antigens selected using the protein microarray. Each protein is assigned to one main cellular role category (Fig. 4). The greatest number (56.78%) of the total positive antigens is categorized as unknown function (hypothetical proteins). Most of the categorized cellular roles of all the identified positive antigens are in close proportion to the percentage represented in the C. trachomatis genome. In contrast, proteins from the cell envelope, cellular processes, protein fate and unknown function categories have a higher representation in the immunodominant antigen selection when compared with the positive antigens, or the entire genome, and for the protein fate group this difference is significant (P<0.05).
Figure 4. Functional roles of the ORFs of the whole C. trachomatis serovar D genome, all the positive antigens identified following infection of the pigtailed macaques and the 20 selected immunodominant antigens.
The distribution of the predicted roles of the 20-immunodominant antigens is compared to the whole C. trachomatis serovar D and to all the reactive chlamydial antigens using the JCVI cellular role categories.
Discussion
Archived serum samples from pigtailed macaques, infected with C. trachomatis in the cervix or in the fallopian tubes, were used to identify chlamydial antigens that elicited a humoral immune response. A protein microarray corresponding to 96.4% of the ORF of the C. trachomatis serovar D was used to screen the samples. We selected 20 chlamydial proteins that were recognized by sera from 68% (17/25) of the macaques. In addition to well-known chlamydial antigens such as MOMP, the 15- and 60-kDa crp and the 10-kDa and 60-kDa hsp, we identified nine antigens not previously recognized as immunodominant. Macaques infected in the fallopian tubes mounted a more robust immune response and reacted with antigens that may be used as markers of upper genital pathology. Furthermore, we showed that antibiotic treatment following C. trachomatis infection decreases the antibody response. The immunodominant antigens we have identified can now be tested in animal models to determine their ability to elicit protection and evaluate their potential role as markers of disease progression.
Implementation of high throughput screening platforms, such as the one utilized here facilitates the characterization of immune responses to multiple antigens. Some of these approaches have being applied to the identification of C. trachomatis vaccine candidates. For example, Wang et al. [24] using recombinant proteins coated in microtiter plates, tested sera from 99 females with acute C. trachomatis urogenital infection. From the 908 C. trachomatis proteins screened, they selected 27 that were recognized by more than 50% of the antisera. Six of the antigens, that Wang et al. [24] identified as dominant are also included in our list of 20 immunodominant proteins (CT110, CT442, CT443, CT456, CT681 and CT841). Similarly, to identify antibody-producing proteins, Finco et al. [25] utilized a panel of 120 serovar D recombinant proteins to screen sera from C. trachomatis infected patients. The 79 proteins that gave positive results were used to stimulate splenocytes from C. trachomatis-infected mice. Five of the proteins (CT119, CT372, CT443, CT681 and CT823) induced CD4+/IFN-γ. Two of the proteins (CT443 and CT681) identified by Finco et al. [25] as having T- and B-cell epitopes are included in the list of 20 immunodominant antigens selected in this study. Also, to select B and T-cell epitopes, Follmann et al. [26] used a panel of 116 recombinant C. trachomatis serovar D proteins to screen serum from 46 and peripheral blood mononuclear cells, from 10 C. trachomatis infected patients. CT082, CT089, CT322, CT396 and CT681 proteins bound antibodies, CT004, CT043, CT184, CT509 and CT611, were recognized exclusively by T cells and CT110 and CT443 were identified by both, antibodies and T cells. The last two proteins, CT110 and CT443, in addition to CT681, are included in our list of immunodominant antigens. From the immunodominant antigens identified in these studies, MOMP (CT681) is the protein that has been shown to induce the most robust protection in various animal models, including non-human primates [7, 27–30]. In a mouse model, Olsen et al. [31] have shown limited protection with a recombinant fusion protein of CT443 and CT521.
