Summary
Primary cilia are solitary nonmotile extensions of the centriole found on nearly all nucleated eukaryotic cells between cell divisions. Only ∼200-300 nm in diameter and a few microns long, they are separated from the cytoplasm by the ciliary neck and basal body. Often called sensory cilia, they are hypothesized to receive chemical and mechanical stimuli and initiate specific cellular signal transduction pathways. When activated by a ligand, Hedgehog (Hh) pathway proteins, such as Gli2 and Smoothened (Smo), translocate from the cell into the cilium1,2. Mutations in primary ciliary proteins are associated with severe developmental defects3. The ionic conditions, permeability of the primary cilia membrane, and effectiveness of the diffusion barriers between the cilia and cell body are unknown. Here we show that cilia are a unique calcium compartment regulated by a heteromeric TRP channel, PKD1-L1/PKD2-L1. In contrast to the hypothesis that polycystin (PKD) channels initiate changes in ciliary calcium that are conducted into the cytoplasm4, we show that changes in ciliary calcium concentration ([Ca2+]cilia) occur without substantially altering global cytoplasmic calcium ([Ca2+]cyto). PKD1-L1/PKD2-L1 acts as a ciliary calcium channel controlling [Ca2+]cilia and thereby modifying Smo-activated Gli2 translocation and Gli1 expression.
We generated a transgenic Arl13B-EGFP mouse (Arl13B-EGFPtg) in which primary and motile cilia show spectacular fluorescence labeling throughout the animal, developed a ratiometric Ca2+ sensor directed specifically to cilia, and patch-clamped individual cilia and measured the resting membrane potential and a calcium-permeant conductance (Icilia; see5). The Arl13B-EGFPtg mice display ubiquitous green fluorescence only in primary and motile cilia but not in microvilli (Fig. 1, Extended Data Fig. 1a-c, Supplementary Video 2), 3D imaging of Arl13B-GFP reveals its exclusive localization to cilia. Staining of wt and Arl13B-EGFPtg retinal pigmented cell epithelia (mRPE) cells and mouse embryonic fibroblasts (MEFs) with ciliary markers confirms that Arl13B-EGFP is indeed localized exclusively to cilia without noticeable alteration of cilia morphology (Extended Data Fig. 2).
Figure 1. ArlGFP localizes to primary cilia and motile cilia in vivo.
(a) Fixed paw of E14.5 embryo, and (b) after incubation in ScaleA2. Insert indicates area where z-stacks from 0 to 1.2 mm depth were acquired. Scale bar, 500 μm. (c) Arl13B-EGFP expression in E14.5 paw. 3 × 4 z-stacks were stitched together, depth color-coded and projected onto a plane. Scale bar, 200μm. (d-f) EGFP-positive cilia are present in fibroblasts within the digit (arrows) that overlap with ACIII labeling in e and merged in f. Scale bar = 10 μm; insert, 3 μm; asterisks indicate autofluorescence. Red and green channels are offset to visualize colocalization. (g-i) Motile cilia in the Fallopian tube express Arl13B-EGFP (g) that overlaps with acetylated tubulin (h). Overlay (i). Insert in (i) is DIC image. Scale bar, 5 μm.
Primary cilia are not reliably loaded with Ca2+-sensitive dyes, forcing experimentalists to rely on changes in [Ca2+]cyto as an indirect indicator of [Ca2+]cilia. To ameliorate this issue, we generated a genetically encoded calcium sensor that is targeted to the cilium by fusing GCaMP37 to the C-terminus of Smoothened (Smo-GCaMP3), enabling monitoring of [Ca2+]cilia and [Ca2+]cyto simultaneously. As shown in Extended Data Fig. 3 and Supplementary Video 3, addition of the Ca2+ ionophore, ionomycin, increased fluorescence in both the cytoplasm and cilium of hRPE1 cells stably expressing Smo-GCaMP3, although variation in ionomycin incorporation precludes precise [Ca2+] comparison between the two compartments.
To quantify [Ca2+] we developed a ratiometric Smo-mCherry-GCaMP3 calcium sensor8 (Fig. 2a,b). In hRPE1 cells stably expressing Smo-mCherry-GCaMP3, the cilia-targeted GCaMP3 and mCherry fluorescence colocalized with the cilia-specific marker, acetylated tubulin (Extended Data Fig. 3e-h). To determine whether [Ca2+]cilia could be increased without affecting [Ca2+]cyto, we ruptured the cilia membrane at the tip of the cilia (circle; Extended Data Fig. 3i, j) with an intense 1-2 s laser pulse (405 nm), leading to a rapid increase in [Ca2+]cilia from the tip that traveled to the ciliary base (Supplementary Video 4). Peak [Ca2+] propagated down the cilia at a rate of 4.6 ± 0.6 μm/s, yielding an apparent diffusion constant (DCa) of 5.3 μm2/s (similar to 5-10 μm2/s for DCa in stellate cell dendrites9).
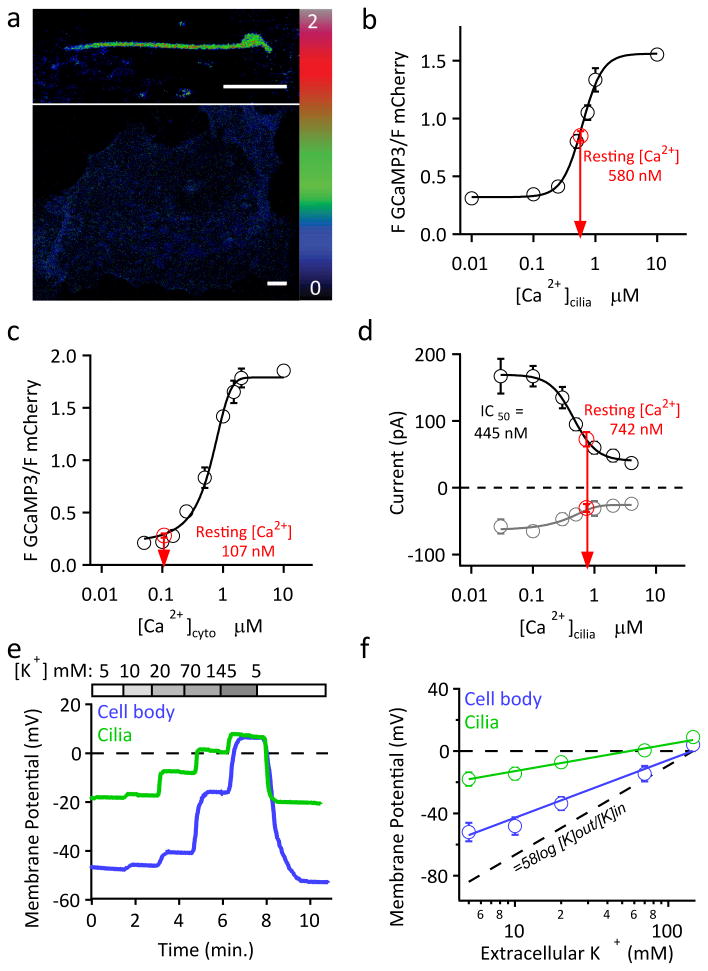
Figure 2. Smo-mCherry-GCAMP3 localizes to the primary cilium and reports ciliary [Ca2+]. [Ca2+]cilia is poorly coupled to [Ca2+]cyto.
