Abstract
Approximately half of all patients with colorectal cancer develop local recurrence or distant metastasis during the course of their illness. Recently, the molecular detection of metastatic cancer cells in various types of clinical samples, such as lymph nodes, bone marrow, peripheral blood, and peritoneal lavage fluid, has been investigated as a potential prognostic marker. The prognostic value of molecular tumor cell detection was independent of the type of detection method used. As assays become more sensitive and quantitative, a more thorough assessment of the cancer status of patients will be based on molecular markers alone. At present, it is difficult to conclude that one specific molecular marker is superior to others. Comparative analyses are recommended to assess the prognostic impact of molecular analyses in the same patient and determine the biomarkers that provide the most accurate prognostic information.
Keywords: Molecular biomarker, Metastatic colorectal cancer cell, Circulating tumor cell, Disseminated tumor cell, Peritoneal lavage fluid, Colorectal cancer
Core tip: We focus on methods of the detection of molecular changes in metastatic colorectal cancer cells, and describe the characteristics for the methods, such as DNA methylation, mRNA, microRNA, immunomagnetic separation, protein and cancer-associated mutations. Moreover, we review the clinical significance according to the type of samples, such as blood, lymph node, bone marrow and peritoneal lavage fluid. At present, it is difficult to conclude that one specific molecular marker is superior to others. Comparative analyses are recommended to assess the prognostic impact of molecular analyses in the same patient and determine the biomarkers that provide the most accurate prognostic information.
INTRODUCTION
Colorectal cancer (CRC) is a common cause of morbidity and mortality[1]. The 5-year survival rate of this disease is approximately 90% for patients with localized disease and approximately 66% for patients with regional disease, as determined at diagnosis[2,3]. The incidence of disease recurrence is 25% in the absence of regional node involvement, suggesting that conventional pathology may fail to detect occult nodal metastasis[2,4]. Most deaths from cancer are caused by metastatic disease, and the prevention of subsequent metastasis is now the focus of clinical research in this field[5]. The early spread of tumor cells is usually not detected by the imaging technologies currently available. Metastasis-specific markers are also urgently required to help in delineating the spread of disease to neighboring tissues, lymph nodes (LNs), and distant parts of the body[6].
Molecular biomarkers have also been used to detect metastatic cancer cells in peripheral blood samples, bone marrow (BM), LNs, or peritoneal fluid[6,7]. Tumor cells detected in the peripheral blood are called circulating tumor cells (CTCs). The precise role of CTCs detected in patients with metastases remains unknown. Some of these CTCs may be transiting from the primary tumor to sites of future metastasis, indicating that metastasis is in progress. Alternatively, the detected CTCs may primarily be innocent bystanders that simply reflect a high metastatic burden or aggressive disease. The latter theory would explain the fact that the detection of CTCs is associated with poor outcome in patients with metastatic CRC[8]. Tumor cells located in the BM are termed disseminated tumor cells (DTCs). Evidence indicates that the BM is the common organ to which tumor cells from many types of carcinomas migrate. It can be speculated that the BM also forms an important reservoir of tumor cells, from which these cells may recirculate into other distant organs where better growth conditions may exist (such as the liver or lungs)[9,10]. The pivotal roles of biomarkers of LNs may help in identifying patients with node-negative CRC who are at a high risk of tumor recurrence and who may benefit from adjuvant therapy[11]. Peritoneal dissemination in patients with CRC is less frequent; therefore, from a prognostic perspective, it is considered less important compared with LN and liver metastasis[12]. However, the incidence of peritoneal seeding during potentially curative surgery for primary CRC reported in a series of 12 patients varied widely from 3% to 28%, which may be explained by differences in the methods used for tumor cell detection[13,14].
In this review, we will focus on the different types of molecular biomarkers of CRC that can be used for the detection of metastatic cancer cells and discuss their potential as prognostic markers of CRC.