Trying to identify markers of disease progression Rodgers et al. [32] used an array of 30 serovar D recombinant proteins to screen sera from 31 patients with tubal factor infertility (TFI) and 23 sera from infertile patients. Based on the ability of sera to react with the 30 chlamydial antigens, they concluded that using a combination of CT381 and CT443, as reactive markers, they had 67% sensitivity and 100% specificity to identify patients with TFI. In our study, CT111, CT443, CT461 and CT791 were detected by the majority (~90%) of samples from monkeys infected in the fallopian tube while less than 50% of sera from animals infected in the cervix gave positive signals. Therefore, our analysis confirms the finding by Rodgers et al. [32] that reactivity with the CT443 (60kDa crp) is preferentially associated with upper genital pathology. Interestingly, it has previously being shown that CT443, through antigenic mimicry on the signal peptide, may be involved in the pathogenesis of Chlamydia-related heart diseases [33]. It is possible that a similar immune mechanism may account for the association between antibodies to this protein and upper genital pathology. Neither Rodgers et al. [32], nor this study, found an association between upper genital infection and the 60kDa hsp (CT110). It has been proposed that this protein is the antigen responsible for the hypersensitivity reaction observed during the trachoma vaccine trials and some studies, have also found an association of reactivity to the 60kDa hsp and upper genital tract pathology [34–36].
We also analyzed the effect of antibiotic treatment on the antibody response in pigtailed macaques infected in the fallopian tubes with C. trachomatis serovar D. Countries that have implemented screening programs for C. trachomatis, followed by antibiotic treatment, have noticed an increase in the prevalence of the infection [3, 4]. Treatment with antibiotics was proposed to result in weakening of the immune response to Chlamydia and therefore, possibly increased susceptibility to reinfection and/or recurrences [4, 37]. There is evidence in humans and animal models to support this hypothesis [38, 39]. Our results confirm that there is decline in antibody levels following antibiotic treatment therefore, supporting this hypothesis. Antibodies are known to play a critical role in protection against Chlamydia and therefore, reduction in antibody levels may render an individual more susceptible to infection [40, 41].
Our study has several limitations. The microarray used here is a bacterial-based expression system that does not control for post-translational modifications, such as phosphorylation, glycosylation or lipidation. Therefore, antibodies that recognized those structures may not be identified. However, using vaccinia virus arrays produced by this method, all the known glycosylated proteins were recognized by sera from immunized humans and animals [20]. Since proteins on these arrays were not glycosylated, these findings suggest that at least some antibody responses are directed against epitopes not post-translationally modified.
Similar limitations may occur with conformational epitopes and those formed by disulfide bonds. The Chlamydia MOMP (CT681) has disulfide bonds and forms trimers in the outer membrane [42, 43]. Monoclonal antibodies that only bind to the MOMP trimer do not recognize the MOMP printed on this array, but monoclonals to linear epitopes do react (unpublished data). However, as shown here, anti-MOMP antibodies produced in monkeys following infection with live Chlamydia were detected with this array.
The limitations of all the screening assays may explain, in addition to the different types of samples analyzed, the apparent discrepancies between some studies. In this respect, most investigators have chosen to use proteins from the C. trachomatis serovar D in their platforms since its genome sequence is available [44]. Although, there is more than 98% DNA homology between the 15 C. trachomatis serovars, single nucleotide polymorphism and mutations may result in antigenic variation that may be reflected in the immune response to certain proteins. Therefore, screening human samples with proteins from a single C. trachomatis serovar may bias the results since multiple serovars cause infections. For example, our results will suggest that serovar D elicits a more robust humoral immune response than serovar E. However, we cannot be certain of this conclusion since our microarray utilized serovar D proteins. However, certain Chlamydia proteins are consistently identified by different methodologies using various types of samples and therefore, these antigens may be the best candidates for further testing.