(a) hRPE1 cells expressing Smo-mCherry-GCaMP3 were treated with 5 μM ionomycin and changes in fluorescence were measured in the cilium and cytoplasm. (b) Schematic of ratiometric calcium sensor and quantified fluorescence. (c, d) After rupture of the ciliary tip with a laser pulse (t = 10 s), [Ca2+]cilia rapidly increases, while [Ca2+]cyto near the base of the cilium (circle pos. 2)increases only slightly 40 s later (t = 60 s). Ionomycin added for normalization. Circles in (c) indicate areas where fluorescence was measured. (e) Distribution of lag times between ciliary and cytoplasmic [Ca2+] increases. (f) Calcium diffusion from the cytoplasm to the cilium is not restricted. Red dot indicates the position of calcium uncaging while the white line indicates length and position of line scan. Arrows indicate cytoplasm to cilium transition. Scale bar, 5 μm. See also Supplementary Video 6.
In order to closely monitor changes in [Ca2+]cyto at the cilia-cytoplasm junction, we loaded the calcium indicator Fluo-4 into hRPE1 cells stably expressing Smo-mCherry-GCaMP3. Ciliary membrane rupture increased [Ca2+]cilia and was detectable at the ciliary base after a ∼40 s delay (Fig. 2c-e and Supplementary Video 5). More distant parts of the cytoplasm remain unaffected by fluctuations at the base of the cilium. Elevation of [Ca2+]cyto after ciliary rupture occurs infrequently and with varying delay; ∼40% of the cells did not show any elevation in the cytoplasmic side of the cilia-cell junction even 60 s after [Ca2+]cilia had saturated (Fig. 2e). Apparently the Ca2+-sensor does not hinder the diffusion of Ca2+ from cilium to cytoplasm after rupture: in this case the cilium fills with 2 mM [Ca2+], which saturates the Ca2+-sensor.
Next, we asked whether the protein-dense structure at the base of the cilium might act as a localized [Ca2+] buffer and/or impermeant physical barrier to [Ca2+] entry into the cytoplasm. We loaded Smo-mCherry-GCaMP3-expressing cells with the caged calcium chelator NP-EGTA-AM and uncaged [Ca2+] in the cytoplasm in close proximity to the ciliary base. The velocity of this Ca2+ wave was 16 ± 2 μm/s in the cytoplasm and continued at 15 ± 2 μm/s in the cilium, indicating no significant delay of [Ca2+] entering the cilium from the cytoplasm (Fig. 2f and Supplementary Video 6). Since the ratio of cilium/cytoplasmic volume is exceedingly small (1:30,000), the tiny ciliary Ca2+ rivulet is rapidly diluted in the large cytoplasmic volume. Since the number of free calcium ions at 1 μM concentration within the ∼ 0.5 fL cilioplasm is ∼200 ions at an instant in time, calcium does not perturb cytoplasmic [Ca2+] substantially, nor does it initiate a measurable Ca2+-induced Ca2+ release wave in the cytoplasm. These considerations led us to ask whether resting [Ca2+]cilia differs from resting [Ca2+]cyto.
The KD for the ratiometric ciliary Ca2+ sensor in situ was comparable to that in solution (625 nM vs 660 nM7; Extended Data Fig. 4). In 2 mM [Ca2+]e, resting [Ca2+]cilia was 580 nM in hRPE cells (Fig. 3b). We also calibrated the ratiometric Ca2+ sensor in the cytoplasm using cells that had not yet formed a cilium (Smoothened in the plasma membrane; Fig. 3a bottom), and obtained a similar calibration curve and KD (550 nM), suggesting that the sensor reported [Ca2+] similarly in cilia and cytoplasm. The average ratio for the sensor in the cytoplasm of 0.28 ± 0.02 corresponding to a resting [Ca2+]cyto of < 110 nM, a normal resting cytoplasmic [Ca2+]10 (Fig. 3c). To test this surprisingly high resting [Ca2+]cilia using an independent measurement, we patch-clamped Smo-EGFP-labeled cilia in the whole-cell configuration and varied free [Ca2+]cilia. High [Ca2+]cilia (≥ 400 nM; IC50 = 445 nM, Fig. 3d) inactivates Icilia, enabling an independent calibration of [Ca2+]cilia. We then estimated the undisturbed resting [Ca2+]cilia by comparing the current amplitude from perforated-patch measurements with the [Ca2+] calibration curve. The magnitude of Icilia confirmed that [Ca2+]cilia was ∼7-fold higher than resting [Ca2+]cyto (Fig. 3c).
Figure 3. Resting cilium [Ca2+] is substantially higher than resting cytoplasmic [Ca2+].
(a) Live hRPE1 cell F GCaMP3/F mCherry ratios in 2mM [Ca2+]e in the cilium (top) and cytoplasm (bottom). Scale bar, 5 μm. (b) Plot of measured ciliary fluorescence ratio (avg. = 0.8 ± 0.12, n = 16 cilia) vs. estimated [Ca2+]cilia. Resting [Ca2+]cilia = 580 nM. (c) Plot of measured cytoplasmic fluorescence ratio (avg. = 0.28 ± 0.02, n = 20 cells) vs. estimated [Ca2+]cyto. Resting [Ca2+]cyto is 110 nM. (d) By measuring current amplitudes in perforated patches (Methods), we estimated resting free [Ca2+]cilia as 742 nM. Black circles = current at +100 mV; grey circles = current at -100 mV. (e) Changes in cell (Vm) and cilia (Vcilia) potentials in response to external [K+]. (f) Average potential of the cell body and cilia plotted as a function of external [K+]. Vm differs from Vcilia at all [K+] other than [K+]e = 145mM; p < 0.05). The measured resting membrane potential is -18 mV for the cilia and -53 mV for the cell (± SEM, n = 5 cells and 4 cilia). The grey dashed line is the K+ Nernst potential.
We next determined the cilia's resting membrane potential (Ecilia) by measuring changes in response to depolarizing concentrations of extracellular potassium ([K+]e) from hRPE1 Smo-EGFP cytoplasm and from its primary cilia. Consistently, Ecilia was >30 mV positive to the cytoplasm (Ecilia =–18 ± 1 mV; Ecyto = –54 ± 2 mV, respectively; Fig. 3e) and significantly less [K+]e (70 mM vs. 129 mM) was required to depolarize the ciliary membrane potential to 0 mV (Fig. 3f). In summary, the cilium is a functionally distinct cell compartment with respect to ions. This calcium compartment is maintained by a favorable influx/efflux ratio: the large number of calcium-permeant channels5 or other ion channels/transporters can easily maintain high [Ca2+] in the small volume of the cilia, despite steady diffusion of Ca2+ into the cytoplasm at its base. An analogy is a water tower (Ca2+ within the cilia) connected to a large lake (cytoplasm) by a small pipe (basal body). Because ciliary [Ca2+] is high (∼600 nM) compared to the cytoplasm (∼100 nM) calcium flows from cilium to cytoplasm, but not cytoplasm to cilia. In addition, an ∼ –30 mV gradient from cilia to cytoplasm further insures asymmetry of Ca2+ flux. We speculate that these factors insulate cilia from the < 1 μM Ca2+ fluctuations that characterize cytoplasmic Ca2+ signaling.