MOLECULAR BIOMARKERS OF CANCER
Alteration in gene sequence and expression levels and protein structure or function can be used as molecular biomarkers to detect cancers at an early stage, determine prognosis, and monitor disease progression or therapeutic response[6]. Molecular biomarkers are defined as markers detected using molecular detection techniques such as immunohistochemistry or polymerase chain reaction (PCR).
Molecular tests usually begin with the preparation of a DNA, RNA, or protein extract from a clinical sample. The ratio of neoplastic cells to normal cells varies considerably from one clinical sample to another. It is difficult to isolate specifically neoplastic cells on the basis of clinical sample analysis, considering that samples are often composed primarily of cellular debris and free substrates (such as DNA, RNA, and protein). Therefore, clinical samples are frequently a heterogeneous mix of normal and cancer cells, DNA, RNA, and protein[15]. Two of the most important factors in determining the efficiency of a molecular marker assay are sensitivity (i.e., the minimal amount of the substrate that can be detected) and specificity (i.e., the percentage of assays that correctly distinguish normal from cancer-containing samples). The sensitivities and specificities reported in various studies that used molecular methods to analyze blood samples are shown in Table 1. In general, there is a trade-off between sensitivity and specificity[6,15].
Table 1.
Sensitivities and specificities of various studies that took blood samples n (%)
| Source | Method | Marker | Sensitivity | Specificity | Ref. |
| Serum | MSP | CDKN2A (p16) | 14/52 (27) | 44/44 (100) | [32] |
| Serum | RT-PCR | CEA | 51/121 (42) | 27/33 (82) | [56] |
| Serum | RT-PCR | CK20 | 22/99 (22) | 150/150 (100) | [38] |
| Serum | qPCR | miR-92 | 80/90 (89) | 35/50 (70) | [84] |
| Serum | CellSearch | EpCAM | 20/74 (27) | 228/246 (93) | [88] |
| Serum | CLIA | CEA | 188/429 (44) | 184/201 (92) | [112] |
MSP: Methylation-specific polymerase chain reaction; qPCR: Quantitative reverse transcription polymerase chain reaction (RT-PCR); CLIA: Chemiluminescence immunoassay; CEA: Carcinoembryonic antigen; CK20: Cytokeratin 20; EpCAM: Epithelial cell adhesion molecule.
METHODS OF DETECTION OF MOLECULAR CHANGES IN METASTATIC CRC CELLS AND THEIR CLINICAL SIGNIFICANCE
DNA methylation
One of the most promising types of markers is based on the detection of hypermethylation of promoter regions of cancer-associated genes[6,16-18]. Many types of cancer cells use this mechanism to inactivate tumor suppressor genes[16,19-22]. This assay can detect approximately one cancer cell among 1000 normal cells, a sensitivity that is sufficient to detect tumor DNA in most body fluids[23,24]. DNA methylation profiles represent a more chemically and biologically stable source of molecular diagnostic information compared with RNA or most proteins[16,25]. Cancer-specific DNA methylation patterns can be found in detached tumor cells in bodily fluids and biopsies and in free-floating DNA released from cancer cells[6,16]. The methylation biomarker studies performed till date varied in methylation targets, source of DNA, and type of tumor[15-17]. If a patient with CRC tests negative for a methylation marker in a tumor as well as in a remote sample, the result is classified as false-negative for disease detection. Therefore, researchers need to choose multiple target genes that are reportedly methylated with different frequencies in tumors and rarely in normal tissues[18,26].
Serum (peripheral blood): Cancer patients exhibit elevated levels of free DNA in their blood because of a high cellular turnover[25,27]. The circulatory DNA from blood or other body fluids can be captured easily, and the status of DNA methylation at various gene promoters can be determined using various methods[16,21,25]. Nakayama et al[28] demonstrated that methylated p16 is detectable in the serum of 69% patients with recurrent CRC. Several studies have addressed whether gene promoter methylation can be used to detect prevalent CRC using DNA recovered from the plasma or serum[19,29-32]. The presence of detectable tumor DNA in the plasma or serum is generally associated with a poor prognosis[26,28,29,32]. Wallner et al[26] identified HPP1, HLTF, and hMLH1 as promising methylation markers in the serum of patients with CRC because these genes are not methylated in the serum of healthy controls and are methylated more frequently in metastatic disease than in local disease.