In conclusion, using sera from C. trachomatis-infected pigtailed macaques we have identified immunodominant antigens that can be tested in animal models for their ability to protect against this pathogen. Amongst the immunodominant antigens some were predominantly recognized by sera from macaques inoculated in fallopian tubes rather than in the cervix and therefore, may be markers for upper genital tract pathology. In addition, we have shown that antibiotic treatment following infection weakens antibody responses and thus, may make the individual more susceptible to reinfection. Screening patients infected with C. trachomatis with whole proteome arrays may help stage the infection and the response to therapy.
Supplementary Material
The figure provides a global view of the increase in reactivity following inoculation. The x-axis represents the mean of pre-inoculation samples and the y-axis represents the mean of post-inoculation samples. The pre-inoculation samples (n=25) were compared to the set of the latest post-inoculation samples for the subjects (n=25). The larger black dots indicate the 20 selected immunodominant antigens. The identify line (y=x) is shown with dashes.
Significance.
This is the first time that Chlamydia trachomatis immunodominant antigens have been identified in pigtailed macaques following a uterine cervix or a fallopian tubes infection. These immunodominant antigens can now be used to vaccinate non-human primates and determine their ability to protect against a C. trachomatis genital infection. Proteins that are protective can subsequently be tested in humans. Amongst the immunodominant antigens some were predominantly recognized by sera from macaques inoculated in the fallopian tubes rather than in the cervix and therefore, may be markers for upper genital tract pathology. In addition, treatment with doxycycline following infection significantly decreased Chlamydia-specific antibody levels. This information can be used to evaluate the efficacy of antibiotic treatment and potentially susceptibility to reinfection.
Highlights.
-
-
Pigtailed macaques were infected with Chlamydia trachomatis in the cervix and fallopian tubes.
-
-
A protein microarray with all the open reading frames of C. trachomatis was probed with sera.
-
-
Twenty chlamydial antigens reacted with sera from at least 68% (17/25) of the macaques.
-
-
Treatment with doxycycline significantly decreased Chlamydia-specific antibody levels.
-
-
Immudominant antigens can be tested as vaccine antigens and to evaluate antibiotic therapy.
Acknowledgments
This work was supported by grants N01-AI95388 (DP), AI40307 (DP), AI33118 (DP), AI72847 (DP), R41AI072847 (LM), R42AI072847 (LM) and RO1AI067888 (LM) from NIH/NIAID and the Washington National Primate Research Center grant RR00166 (DP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chlamydia screening among sexually active young female enrollees of health plans--United States, 2000–2007. MMWR Morb Mortal Wkly Rep. 2009;58:362–365. [PubMed] [Google Scholar]
- 2.Schachter J, Dawson CR. Human chlamydial infections. Littleton, Mass: PSG Pub. Co; 1978. [Google Scholar]
- 3.Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J Infect Dis. 2005;192:1836–1844. doi: 10.1086/497341. [DOI] [PubMed] [Google Scholar]
- 4.Gotz H, Lindback J, Ripa T, Arneborn M, Ramsted K, Ekdahl K. Is the increase in notifications of Chlamydia trachomatis infections in Sweden the result of changes in prevalence, sampling frequency or diagnostic methods? Scand J Infect Dis. 2002;34:28–34. doi: 10.1080/00365540110077001. [DOI] [PubMed] [Google Scholar]
- 5.Nichols RL, Bell SD, Jr, Murray ES, Haddad NA, Bobb AA. Studies on trachoma. V. Clinical observations in a field trial of bivalent trachoma vaccine at three dosage levels in Saudi Arabia. Am J Trop Med Hyg. 1966;15:639–647. [PubMed] [Google Scholar]
- 6.Dawson C, Wood TR, Rose L, Hanna L. Experimental inclusion conjunctivitis in man. 3. Keratitis and other complications. Archives of ophthalmology. 1967;78:341–349. doi: 10.1001/archopht.1967.00980030343015. [DOI] [PubMed] [Google Scholar]