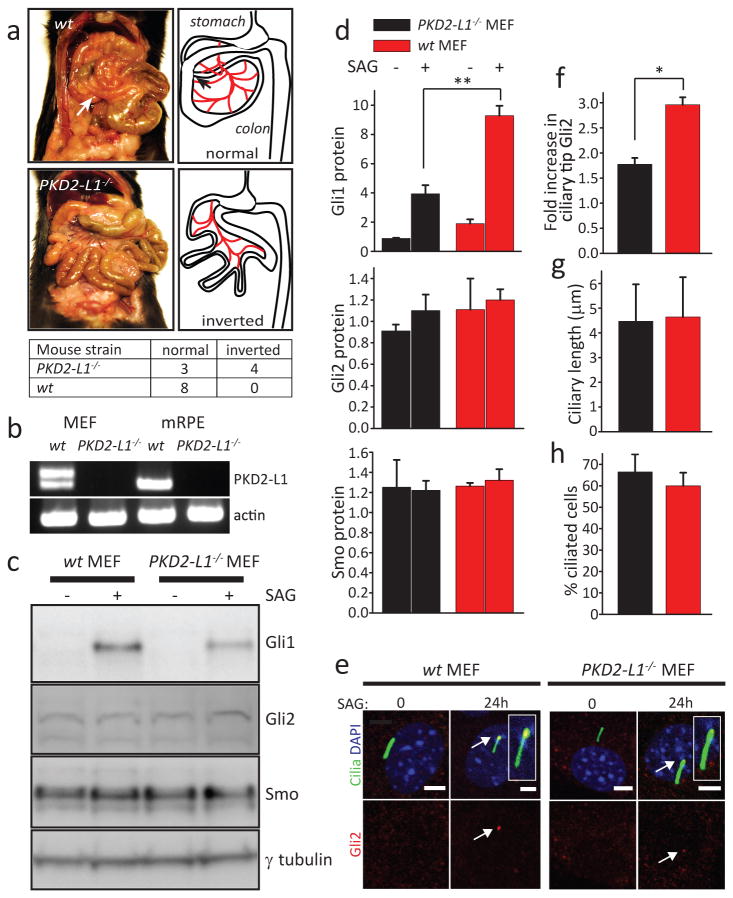
What are the consequences to cell function of ciliary ion independence from the cytoplasm? As we show in DeCaen et al.5, Icilia is mediated predominantly by the heteromeric channel, PKD1-L1 and PKD2-L1. PKD1-L1-/- mice have a ciliary phenotype11,12, but in this study we focused on the pore-forming subunit of Icilia, PKD2-L1. Although PKD2-L1 was initially proposed to be a sour taste receptor, PKD2-L1-/- mice display no sour taste deficit13. Thus, we tested PKD2-L1-/- mice for a potential ciliary defect. While we did not observe any major organ laterality defects, we observed intestinal malrotation in about 50% of the PKD2-L1-/- knockout animals that was not observed in wt mice (Fig. 4a), indicating a mild penetrance of the phenotype. Intestinal malformations are associated with Sonic hedgehog (Shh) pathway defects during early development14,15.
Figure 4. PKD2-L1-/- mice exhibit defects in Smo-mediated Gli1 activation.
(a) Intestinal malrotation in PKD2-L1-/- mice, and comparison with wt. Arrow indicates correct rotation of intestine. Schematic orientation of intestines is on right, red = mesentery. (b) RT-PCR of MEF and mRPE-derived cDNAs shows that PKD2-L1 is expressed in both cell types. (c) Western blot of Gli1, Smo, Gli2, and γ-tubulin expression in wt and PKD2-L1-/- MEFs after stimulation for 24 h with 400 nM SAG. (d) upper panel: Gli1 in wt (1.9 ± 0.3 vs. 9.3 ± 0.7 after stimulation) and PKD2-L1-/- MEFs (0.9 ± 0.04 vs. 4.0 ± 0.6 after stimulation). p < 0.01. middle panel: Gli2 in wt (0.9 ± 0.1 vs. 1.1 ± 0.2 after stimulation) vs. PKD2-L1-/- MEFs (1.1 ± 0.3 vs. 1.2 ± 0.1 after stimulation). lower panel: Smo in wt (1.3 ± 0.3 vs. 1.2 ± 0.09 after stimulation) vs. PKD2-L1-/- MEFs (1.3 ± 0.04 vs. 1.3 ± 0.1 after stimulation; n = 3 wt embryos, 5 PKD2-L1-/- embryos). (e) Localization of Gli2 at the distal tip of Arl13B-EGFPtg and PKD2-L1-/- × Arl13B-EGFPtg MEF cilia. Scale bar, 5 μm. Arrows point to Gli2 at cilia tips. Cilia are magnified in inserts. (f) Quantitation of Gli2 protein accumulation at the ciliary tip after SAG stimulation; wt=3.0 ± 0.2 (n=40 cilia each from 3 embryos) vs. PKD2-L1-/- MEF = 1.8 ± 0.1 fold increase (n=40 cilia each from 5 embryos). p < 0.05. (g, h) quantification of ciliary length (4.5 ± 1.5μm for wt vs. 4.6 ± 1.6μm for PKD2-L1-/- MEFs) and percentage of ciliated cells (67 ± 8% for wt vs. 60 ± 6% for PKD2-L1-/- MEFs; n=40 cilia each from 3 embryos).
Treatment of mouse embryonic fibroblasts (MEF) with the Smo agonist, SAG, directly activates the Smo receptor, leading to an upregulation of Gli1 and Ptch1 expression16,17. As PKD2-L1 is transcribed and localizes to cilia in wt MEF cells (Fig. 4b, Extended Data Fig. 5), we next asked whether the Shh pathway might be affected in PKD2-L1-/- mice by measuring the upregulation of Gli1 in response to stimulation with SAG. As shown in Fig. 4c,d, Gli1 protein increased from 1.9 ± 0.3 to 9.3 ± 0.7 in wt MEF cells stimulated for 24 h with 400 nM SAG. In contrast, SAG upregulated Gli1 from 0.9 ± 0.04 to only 3.9 ± 0.6 in PKD2-L1-/- embryonic MEF cells. As shown in Fig 4d, key members of the Shh pathway were not altered. However, Arl13B-EGFPtg MEFs accumulate significant amounts of Gli2 at the distal tip of the cilium (Fig. 4e,f), as is required for activation of Gli2 and full activation of downstream transcription events18. In PKD2-L1-/- × Arl13B-EGFPtg mutant cells, Gli2 accumulation at the ciliary tip was reduced by ∼50% compared to wt cells. PKD2-L1 is not required for cilia formation as ciliary length and the percentage of ciliated cells did not differ significantly between genotypes (Fig. 4g,h). Finally, we asked whether SAG stimulation itself regulates Icilia and thus ciliary [Ca2+]. Although there was no immediate effect on ciliary current stimulation with SAG, after 24 h Icilia amplitude and resting [Ca2+]cilia increased in the cilium of wt MEFs (Extended Data Fig. 6). This data suggests that SAG initiates recruitment of PKD2-L1 channels into the cilium rather than activating the channel.