Peritoneal lavage fluid: Few studies have addressed gene methylation for the detection of micrometastasis to the peritoneal fluid in patients with gastrointestinal cancer[33]. We first reported the prognostic relevance of the detection of methylation of tumor-related genes in the peritoneal lavage fluid (PLF) of patients undergoing resection of CRC (Figure 1). The methylation pattern of the promoter of four target genes, CDH1, CDKN2A (p16), MGMT, and APC, was examined in 51 primary CRCs and the corresponding matched PLF DNA. The relative methylation levels of these genes in primary CRC tissues and paired PLF samples were assessed by quantitative methylation-specific PCR. An aberrant methylation of at least one gene was found in 45 out of the 51 (88%) primary tumors. In the PLF, the frequencies of aberrant promoter methylation were 16% for CDH1, 2% for p16, 4% for MGMT, and 24% for APC. Patients with PLF samples that exhibited methylation of more than one of these four target genes had a significantly shorter relapse-free survival[34].
Figure 1.
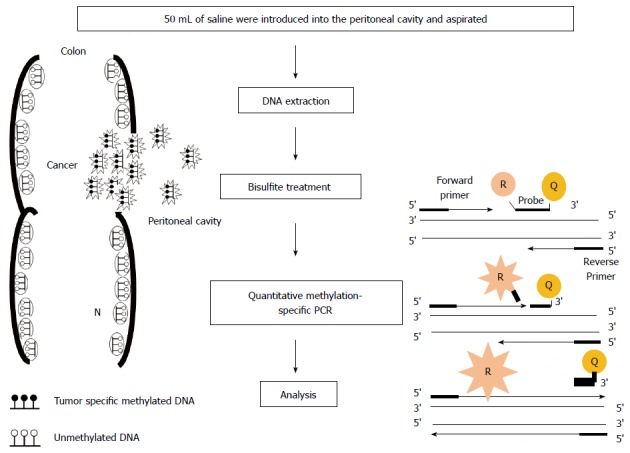
Outline of quantitative methylation-specific polymerase chain reaction analysis in peritoneal lavage. The depth of invasion and subsequent increased likelihood of tumor cells exfoliated from colonic serosa is reflected in the presence of tumor-related methylated DNA in peritoneal cavity. In quantitative methylation-specific polymerase chain reaction (PCR) step, the DNA polymerase cleaves only probes that are hybridized to the target. Cleavage separates the reporter dye (R) from the quencher dye (Q); resulting in increased fluorescence by the reporter. The increase in fluorescence signal occurs only if the target sequence is complementary to the probe and is amplified during PCR.
mRNA (cDNA)
CRC cells show marked changes in the expression of many genes at the mRNA level[6]. One of the most common approaches used to identify and quantify mRNA levels in clinical samples is reverse transcription PCR (RT-PCR)[7,35]. Cytokeratin (CK) mRNA is a common marker of epithelial cells[36]. Many target genes have been previously used to detect micrometastasis from CRC, including carcinoembryonic antigen (CEA), MUC1, CK-8, CK18, CK19, and CK20. There is no specific marker of CRC, and the detection of disseminated neoplastic cells is based on epithelial markers such as CK20 and CEA. CK20 mRNA is considered to be a reliable target for the detection of disseminated CRC cells, and the frequency of false-positive results is reportedly lower (0%-8%) for CK20 mRNA than for CEA mRNA (0%-33%)[37]. It is important to remember that the altered expression of some of these genes has also been reported in normal cells, leading to false-positive results[2,7]. To solve this problem, more quantitative analysis may eventually determine a cut-off level for differentiating between cancer and normal cells[38]. Although the isolation of intact RNA from bodily fluids and tissue samples is also possible, it generally requires cumbersome efforts to neutralize ubiquitous RNase enzymes[6].
