Abstract
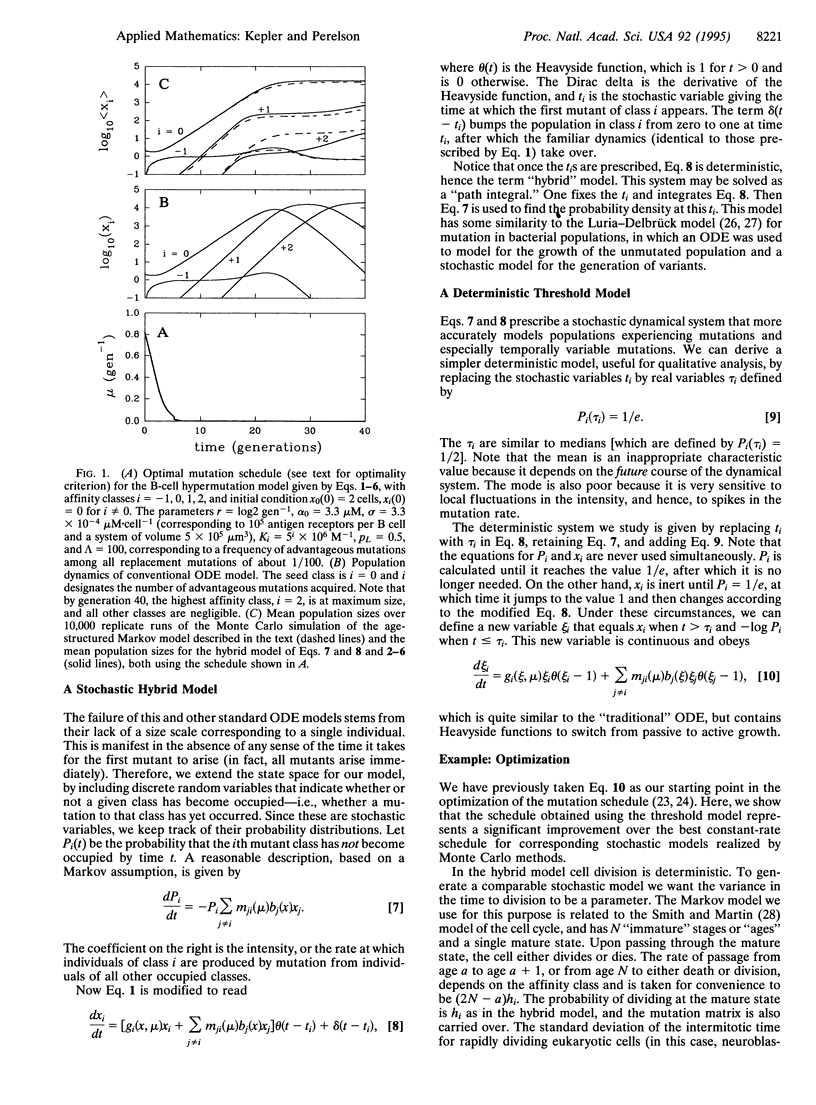
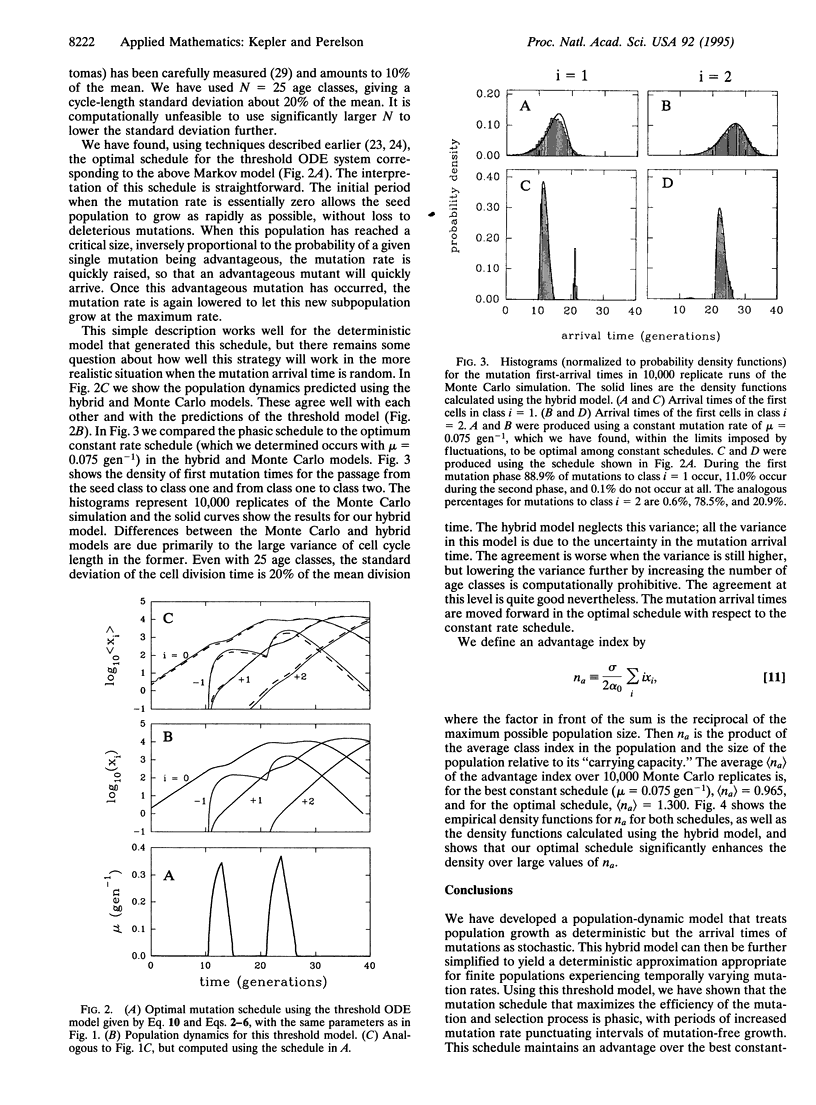
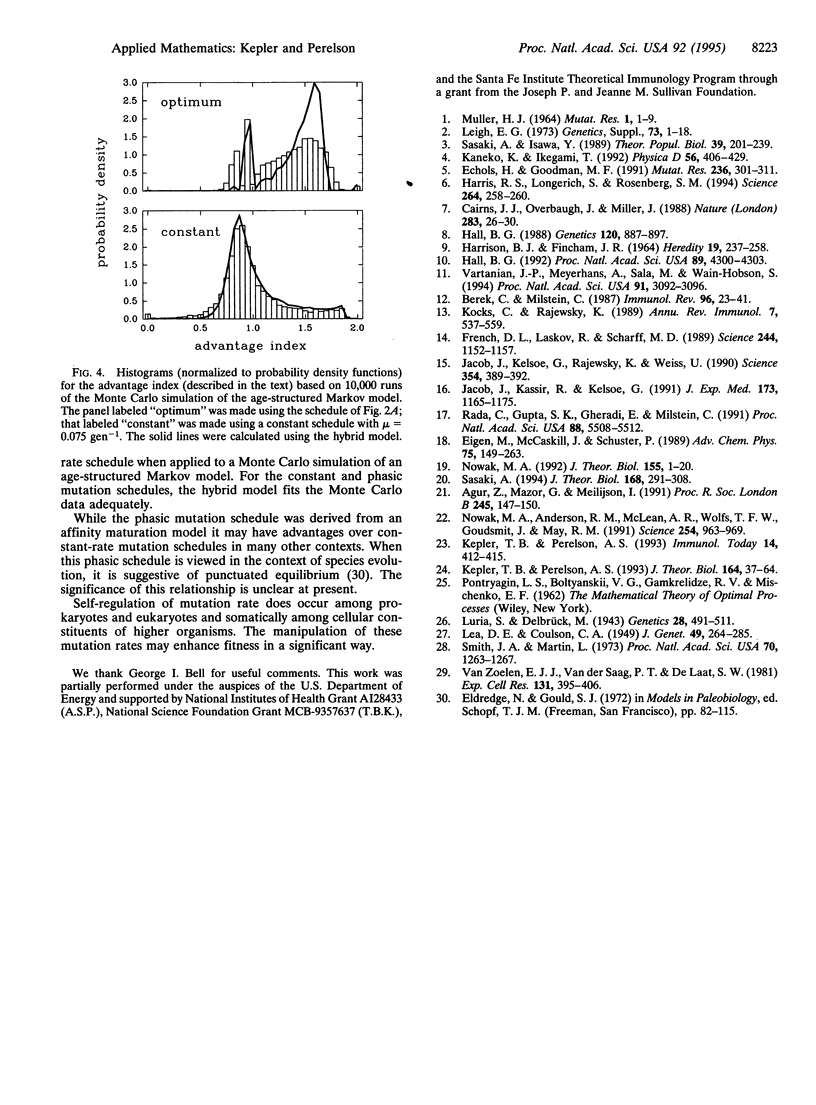
It has become clear that many organisms possess the ability to regulate their mutation rate in response to environmental conditions. So the question of finding an optimal mutation rate must be replaced by that of finding an optimal mutation schedule. We show that this task cannot be accomplished with standard population-dynamic models. We then develop a "hybrid" model for populations experiencing time-dependent mutation that treats population growth as deterministic but the time of first appearance of new variants as stochastic. We show that the hybrid model agrees well with a Monte Carlo simulation. From this model, we derive a deterministic approximation, a "threshold" model, that is similar to standard population dynamic models but differs in the initial rate of generation of new mutants. We use these techniques to model antibody affinity maturation by somatic hypermutation. We had previously shown that the optimal mutation schedule for the deterministic threshold model is phasic, with periods of mutation between intervals of mutation-free growth. To establish the validity of this schedule, we now show that the phasic schedule that optimizes the deterministic threshold model significantly improves upon the best constant-rate schedule for the hybrid and Monte Carlo models.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agur Z., Mazor G., Meilijson I. Maturation of the humoral immune response as an optimization problem. Proc Biol Sci. 1991 Aug 22;245(1313):147–150. doi: 10.1098/rspb.1991.0101. [DOI] [PubMed] [Google Scholar]