Our results suggest that primary cilia are functionally distinct from the cytoplasm. Icilia is encoded by a heteromeric PKD1-L1/PKD2-L1 channel and resting [Ca2+]cilia is at least 0.4 μM higher than resting [Ca2+]cyto, which regulates trafficking of Hh-mediated transcription factors in the cilium. PKD2-L1 is increased by Shh pathway stimulation, most likely by channel recruitment to the cilium. Interestingly, the IFT25/IFT27 complex seems to be specific for transporting Shh components and an IFT25 mutant MEF showed an impaired Gli2 ciliary trafficking phenotype17 similar to PKD2-L1 mutant MEFs. Since IFT25 has a unique Ca2+ binding site19, the observed Shh defect of PKD2-L1 nulls implies that PKD2-L1 may ‘tune’ [Ca2+]cilium to optimize IFT25 function. [Ca2+]cilium is likely also adjusted by other PKD members or even other as yet unidentified ion pumps or transporters in the cilium during development, which may explain the mild phenotype of the PKD2-L1 mutant mouse compared to other Shh-deficient mutant mice (IFT25-/- and Shh-/- mice15,17). An alternative, but not mutually exclusive, hypothesis is that acute Icilia regulation by G protein-coupled receptors and growth factors dynamically regulate ciliary trafficking. Further studies are necessary to determine whether other intermediates in the Shh pathway, such as SUFU-Gli dissociation or Gli proteolytic processing, are [Ca2+]-dependent. A second conclusion from this work is that the cilium funnels a small but steady Ca2+ load into the peri-ciliary cytoplasm.
Methods Summary
A ratiometric GCAMP3-based calcium sensor was expressed in cilia to measure ciliary calcium in hRPE1 cells. A transgenic mouse model expressing GFP in cilia was generated to measure ciliary calcium and membrane potential by patch clamp recordings of cilia. Defects in Shh signaling of MEF isolated from PKD2-L1-/- mutant mice were quantified by Western blotting and ciliary localization of Gli1 protein.
Online Methods
Molecular biology
The hArl13B clone was obtained from Open Biosystems and cloned into the pEGFP-N1 vector. Smo-EGFP, GECO1.2 and GCaMP3 were obtained from Addgene. GECO1.2 is an improved version of GCaMP320. GCaMP3 was spliced in frame to the 3′ end of Smo. The ratiometric sensor Smo-mCherry-GCaMP3 was obtained by adding short glycine-serine linkers to the 5′ and 3′ ends of mCherry by PCR amplification. mCherry was cloned nondirectionally into mSmo-GCaMP3 after linearization with AgeI. Correct orientation of mCherry was confirmed by sequencing. The same approach was used to add mCherry-GECO1.2 to hArl13B.
Transgenic and PKD2-L1 knockout animals
We injected both a human Arl13B-EGFP cDNA construct and a hArl13B-mCherry-GECO1.2 cDNA construct under the control of a chicken actin promoter (CAG) into the pronucleus of mouse C57/B6 oocytes and obtained 2 independent founder lines for Arl13B-EGFP and 4 independent founder lines for Arl13B-mCherry-GECO1.2. Transgenic males and females were viable and fertile. The ScaI/HindIII linearized plasmid was gel-purified and injected into the pronucleus of C57/BL6 oocytes at the transgenic animal core facility at Boston Children's Hospital. Oviducts of Arl13B-EGFPtg mice were isolated as described previously21. All animal protocols in this work were approved by the IUACUC of Boston Children's Hospital. PKD2-L1 knockout animals were obtained from Jackson laboratories and have been described previously22. P21 PKD2L1-/- and age matched C57bl/6 wt mice were used for morphological analysis.
Isolation of primary mouse embryonic fibroblasts (MEFs) and primary mouse retinal pigmented epithelial cells (mRPE) cells
MEFs were isolated from E15 wt, Arl13B-EGFPtg, Arl13B-mCherry-GECO1.2tg or PKD2-L1-/-, or PKD2-L1-/- × Arl13B-EGFPtg embryos. Freshly harvested embryos were placed in a 10 cm cell culture dish in sterile PBS. The head and inner organs were removed and used for genotyping. The body was minced in 1 ml of 0.5% trypsin (Life Technologies) with a razor blade and incubated at 37°C for 30 min. Tissue was triturated by pipetting 10-20× and the trypsin reaction was quenched by addition of 10 ml complete medium with serum (DME w/ 10% FCS). The suspension was passed through a cell strainer and plated. MEFs were seeded onto coverslips at 50% cell density and serum-starved the next day for 24 h in DMEM + 0.2% FCS for electrophysiological recordings, immunostaining, biochemistry and Gli2 translocation assays. MEF cells were not used beyond passage 4. mRPE cells were isolated as described previously23.
Antibodies
Rabbit anti-adenylyl cyclase III, Santa Cruz; mouse anti-acetylated tubulin, Sigma-Aldrich; rabbit monoclonal anti-villin antibody, Abcam; mouse HMB45 and rabbit PKD2-L1, Thermo Scientific. Goat anti-Smoothened and anti-Gli2 antibody was a gift from A. Salic, Harvard Medical School; Mouse monoclonal anti γ-tubulin clone GTU 88, Sigma-Aldrich; rabbit anti Gli2, Sigma Aldrich; rabbit anti-Gli1 V812, Cell Signaling.
Immunocytochemistry and confocal microscopy
Cells were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked by 10% goat serum in PBS. Cells were labeled with the indicated antibody and secondary goat anti-rabbit or anti-mouse fluorescently labeled IgG (Life Technologies) and Hoechst 33342 (Life Technologies). Confocal images were obtained using an Olympus FV1000 with a 60× water immersion, 1.2 N.A. objective. Images were further processed using ImageJ (NIH). Differential interference contrast images (DIC) are shown to outline cells.
3D reconstruction of cilia
E14.5 embryos were fixed in 4% PFA for 48 h and dissected. Tissue was treated with Scale solutions as described24. In brief, samples were incubated in ScaleA2 and ScaleB4 until they appeared transparent (3 weeks-3 months). Samples were mounted in 2% agarose in ScaleA2 on a Petri dish. Samples were imaged in ScaleA2 with an XLUMPLFL 25× water 1.05 N.A. MPE objective. Tiles of stacks were acquired with an Olympus FV1000 MPE Exclusive with SpectraPhysics MaiTai DeepSee laser set at 915 nm. Images were acquired with single excitation in GFP and RFP channels. Subtraction eliminated autofluorescence. Tiles were stitched together using an ImageJ plug-in25. 3D reconstruction was done using Imaris software (Bitplane AG, Switzerland).