Serum (peripheral blood): A meta-analysis of nine studies performed between 1998 and 2006 showed that patients with CTC positivity detected using RT-PCR of blood samples collected from the tumors’ drainage veins correlated more with LN positivity (50%) than with LN negativity (21%). Furthermore, hepatic metastasis was found more often in CTC-positive patients (21%) than in CTC-negative patients (8%)[39,40]. A systematic review that evaluated CTCs after surgical resection of CRC and summarized its characteristics found that 14 reports fulfilled the inclusion criteria[7,36,41-53]. The mean CTC detection rate was 33.4%. Moreover, there were no differences among studies that obtained perioperative, early postoperative, and late postoperative samples or among studies that included patients with early-stage disease only, curative patients only, and patients with disease in all stages. The reported studies showed that perioperative CTC levels were not useful for predicting CRC recurrence[7]. The presence of CTCs in the peripheral blood at least 24 h after CRC resection is an independent prognostic marker of recurrence[7]. The sensitivity and specificity of target PCR amplification in CRC patients and control subjects were 22%-83% and 76%-100%, respectively[38,54-61].
LNs: To determine the spread of disease, many studies have attempted to detect CK mRNA in the LNs of patients with cancer using RT-PCR[2,62-66]. Isolated tumor cells or micrometastases within regional LNs that are not detected via conventional histopathological examination (hematoxylin and eosin staining) have been suspected to be markers of systemic tumor spread in these patients[11,67]. Tumor-specific changes in mRNA expression were detected in one or more LNs in 20%-54% patients with node-negative tumors that could potentially be upstaged to Dukes’ grade C[2,15,62-66,68]. The detection of mRNA transcripts encoding CEA in the LNs of patients with stage II CRC has been reported to predict outcome; the adjusted 5-year survival rate was 41% lower in the group with nodal micrometastases[62]. Micrometastatic LNs metastases identified by RT-PCR were consistently found to be prognostically significant[69].
BM: CK20 is the most commonly used marker in RT-PCR analyses of BM samples from CRC patients, with six studies published till date[46,70-74]. In three of those studies, the number of patients investigated was more than 100, with DTC detection rates of 11%-35%[73,75-77]. The CK20 detection rate in healthy controls ranged between 0% and 10%. Four groups found an association between the presence of CK20 transcripts and poor overall survival (OS)[78].
PLF: Guller et al[37] evaluated the clinical relevance of real-time quantitative PCR (qPCR) detection of CEA and CK20 transcripts in the PLF and blood from patients undergoing surgery for CRC and found that it was potentially related to tumor-cell dissemination. Among 39 patients with CRC, 11 had at least one sample that was positive for CEA or CK20. Six patients had evidence of disseminated CRCs before resection, whereas 10 had evidence of disseminated CRCs after resection. CEA qPCR amplification was detected in eight patients, and CK20 qPCR amplification was detected in 10 patients. Nine of the 11 PCR-positive patients developed recurrence (five distal metastases and four local metastases) after an average follow-up of 12 mo, whereas only two of the 28 qPCR-negative patients developed recurrence. In seven patients, disseminated CRCs were found in the PLF but not in the blood; five of these patients (71%) developed recurrence.
MicroRNAs
MicroRNAs (miRNAs) are small (18-24 nucleotides) RNAs that regulate the translation and stability of specific target mRNAs[79]. The deregulation of specific miRNAs contributes to a variety of diseases, most notably the development and progression of cancer, including CRC[80]. Once thought to be unstable RNA molecules, miRNAs are now known to be stably expressed in serum, plasma, urine, saliva, and other body fluids[81]. Biochemical analyses indicate that miRNAs are resistant to RNase activity as well as extreme pH and temperature[81-83]. The enormous potential of circulating miRNAs as an ideal class of cancer biomarkers is based on certain facts. First, they are remarkably stable molecules, well-preserved in harsh conditions, and resistant to RNase activity. Second, their expression profiles are specifically correlated with certain types of cancer or pathognomonic conditions. Third, they are easily accessible and can be sampled in a relatively noninvasive manner and readily detected using various techniques[81].