- Berek C., Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987 Apr;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990 Sep-Nov;236(2-3):301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- French D. L., Laskov R., Scharff M. D. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989 Jun 9;244(4909):1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Adaptive evolution that requires multiple spontaneous mutations. I. Mutations involving an insertion sequence. Genetics. 1988 Dec;120(4):887–897. doi: 10.1093/genetics/120.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Selection-induced mutations occur in yeast. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4300–4303. doi: 10.1073/pnas.89.10.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Longerich S., Rosenberg S. M. Recombination in adaptive mutation. Science. 1994 Apr 8;264(5156):258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Jacob J., Kassir R., Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991 May 1;173(5):1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Kelsoe G., Rajewsky K., Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991 Dec 5;354(6352):389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- Kepler T. B., Perelson A. S. Cyclic re-entry of germinal center B cells and the efficiency of affinity maturation. Immunol Today. 1993 Aug;14(8):412–415. doi: 10.1016/0167-5699(93)90145-B. [DOI] [PubMed] [Google Scholar]
- Kepler T. B., Perelson A. S. Somatic hypermutation in B cells: an optimal control treatment. J Theor Biol. 1993 Sep 7;164(1):37–64. doi: 10.1006/jtbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- Kocks C., Rajewsky K. Stable expression and somatic hypermutation of antibody V regions in B-cell developmental pathways. Annu Rev Immunol. 