- 7.Farris CM, Morrison RP. Vaccination against Chlamydia Genital Infection Utilizing the Murine C. muridarum Model. Infect Immun. 2011;79:986–996. doi: 10.1128/IAI.00881-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Maza LM, Peterson EM. Vaccines for Chlamydia trachomatis infections. Curr Opin Investig Drugs. 2002;3:980–986. [PubMed] [Google Scholar]
- 9.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 10.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Fisher MI, Cheng C, Sun G, Pal S, Teng A, Molina DM, et al. Identification of immunodominant antigens by probing a whole Chlamydia ORFome microarray using sera from immunized mice. Infect Immun. 2011 doi: 10.1128/IAI.00626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patton DL, Sweeney YC, Bohannon NJ, Clark AM, Hughes JP, Cappuccio A, et al. Effects of doxycycline and antiinflammatory agents on experimentally induced chlamydial upper genital tract infection in female macaques. J Infect Dis. 1997;175:648–654. doi: 10.1093/infdis/175.3.648. [DOI] [PubMed] [Google Scholar]
- 14.Patton DL, Sweeney YT, Stamm WE. Significant reduction in inflammatory response in the macaque model of chlamydial pelvic inflammatory disease with azithromycin treatment. J Infect Dis. 2005;192:129–135. doi: 10.1086/431365. [DOI] [PubMed] [Google Scholar]
- 15.Patton DL, Halbert SA, Kuo CC, Wang SP, Holmes KK. Host response to primary Chlamydia trachomatis infection of the fallopian tube in pig-tailed monkeys. Fertil Steril. 1983;40:829–840. [PubMed] [Google Scholar]
- 16.Molina DM, Pal S, Kayala MA, Teng A, Kim PJ, Baldi P, et al. Identification of immunodominant antigens of Chlamydia trachomatis using proteome microarrays. Vaccine. 2010;28:3014–3024. doi: 10.1016/j.vaccine.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng A, Cruz-Fisher MI, Cheng C, Pal S, Sun G, Ralli-Jain P, et al. Proteomic identification of immunodominant chlamydial antigens in a mouse model. Journal of proteomics. 2012;77:176–186. doi: 10.1016/j.jprot.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Statistical applications in genetics and molecular biology. 2003;2 doi: 10.2202/1544-6115.1008. Article3. [DOI] [PubMed] [Google Scholar]
- 19.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 20.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 21.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012;40:W553–W559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol. 2010;185:1670–1680. doi: 10.4049/jimmunol.1001240. [DOI] [PubMed] [Google Scholar]
- 25.Finco O, Frigimelica E, Buricchi F, Petracca R, Galli G, Faenzi E, et al. Approach to discover T- and B-cell antigens of intracellular pathogens applied to the design of Chlamydia trachomatis vaccines. Proc Natl Acad Sci U S A. 2011;108:9969–9974. doi: 10.1073/pnas.1101756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Follmann F, Olsen AW, Jensen KT, Hansen PR, Andersen P, Theisen M. Antigenic profiling of a Chlamydia trachomatis gene-expression library. J Infect Dis. 2008;197:897–905. doi: 10.1086/528378. [DOI] [PubMed] [Google Scholar]
- 27.Pal S, Peterson EM, de la Maza LM. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect Immun. 2005;73:8153–8160. doi: 10.1128/IAI.73.12.8153-8160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farris CM, Morrison SG, Morrison RP. CD4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infect Immun. 2010;78:4374–4383. doi: 10.1128/IAI.00622-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen J, Jensen KT, Follmann F, Agger EM, Theisen M, Andersen P. Liposome delivery of Chlamydia muridarum major outer membrane protein primes a Th1 response that protects against genital chlamydial infection in a mouse model. J Infect Dis. 2008;198:758–767. doi: 10.1086/590670. [DOI] [PubMed] [Google Scholar]