Serum (peripheral blood): Circulating miRNAs can be detected in the serum[79-81]. Ng et al[84] were the first to report that circulating miRNA levels in plasma are different between CRC patients and controls. In a population of 90 patients and 50 controls, the authors found that miR-92 was expressed at higher levels in the plasma of patients and that it could distinguish patients from healthy controls with 70% specificity and 89% sensitivity[79,84]. The expression of miR-92 decreased after surgical tumor resection, suggesting that the circulating levels of miRNAs may be a useful marker of disease recurrence[84]. A similar study found that the circulating levels of miR-141 were elevated in metastatic CRC and that its expression was associated with poor prognosis, suggesting that this miRNA may be used in conjunction with CEA to detect CRC with distant metastasis[85].
Immunomagnetic separation
The immunomagnetic separation method using the CellSearch® System (Veridex LLC, Raritan, NJ, United States) gained approval from the United States Food and Drug Administration in 2004 for application in cases of metastatic breast cancer, and it has now been approved for application in cases of metastatic prostate cancer and CRC[85-87]. The CellSearch system detects CTCs according to the presence of the following characteristics: a round-to-oval shape by light scatter; an evident nucleus by 4′,6-diamidino-2-phenylindole staining; epithelial cell adhesion molecule positivity (EpCAM+); and CK8+, CK18+, CK19+, and CD45- status by immunofluorescence. This method is more efficient in terms of sample size and processing time compared with other CTC enrichment methods. However, it is limited by its requirement of EpCAM expression; therefore, it potentiates false-negative results[39].
Plasma (peripheral blood): The CellSearch® system is the most advanced commercially available technology[9,39]. Cohen et al[88] published one of the largest studies of CTCs in metastatic CRC (mCRC). This study included 430 patients with mCRC who were recruited at 55 clinical centers in the United Kingdom, Netherlands, and United States, and it was performed using the CellSearch® system[88]. Patients were eligible to participate in the study if they were being administered a new first-, second-, or third-line systemic chemotherapy regimen. The peripheral blood of patients was collected before treatment initiation and at four time points after treatment initiation. For analysis, patients were categorized into favorable (< 3 CTCs/7.5 mL of blood) or unfavorable (> 3 CTCs/7.5 mL of blood) groups[39,88]. The study showed that, relative to the baseline values, the median progression-free survival (PFS) and OS rates of patients in the favorable group (PFS = 7.9 mo; OS = 18.5 mo) were approximately twice those of patients in the unfavorable group (PFS = 4.5 mo; OS = 9.4 mo)[39].
Protein
Several protein-based assays have also been developed to detect cancer cells[89,90]. Most of these are antibody-based assays, although many new approaches are being developed. Protein-based assays typically detect proteins that are overexpressed or structurally altered in cancer cells compared with those in normal cells. These approaches are generally used in research settings and are not yet applicable to larger clinical studies. The expression levels of CEA are commonly used to monitor colon cancer progression[6]. The expression of protein markers for CRC is increased in the serum of patients with other cancers and is occasionally increased in patients without disease, precluding their use as individual agents for cancer screening[6].
Serum (peripheral blood): CEA is a high molecular-weight glycoprotein that belongs to the immunoglobulin superfamily[89]. The carboxy terminal of CEA contains a hydrophobic region that is modified to provide a glycosyl phosphatidylinositol link to the cell membrane. Although its presence can be determined in biopsy samples, it is usually identified in the serum. Specifically, high CEA levels are associated with cancer progression, and the levels of this marker are expected to fall after cancer surgery[89,91]. However, in the absence of cancer, high CEA levels may also be observed in response to other conditions such as hepatitis, inflammatory bowel disease, pancreatitis, and obstructive pulmonary disease. Clinically, the potential value of the CEA test lies in its use as a prognostic marker that can be used to measure the course of cancer progression after diagnosis, with higher CEA levels being indicative of greater disease severity and poorer prognosis[89,92].