Cell culture and generation of stable cell lines
hRPE1 cells were transfected using TransIT-LT1 (Mirus Bio). Smo-EGFP expressing hRPE1 cells have been described previously26. Stable lines were selected in DMEM/F12 (Life Technologies) containing G418 (400 μg/mL) added 24 h after transfection. Stable cell lines expressing the protein were enriched by fluorescence-activated cell sorting (FACS) 2-4 weeks after initial selection. For SAG (Calbiochem) stimulation experiments, MEFs were serum-starved at 80% confluency in 3.5 cm dishes or on 12 mm coverslips for 24 h in DMEM/0.2% FCS. MEF cells were stimulated and protein levels were analysed as described27. In brief, MEFs were stimulated with 400 nm SAG for 24 h at 37°C. Cells were washed ×1 with PBS and lysed directly in 2× sample buffer (Life Technologies). Western blots were developed using chemiluminescence (Super Signal West Dura, Pierce Thermo) and a LAS-3000 imaging system (Fujifilm). Detection of RNA levels was performed as described previously22.
Immunohistochemistry
15 μm formalin-fixed frozen tissue sections were permeabilized with 0.5% TX100 / PBS pH 7.4 for 15 min. Sections were blocked with 5% goat serum, 1% BSA, 0.1% fish gelatin, 0.1% TX-100 and 0.05% Tween20. For primary mouse antibodies, endogenous mouse IgGs were blocked by incubating sections with the unconjugated Fab fragment goat anti-mouse IgG for 1 h at room temperature (RT). Sections were washed twice in PBS-T and primary antibodies were diluted in blocking solution and incubated overnight at 4°C. Slides were washed twice in PBS-T and goat anti-rabbit/anti-mouse fluorescent-labeled secondary antibodies were applied at RT for 1 h together with Hoechst 33342 nuclear dye. Sections were washed twice in PBS-T, mounted in Prolong Gold Antifade (Life Technologies) and imaged with an Olympus FV1000; water immersion 60×, 1.2 N.A. objective. Images were further processed using ImageJ (NIH).
Electrophysiology
Unless otherwise stated, all experimental conditions and methods are described in DeCaen et al28. For the experiments in Fig. 3d, intracellular free [Ca2+] was calculated using the Maxchelator website (maxchelator.stanford.edu) and formulated by titrating CaCl2 in 10 mM BAPTA-Cs buffering conditions. The resting membrane potential measurements from the cell and cilia were made in current-clamp mode. Electrical access was established in the perforated-patch configuration using Amphotericin B. Electrodes contained (in mM): 95 K-Aspartate, 30 KCl, 10 HEPES, 10 NaCl, 2 MgCl2, 5 EGTA, 100 nm free Ca2+. Extracellular solutions in Fig. 3e contained one of following ratios of NaCl / KCl (mM): 140/5, 135/10, 125/20, 75/70, 5/145; and 10 HEPES, 1 MgCl2 and 1.8 CaCl2. Intraciliary [Ca2+] was equilibrated with the known pipette [Ca2+] in whole-cilia recordings. Note that increasing [Ca2+]cilia inhibits Icilia. The dose-response curve was fit to the Hill equation (IC50 = 445 ± 18 nm) and used as a calibration curve. We estimated resting free [Ca2+]cilia by comparing this curve to the current amplitude measured in perforated patches (where internal cilia calcium levels are unperturbed).
Ca2+ imaging in cilia
Smo-GCaMP3 and Smo-mCherry-GCaMP3 were imaged on an Olympus FV1000 at a frame interval of 250 or 64 ms. Ionomycin incorporates into both the plasma membrane and cilia; the ratios of F GCaMP3/F mCherry, reflecting changes in [Ca2+], rise simultaneously in the cytoplasm and cilium. Imaging of [Ca2+] transients from cilium to cytoplasm: Smo-mCherry-GCaMP3 hRPE1 cells were serum-starved for 4-5 days and loaded with Fluo4-AM at 25°C for 60 min in imaging buffer (150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 2 mM HEPES, pH 7.4) and then washed for 10–30 min. Cells with cilia extending beyond the cell body were chosen for experiments to avoid possible induction of cytoplasmic calcium signals by membrane rupture. Images were acquired as sequential scans at a frame interval of 2.20 s with spectral detectors set for GFP and mCherry emission on an Olympus FV1000 equipped with a SIM scanner. PMT and offset were adjusted for each channel such that the 16-bit pixel intensity was in the 1000 range in the GFP channel and 2500-3000 for the mCherry channel before each experiment. Under these settings GCaMP3 in the GFP channel did not reach pixel saturation after ionomycin stimulation. The ciliary membrane was ruptured using the SIM scanner coupled to a 405 nm laser. The region of interest (ROI) for photobleaching was chosen to cover the tip of the cilium; 405 nm laser intensity was set to 25% and pulse duration set to 2 s. Changes of ΔF/F >10% for three consecutive scans in the cytoplasm were considered significant. Cytoplasmic [Ca2+] was raised by loading hRPE1 Smo-mCherry-GCaMP3 cells with o-Nitrophenyl EGTA AM (NP-EGTA AM) for 30 min at RT. Calcium was uncaged in the cytoplasm near the base of the cilium with a SIM scanner by a short (30 ms) 405 laser pulse. Diffusion of cytoplasmic Ca2+ to the cilium was measured by imaging changes in GCaMP3 fluorescence of Smo-mCherry-GCaMP3 in the line-scanning mode – the line scanned the cytoplasm and the proximal side of the cilium. Apparent [Ca2+] diffusion speed was 15.8 ± 1.7 μm/sec for the cytoplasm vs. 14.9 ± 2.1 μm/sec in the cilium, indicating no significant delay of Ca2+ entering the cilium from the cytoplasm. Representative traces are shown in figures. All images were analysed using ImageJ (NIH) and Origin 8 (OriginLab).
Diffusion coefficient, DCa
The 405 nm SIM laser was used to rupture the tip of a cilium and allow Ca2+ from the external solution (2 mM) to diffuse along the shaft towards the basal barrier. The cilium is modeled with one-dimensional diffusion assuming a constant supply at the boundary (ruptured tip), with diffusion length given by L = 2(Dt)½. The measured fluorescence increase (GCaMP3) along the ciliary shaft immediately following tip rupture progresses at 4.6 ± 0.6 μm/s, thus providing an estimate for DCa of 5.3 μm2/s. The volume of a cilium is roughly 0.5 fL, containing <200 free Ca2+ ions at 1 μM concentration. We are aware that the number of Ca2+ sensor proteins is thus a significant fraction of Ca2+ ions and will tend to ‘clamp’ reported levels to the KD of the indicator. For this reason we estimated ciliary [Ca2+] by an independent method that does not depend on calcium-binding proteins as calcium sensors. However, DCa is probably a low estimate since the Ca2+-sensor protein may be a significant immobile buffer.
Calibration of the ratiometric Smo-mCherry-GCaMP3 sensor
Standard solutions of various [Ca2+] concentrations were prepared ranging from ∼10 nm to 50 μM by adjusting the ratio of EGTA and CaCl2 in each preparation to clamp free [Ca2+] at the desired value. The solutions contained 137 mM NaCl, 5.4 mM KCl, 10 mM HEPES, 5 mM EGTA and CaCl2 ranging from 0 to 5.04 mM (corresponding to 50 μM free [Ca2+]).