- 30.Kari L, Whitmire WM, Crane DD, Reveneau N, Carlson JH, Goheen MM, et al. Chlamydia trachomatis native major outer membrane protein induces partial protection in nonhuman primates: implication for a trachoma transmission-blocking vaccine. J Immunol. 2009;182:8063–8070. doi: 10.4049/jimmunol.0804375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen AW, Theisen M, Christensen D, Follmann F, Andersen P. Protection against Chlamydia promoted by a subunit vaccine (CTH1) compared with a primary intranasal infection in a mouse genital challenge model. PLoS One. 2010;5:e10768. doi: 10.1371/journal.pone.0010768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodgers AK, Budrys NM, Gong S, Wang J, Holden A, Schenken RS, et al. Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertil Steril. 2011;96:715–721. doi: 10.1016/j.fertnstert.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachmaier K, Neu N, de la Maza LM, Pal S, Hessel A, Penninger JM. Chlamydia infections and heart disease linked through antigenic mimicry. Science. 1999;283:1335–1339. doi: 10.1126/science.283.5406.1335. [DOI] [PubMed] [Google Scholar]
- 34.Morrison RP, Lyng K, Caldwell HD. Chlamydial disease pathogenesis. Ocular hypersensitivity elicited by a genus-specific 57-kD protein. J Exp Med. 1989;169:663–675. doi: 10.1084/jem.169.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison RP, Belland RJ, Lyng K, Caldwell HD. Chlamydial disease pathogenesis. The 57-kD chlamydial hypersensitivity antigen is a stress response protein. J Exp Med. 1989;170:1271–1283. doi: 10.1084/jem.170.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinnunen A, Molander P, Morrison R, Lehtinen M, Karttunen R, Tiitinen A, et al. Chlamydial heat shock protein 60--specific T cells in inflamed salpingeal tissue. Fertil Steril. 2002;77:162–166. doi: 10.1016/s0015-0282(01)02922-3. [DOI] [PubMed] [Google Scholar]
- 37.Geisler WM, Suchland RJ, Rockey DD, Stamm WE. Epidemiology and clinical manifestations of unique Chlamydia trachomatis isolates that occupy nonfusogenic inclusions. J Infect Dis. 2001;184:879–884. doi: 10.1086/323340. [DOI] [PubMed] [Google Scholar]
- 38.Geisler WM, Lensing SY, Press CG, Hook EW., 3rd Spontaneous Resolution of Genital Chlamydia trachomatis Infection in Women and Protection from Reinfection. J Infect Dis. 2013 doi: 10.1093/infdis/jit094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su H, Morrison R, Messer R, Whitmire W, Hughes S, Caldwell HD. The effect of doxycycline treatment on the development of protective immunity in a murine model of chlamydial genital infection. J Infect Dis. 1999;180:1252–1258. doi: 10.1086/315046. [DOI] [PubMed] [Google Scholar]
- 40.Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol. 2005;175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal S, Bravo J, Peterson EM, de la Maza LM. Protection of wild-type and severe combined immunodeficiency mice against an intranasal challenge by passive immunization with monoclonal antibodies to the Chlamydia trachomatis mouse pneumonitis major outer membrane protein. Infect Immun. 2008;76:5581–5587. doi: 10.1128/IAI.00574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun G, Pal S, Sarcon AK, Kim S, Sugawara E, Nikaido H, et al. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J Bacteriol. 2007;189:6222–6235. doi: 10.1128/JB.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Maranon MJ, Bush RM, Peterson EM, Schirmer T, de la Maza LM. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein science : a publication of the Protein Society. 2002;11:1854–1861. doi: 10.1110/ps.3650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The figure provides a global view of the increase in reactivity following inoculation. The x-axis represents the mean of pre-inoculation samples and the y-axis represents the mean of post-inoculation samples. The pre-inoculation samples (n=25) were compared to the set of the latest post-inoculation samples for the subjects (n=25). The larger black dots indicate the 20 selected immunodominant antigens. The identify line (y=x) is shown with dashes.