LNs: The choice of antibody used for IHC is an important factor for the accurate identification of occult disease. AE1/AE3 (DAKO, Carpinteria, CA, United States) is the most widely used antibody for IHC analysis of LNs from CRC patients[69]. This polyclonal antibody is raised against several CKs, including CK19[69]. Studies that used IHC to detect occult disease reported diverse methodologies and design. The sample size included in these studies ranged from 32 to 147 patients; therefore, none were well powered to detect smaller but potentially significant metastases[69].
BM: Most studies describing ICC for the detection of DTCs in CRC patients have either used the monoclonal antibody CK2 against CK18 or the pancytokeratin antibody A45-B/B3[93-95]. The DTC detection rate in studies that used CK2 was between 16% and 32%[96-100], whereas the detection rate was higher (24%-55%) in studies that used the A45-B/B3 antibody[101-104]. Both antibodies rarely detected CK-positive cells in the BM of noncancer controls (0%-5.5%)[78]. Flatmark et al[75] reported the detection of DTCs in 41 (17%) and 28 (12%) of the 235 BM samples examined by immunomagnetic selection and ICC, respectively.
PLF: A variety of monoclonal antibodies have been used for the detection of disseminated single CRCs in the peritoneal cavity, including Ber-Ep4, CA19-9, CAM5.2, CIP83, Ra96, and C54-0. Bosch et al[105] used a combination of three monoclonal antibodies (Ks20.8, Lu5, and Ber-Ep4) to perform ICC. The rate of detection of disseminated CRCs in the PLF ranges from 10% to 67% and from 17% to 29% using ICC alone or in combination with CYT, respectively. Bosch et al[105] showed that 15% and 11% washing samples taken before and after resection, respectively, tested positive by CYT, and that 17% and 13% samples, respectively, tested positive by the combined method[105]. The best monoclonal antibody (or combination of antibodies) for the detection of disseminated CRCs has not been defined.
Cancer-associated mutations
Genetic mutation analysis is useful not only for detecting cancer in patients but also for monitoring disease spread and determining prognosis. In 1995, it was first reported that TP53 mutations could be used to follow tumor spread into margins and the draining LNs of patients with head and neck cancer[6]. K-RAS and TP53 mutations are detected in the LNs of CRC patients without histological evidence of nodal metastasis. Mutations in K-RAS are observed in approximately 40% patients with colon tumors. Currently, mutations in K-RAS are not being used for the early detection of metastatic cancer cells; however, they have significant implications in predicting the likelihood of response to antibody-based EGFR inhibitor therapy[90,106].
LNs: Studies performed on patients with CRC showed that the presence of a tumor-specific KRAS or TP53 mutation in LN samples predicted poor outcome[5]. Hayashi et al[107,108] screened the LNs of patients without histological evidence of nodal metastasis for KRAS and p53 mutations[15].
DISCUSSION
Once primary tumors are resected, metachronous metastases must arise from tumor cells that disseminate to ectopic sites before surgery. Systemic therapy primarily targets tumor cells that have detached from the primary lesion, have lodged elsewhere, are undetectable by clinical imaging, and are inaccessible to excision[5]. Therefore, metastasis-specific markers are required to accurately diagnose the existence of metastatic cancer cells. Numerous studies have demonstrated a more accurate prediction of the prognosis of patients using various immunological and PCR-based assays[109]. Each molecular method has advantages and disadvantages (Table 2). At present, it is difficult to conclude that one specific method is superior to others. Assessment of the independent prognostic impact of molecular analyses in different compartments within the same populations using comparative analyses to determine the biomarkers that provide the most accurate prognostic information remains a subject of additional investigation[11].
Table 2.
Comparison of biomarkers for detection of metastatic colorectal cancer cells
| Marker | Stable | Amplifiable | Widely reported | FDA approval | False negative | False positive | Ref. |
| DNA methylation | ● | ● | ● | [16] | |||
| mRNA | ● | ● | ● | [7] | |||
| Micro RNA | ● | ● | [81] | ||||
| CellSearch (EpCAM) | ● | ● | ● | [39] | |||
| Genetic mutation | ● | ● | ● | [15] |
EpCAM: Epithelial cell adhesion molecule; FDA: Food and Drug Administration.