For imaging, hRPE1 cells were plated onto 12 mm glass coverslips, serum-starved for 4-5 days to allow for cilia formation, and imaged in the various standard solutions following digitonin membrane permeabilisation. Briefly, coverslips were washed ×2 in Ca2+-free solution to remove residual Ca2+ and incubated in the Ca2+ standard solution. The samples were then imaged using an Olympus Fluoview FV1000 laser point-scanning confocal microscope (60× water immersion, 1.2 N.A. objective) with spectral detectors set up for optimal detection of GFP and mCherry fluorescence with sequential excitation with 488 nm and 543 nm lasers, respectively. The settings were adjusted for a GCaMP3 signal in the 1000 sub-range and mCherry signal in the 2000 sub-range of the 16-bit intensity range. Identical settings were used for all Ca2+ standard solutions. Subsequently, the cells were permeabilized on the microscope stage by addition of an identical volume (0.5 mL) of 32 μM digitonin dissolved in the same Ca2+ standard, resulting in a final concentration of ∼16 μM digitonin. Images were acquired for multiple fields of view after allowing permeabilisation to occur for ∼1 min. Cytoplasmic [Ca2+] was measured in RPE cells that had not formed a cilium and thus had significant levels of Smoothened-mCherry-GCaMP3 protein in the plasma membrane.
To obtain the standard calibration curve, the acquired images were processed with ImageJ in the following way. Briefly, after background subtraction, the images were thresholded in the mCherry channel to only take into account pixels with a minimum expression level of the sensor (a threshold of 20× background signal was generally used). Dividing the GCaMP3 fluorescence image by the mCherry channel intensity generated ratio images. ROIs exclusively located in cilia of multiple cells were selected, and the average ratios measured for multiple cells and coverslips were reported. The average ratios obtained were plotted as a function of free [Ca2+] and the resulting points were fitted with a sigmoid curve. Images in Figure 3a-d were acquired using the same acquisition settings as in Extended Data Figure 5.
For quantification of Gli2 at ciliary tips, MEF isolated from Arl13B EGFPtg and PKD2-L1-/- × Arl13B EGFPtg mice were stained with anti-Gli2 antibody29. Cilia were outlined in ImageJ based on the EGFP signal. Mean background signal in the Gli2 channel was determined using ImageJ, multiplied by 1.2 and subtracted from the image. Integrated fluorescence intensity was measured within the ciliary outline. Gli2 quantitation was compiled from measurements of 40 cilia from MEF each isolated from 3 wt and 5 PKD2-L1-/- embryos.
Data analysis
Group data are presented as mean ± SEM. Statistical comparisons were made using unpaired t tests (Origin 8). Statistical significance is denoted with asterisk (* p <0.05; ** p <0.01)
Extended Data
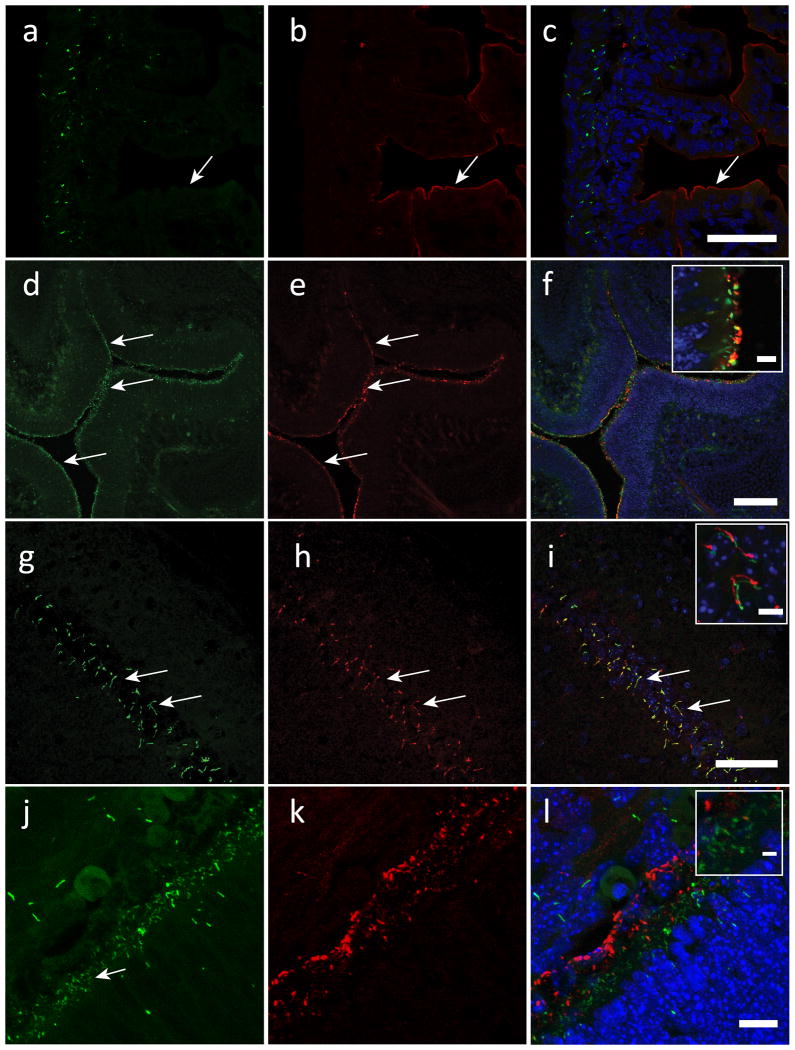
Extended Data Figure 1. Arl13B-EGFP identifies primary and motile cilia in transgenic mouse tissue.
Frozen tissue sections were prepared from (a-c) P0 intestine; (d-f) E15 nasal cavity; (g-i) 6-week-old hippocampus; (j-l) P0 retina from Arl13B-EGFPtg mice. First column, Arl13B-EGFPtg (green) fluorescence; second column (red) labels villin in b, adenylyl cyclase III (ACIII) in e, h, or HMB-45 (human melanoma black antibody, retinal pigmented epithelial cells) in k.
a-c. Intestine: EGFP fluorescence in a does not overlap with the anti-villin staining in b as shown in the merged image in c, indicating that Arl13B-EGFP is absent from microvilli (arrows). Scale bar, 100 μm.
d-f. Nasal cavity, (d) several Arl13B-EGFP-positive cilia (arrows) that face the lumen of the turbinate colocalize with ACIII (e) as shown in the merged image (f). Scale bar, 100 μm. Insert shows magnification of nasal cavity surface. Scale bar of inset in f, 5 μm.
g-i. CA1 region of hippocampus: (g) Prominent cilia (arrows) overlap with ACIII staining (h) as shown in the merged image (i). Scale bar, 50 μm. Insert shows magnification of hippocampal cilia. Inset scale bar, 5 μm. Merged red and green channels were offset for clarity (inset).
j-l. Retinal pigmented epithelia (RPE): (j) Short cilia (arrow) are visible at the intersection between RPE (labeled with HMB-45 antibody, (k) and developing photoreceptor cells, as shown in the merged image (l). Scale bar, 10 μm. Insert shows magnification of RPE/photoreceptor interface. Inset scale bar, 2 μm.