1989;7:537–559. doi: 10.1146/annurev.iy.07.040189.002541. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. A., Anderson R. M., McLean A. R., Wolfs T. F., Goudsmit J., May R. M. Antigenic diversity thresholds and the development of AIDS. Science. 1991 Nov 15;254(5034):963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- Nowak M. A. Variability of HIV infections. J Theor Biol. 1992 Mar 7;155(1):1–20. doi: 10.1016/s0022-5193(05)80545-4. [DOI] [PubMed] [Google Scholar]
- Rada C., Gupta S. K., Gherardi E., Milstein C. Mutation and selection during the secondary response to 2-phenyloxazolone. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5508–5512. doi: 10.1073/pnas.88.13.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. Evolution of antigen drift/switching: continuously evading pathogens. J Theor Biol. 1994 Jun 7;168(3):291–308. doi: 10.1006/jtbi.1994.1110. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Iwasa Y. Optimal growth schedule of pathogens within a host: switching between lytic and latent cycles. Theor Popul Biol. 1991 Apr;39(2):201–239. doi: 10.1016/0040-5809(91)90036-f. [DOI] [PubMed] [Google Scholar]
- Smith J. A., Martin L. Do cells cycle? Proc Natl Acad Sci U S A. 1973 Apr;70(4):1263–1267. doi: 10.1073/pnas.70.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J. P., Meyerhans A., Sala M., Wain-Hobson S. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zoelen E. J., van der Saag P. T., de Laat S. W. Family tree analysis of a transformed cell line and the transition probability model for the cell cycle. Exp Cell Res. 1981 Feb;131(2):395–406. doi: 10.1016/0014-4827(81)90243-3. [DOI] [PubMed] [Google Scholar]