CTC detection may be useful for CRC patients receiving chemotherapy. Sequential peripheral blood analyses are more acceptable compared with other resources, and many research groups are currently assessing the clinical value of CTC analyses for therapy monitoring in clinical studies[10]. Monitoring of peripheral blood during and after systemic adjuvant therapy for CTCs may provide unique information for the clinical management of individual cancer patients and allow an early change in therapy, years before the appearance of overt metastasis signals that are incurable[10]. That is, if CTC levels do not drop, systemic treatment may not be effective[8].
The molecular detection of tumor cells in regional LNs is associated with an increased risk of disease recurrence and poor survival in patients with node-negative CRC[11]. These data may favor molecular or cellular biomarkers to tailor adjuvant chemotherapy in patients with node-negative disease[11].
In patients with breast cancer, the clinical significance of CTCs in the peripheral blood is less clear than that of DTCs in BM[10]. Conversely, a meta-analysis that targeted CRC patients showed that the significance of CTCs in the peripheral blood is much clearer than that of DTCs in BM[109]. The aspiration of BM is invasive, time consuming, uncomfortable for the patient, and difficult to standardize in terms of sample quality. Another major limitation is that BM aspiration is not easy to perform during control visits at outpatient centers, which hampers repeated analyses[10]. The use of DTCs in patients with CRC may provide limited information for their clinical management.
The potential of peritoneal lavage to improve outcomes is most important for the selection of patients for adjuvant chemotherapy who present with seemingly early-stage disease and who would not otherwise receive chemotherapy. Second, intraperitoneal chemotherapy may be advantageous as a prophylactic therapy for patients with positive peritoneal micrometastases[110].
The existence of cancer stem cells in CRC has been convincingly demonstrated on a functional level. In accordance with this hypothesis, it has been shown that the number of tumor cells located at the invasive front that express high amounts of nuclear beta-catenin, which is a sign of aberrant Wnt signaling activation associated with ongoing epithelial-mesenchymal transition and stem-cell formation, is strongly correlated with metastasis and poor survival in patients with rectal cancer[11,111].
CONCLUSION
The ideal biomarker is one that is found in readily available biological samples and can be noninvasively detected. More reliable molecular markers are required to accurately diagnose the existence of metastatic cancer cells. Although thousands of papers describing genetic mutations or alterations in gene expression levels associated with various types of cancers are published each year, very few of these are translated into reliable molecular markers that can be used routinely in clinical settings. At present, it is difficult to conclude that one specific molecular marker is superior to others. Comparative analyses are recommended to assess the prognostic impact of molecular analyses in the same patient and determine the biomarkers that provide the most accurate prognostic information.
Footnotes
P- Reviewers: Di Lorenzo G, Fang BL, Shen J, Tez M, Zhang KH S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
References
- 1.van der Voort van Zijp J, Hoekstra HJ, Basson MD. Evolving management of colorectal cancer. World J Gastroenterol. 2008;14:3956–3967. doi: 10.3748/wjg.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koyanagi K, Bilchik AJ, Saha S, Turner RR, Wiese D, McCarter M, Shen P, Deacon L, Elashoff D, Hoon DS. Prognostic relevance of occult nodal micrometastases and circulating tumor cells in colorectal cancer in a prospective multicenter trial. Clin Cancer Res. 2008;14:7391–7396. doi: 10.1158/1078-0432.CCR-08-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Stat Fact Sheet. Surveillance Epidemiology and End Results, 2007. Available from: http://seer.cancer.gov/