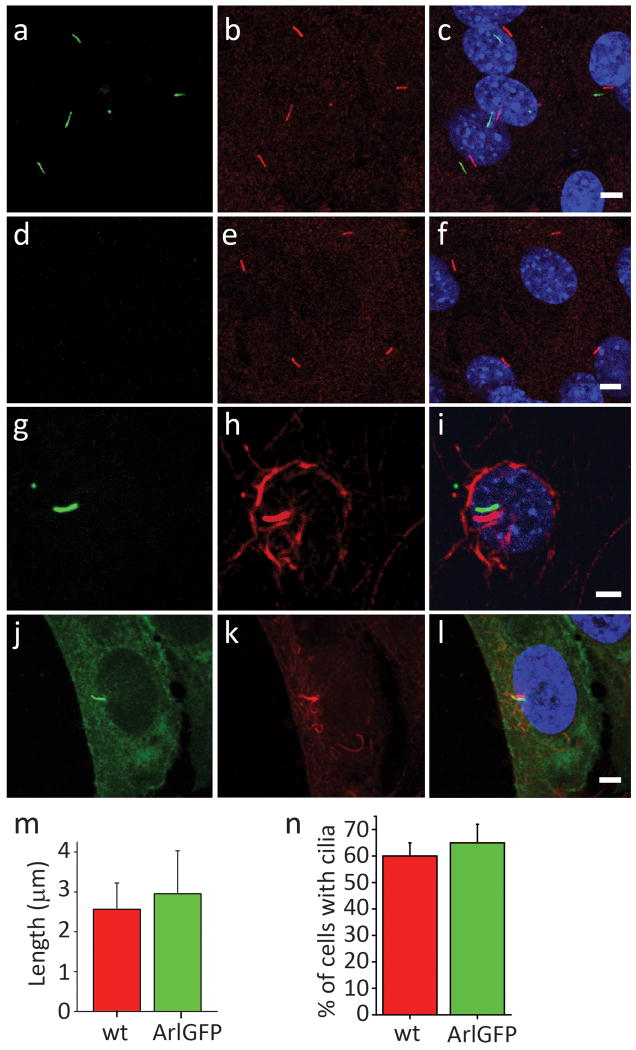
Extended Data Figure 2. Arl13B-EGFP labels a primary cilium in cultured MEF and RPE cells.
First column shows Arl13B-EGFPtg (green) fluorescence; second column labels ACIII (b, e) or (h, k) acetylated tubulin. Third column: merged red and green channels were offset for clarity.
(a-c) Primary mouse embryonic fibroblasts (MEF) of Arl13B-EGFPtg and (d-f) wt mice isolated from E14.5 embryos. Arl13B-EGFP co-localizes with ciliary ACIII in c. MEFs isolated from wt mice show no fluorescence in the cilium (488 nm excitation).
(g-i) Primary RPE cells isolated from P12 Arl13B-EGFPtg mice. Cells were fixed and stained with antibody to acetylated tubulin. Arl13B-EGFP (g) exclusively localized to the primary cilium identified by antibody to acetylated tubulin (h, i).
(j-l) Stable cell line (hRPE1) expressing Smo-EGFP. After 2 days of serum starvation, Smo-EGFP labeled the primary cilium of hRPE1 cells (j) as indicated by acetylated tubulin labeling (k, l). Scale bar in c, f, i, l; 5 μm.
(m, n) To determine whether Arl13B-EGFP expression adversely affected ciliogenesis, ciliary length and percent of cells with cilia were quantified from wt and Arl13B-EGFP-expressing MEFs stained by anti-acetylated tubulin. (m) Ciliary length was similar in wt and Arl13B-EGFP-expressing MEFs (2.6 ± 0.5 μm vs. 2.9 ± 0.8 μm, respectively, n=200). (n) The number of cells with cilia was also comparable between wt and Arl13B-EGFP-expressing MEFs (60.2% ± 5.1% vs. 65.5% ± 7.3%; n=120).
Extended Data Figure 3. Ciliary [Ca2+] changes in stably-transfected RPE cell cilia.
(a-c) Live hRPE1 cells stably expressing Smo-GCaMP3 were treated with 5 μM ionomycin. Fluorescence increases were measured in the cilium and cytoplasm. Image in (c) is DIC. (d) Changes in fluorescence (ΔF/F) of the calcium sensor, GCaMP3, are plotted for both cytoplasm and cilium. (e-h) hRPE1 cells expressing Smo-mCherry-GCaMP3 were stained with acetylated tubulin. GCaMP3 (e) and mCherry (f) fluorescence overlaps with acetylated tubulin staining (g, h). Scale bar, 5 μm; merged channels were offset for clarity. (i) The tip of a cilium was ruptured with an intense 1 s laser pulse (405 nm, hRPE1 cell expressing Smo-mCherry-GCaMP3). Circle indicates area of rupture. Numbered arrowheads indicate positions where changes in fluorescence were measured. Star (*) indicates mCherry fluorescence outside of cilium, indicating that some Smo-mCherry-GCaMP3 is retained in the ER. (j) Quantification of changes in fluorescence at the positions marked in i. Rupture of the ciliary membrane rapidly increases [Ca2+]cilia at the tip and travels along the cilium at 4.6 ± 0.6 μm/s (n=16).
Extended Data Figure 4. [Ca2+]cilia calibration.
(a) Images of hRPE1 cells stably expressing Smo-mCherry-GCaMP3 were acquired after permeabilisation with 15 μM digitonin in varying extracellular [Ca2+]. (b) Averages of several ratios (n = 12-16; ± S.D.) per concentration are plotted against free [Ca2+], yielding the calibration curve for Smo-mCherry-GCaMP3: KD = 625 nM.
Extended Data Figure 5. Overexpressed and endogenous PKD2-L1 localizes to the primary cilium.
(a-c) rabbit anti-PKD2-L1 (Thermo Scientific) recognizes overexpressed PKD2-L1. HEK cells were transfected with hPKD2-L1-IRES mCherry construct and stained with PKD2-L1 ab. PKD2-L1 staining (a) is specific to cells that also express mCherry (b). (c) overlay.
(d-f) overexpressed hPKD2-L1 localizes to the primary cilium in mIMCD3 cells. mIMCD3 cells were transfected with HA-tagged hPKD2-L1 and stained with an anti-HA ab (d) and anti acetylated tubulin ab (e). HA immunoreactivity is visible both in the cytoplasm and cilium. (f) overlay
(g-h) PKD2-L1 antibody labels the primary cilium of mIMCD3 cells. (g) Confluent mIMCD3 cells were stained with anti-PKD2-L1 ab used in a and (h) acetylated tubulin ab to label cilia. PKD2-L1 immunoreactivity is visible in the primary cilium. (i) overlay.
(j-o) Primary mouse embryonic fibroblasts (MEF) of Arl13B-EGFPtg (j-l) and Arl13B-EGFPtg × PKD2-L1-/- mice (m-o) isolated from E14.5 embryos were stained with anti-PKD2-L1 ab used in (a). In Arl13B-EGFPtg MEF PKD2L1 immunoreactivity (j) colocalizes with Arl13B-EGFP signal (k) labeling the primary cilium. (l) overlay. (m-o) PKD2-L1 staining is absent in cilia of Arl13B-EGFPtg × PKD2L1-/- mice. Scale bars = 10 μm.
Extended Data Figure 6. [Ca2+]cilia increases 24 h after SAG stimulation.
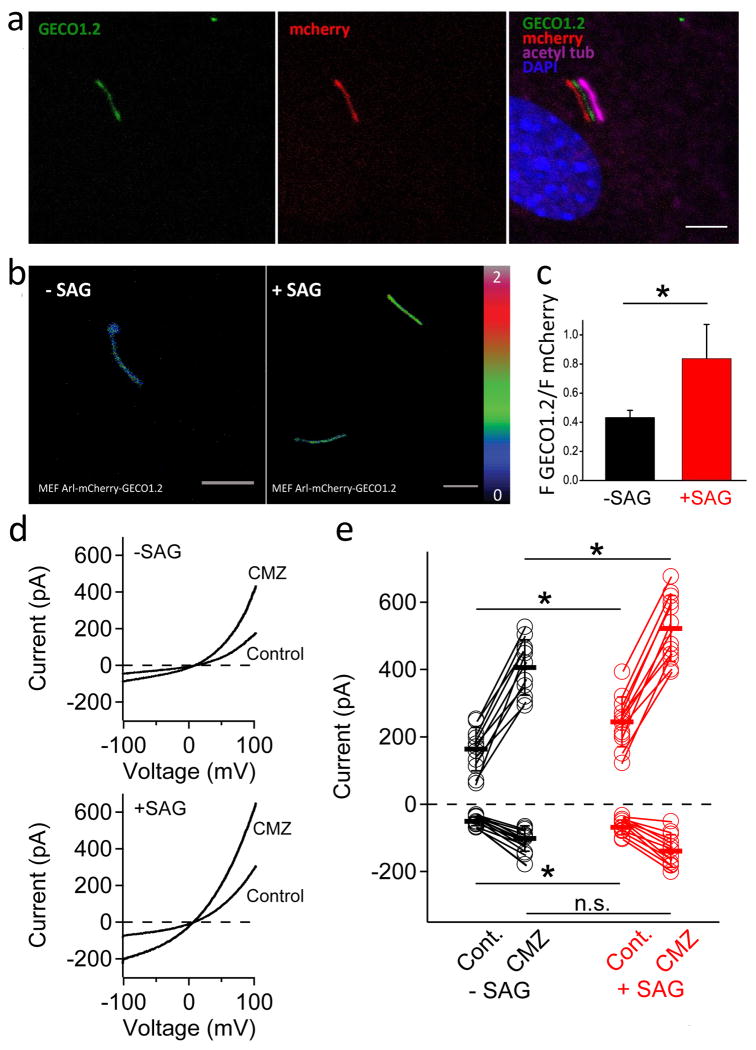
(a) MEF cells expressing Arl13B-mCherry-GECO1.2 were stained with acetylated tubulin. GECO1.2 and mCherry fluorescence overlaps with acetylated tubulin staining. Scale bar, 5 μm; merged channels were offset for clarity. (b) Ratio maps of MEFs isolated from Arl13B-mCherry-GECO1.2 mice stimulated with 0.05% DMSO (left) or 400 nM SAG (right) for 24 h. Scale bar, 5 μm. (c) Quantification of ciliary GECO1.2 / mCherry ratios obtained for MEF cells with and without SAG stimulation. Ratio increases from 0.4 ± 0.05 to 0.8 ± 0.2 after SAG stimulation (*p<0.05; n=20-30 cilia). (d) Example ciliary current measured from MEFs treated with 500 nM SAG (Smo agonist) or with DMSO vehicle (0.05%) in culture for 24-36 h in control conditions and after activation with 10 μM calmidazolium (CMZ). (e) Scatter and whisker (± S.D.) plots from cilia show total outward (+100 mV) and inward (-100 mV) current measured for both treatment groups. Averages are indicated by the thick horizontal lines and individual cilium current magnitudes are represented as circles. P-values resulting from Student's t-test comparing treatment groups are indicated (*< 0.05; n = 11 cilia).
Supplementary Material
Acknowledgments
We thank the Mouse Gene Manipulation Facility of the Boston Children's Hospital Intellectual and Developmental Disabilities Research Center (IDDRC; NIHP30-HD 18655). We thank the Image and Data Analysis Core (IDAC) at Harvard Medical School for help with 3D reconstruction. We also thank Adrian Salic for anti-Smo and anti-Gli2 antibodies. Paul DeCaen was supported by NIH T32-HL007572. We thank Michaela Desai for graphical assistance and Alexander von Gise and Hanna Tukachinsky (Harvard Medical School) and the members of the Clapham laboratory for advice and discussion.
Footnotes
Author Information. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
Online Content Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author Contributions. All authors designed or conducted experiments and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 2.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 5.DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and identification of the calcium channel of primary cilia. Nature. doi: 10.1038/nature12832. xxxxxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hama H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 7.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda R, et al. Imaging calcium concentration dynamics in small neuronal compartments. Sci STKE. 2004:pl5. doi: 10.1126/stke.2192004pl5. [DOI] [PubMed] [Google Scholar]
- 9.Soler-Llavina GJ, Sabatini BL. Synapse-specific plasticity and compartmentalized signaling in cerebellar stellate cells. Nat Neurosci. 9:798–806. doi: 10.1038/nn1698. [DOI] [PubMed] [Google Scholar]
- 10.Clapham DE. Calcium signaling. Cell. 131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Vogel P, et al. Situs inversus in Dpcd/Poll-/ -, Nme7-/ -, and Pkd1l1-/ - mice. Vet Pathol. 47:120–131. doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- 12.Field S, et al. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development. 138:1131–1142. doi: 10.1242/dev.058149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horio N, et al. Sour taste responses in mice lacking PKD channels. PLoS One. 6:e20007. doi: 10.1371/journal.pone.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin V, Shaw-Smith C. Review of genetic factors in intestinal malrotation. Pediatr Surg Int. 26:769–781. doi: 10.1007/s00383-010-2622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 16.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keady BT, et al. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell. 22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhogaraju S, Taschner M, Morawetz M, Basquin C, Lorentzen E. Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30:1907–1918. doi: 10.1038/emboj.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. An expanded palette of genetically encoded Ca(2)(+) indicators. Science. 333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki K, Clapham DE. Rheotaxis Guides Mammalian Sperm. Curr Biol. doi: 10.1016/j.cub.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horio N, et al. Sour taste responses in mice lacking PKD channels. PLoS One. 6:e20007. doi: 10.1371/journal.pone.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs D, Williams DS. Isolation and culture of primary mouse retinal pigmented epithelial cells. Adv Exp Med Biol. 533:347–352. doi: 10.1007/978-1-4615-0067-4_44. [DOI] [PubMed] [Google Scholar]
- 24.Hama H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 25.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keady BT, et al. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell. 22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and identification of the calcium channel of primary cilia. Nature. doi: 10.1038/nature12832. xxxxxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.