- 4.Bilchik AJ, Hoon DS, Saha S, Turner RR, Wiese D, DiNome M, Koyanagi K, McCarter M, Shen P, Iddings D, et al. Prognostic impact of micrometastases in colon cancer: interim results of a prospective multicenter trial. Ann Surg. 2007;246:568–575; discussion 575-577. doi: 10.1097/SLA.0b013e318155a9c7. [DOI] [PubMed] [Google Scholar]
- 5.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 6.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 7.Peach G, Kim C, Zacharakis E, Purkayastha S, Ziprin P. Prognostic significance of circulating tumour cells following surgical resection of colorectal cancers: a systematic review. Br J Cancer. 2010;102:1327–1334. doi: 10.1038/sj.bjc.6605651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groot Koerkamp B, Rahbari NN, Büchler MW, Koch M, Weitz J. Circulating tumor cells and prognosis of patients with resectable colorectal liver metastases or widespread metastatic colorectal cancer: a meta-analysis. Ann Surg Oncol. 2013;20:2156–2165. doi: 10.1245/s10434-013-2907-8. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6:339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 10.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 11.Rahbari NN, Bork U, Motschall E, Thorlund K, Büchler MW, Koch M, Weitz J. Molecular detection of tumor cells in regional lymph nodes is associated with disease recurrence and poor survival in node-negative colorectal cancer: a systematic review and meta-analysis. J Clin Oncol. 2012;30:60–70. doi: 10.1200/JCO.2011.36.9504. [DOI] [PubMed] [Google Scholar]
- 12.Sadahiro S, Suzuki T, Ishikawa K, Nakamura T, Tanaka Y, Masuda T, Mukoyama S, Yasuda S, Tajima T, Makuuchi H, et al. Recurrence patterns after curative resection of colorectal cancer in patients followed for a minimum of ten years. Hepatogastroenterology. 2003;50:1362–1366. [PubMed] [Google Scholar]
- 13.Koppe MJ, Boerman OC, Oyen WJ, Bleichrodt RP. Peritoneal carcinomatosis of colorectal origin: incidence and current treatment strategies. Ann Surg. 2006;243:212–222. doi: 10.1097/01.sla.0000197702.46394.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi H, Kotake K, Sugihara K. Prognostic significance of peritoneal lavage cytology in patients with colorectal cancer. Int J Clin Oncol. 2013;18:411–417. doi: 10.1007/s10147-012-0394-8. [DOI] [PubMed] [Google Scholar]
- 15.Jubb AM, Quirke P, Oates AJ. DNA methylation, a biomarker for colorectal cancer: implications for screening and pathological utility. Ann N Y Acad Sci. 2003;983:251–267. doi: 10.1111/j.1749-6632.2003.tb05980.x. [DOI] [PubMed] [Google Scholar]
- 16.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 17.Cottrell SE. Molecular diagnostic applications of DNA methylation technology. Clin Biochem. 2004;37:595–604. doi: 10.1016/j.clinbiochem.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 20.Noda H, Mashima R, Kamiyama H, Okada S, Kawamura YJ, Konishi F. Promoter hypermethylation of tumor-related genes in sporadic colorectal cancer in young patients. J Exp Clin Cancer Res. 2007;26:521–526. [PubMed] [Google Scholar]
- 21.Noda H, Miyaji Y, Nakanishi A, Konishi F, Miki Y. Frequent reduced expression of alpha-1B-adrenergic receptor caused by aberrant promoter methylation in gastric cancers. Br J Cancer. 2007;96:383–390. doi: 10.1038/sj.bjc.6603555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noda H, Kato Y, Yoshikawa H, Arai M, Togashi K, Nagai H, Konishi F, Miki Y. Microsatellite instability caused by hMLH1 promoter methylation increases with tumor progression in right-sided sporadic colorectal cancer. Oncology. 2005;69:354–362. doi: 10.1159/000089768. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qureshi SA, Bashir MU, Yaqinuddin A. Utility of DNA methylation markers for diagnosing cancer. Int J Surg. 2010;8:194–198. doi: 10.1016/j.ijsu.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Göke B, Lamerz R, Kolligs FT. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res. 2006;12:7347–7352. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 27.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 28.Nakayama H, Hibi K, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A. Molecular detection of p16 promoter methylation in the serum of recurrent colorectal cancer patients. Int J Cancer. 2003;105:491–493. doi: 10.1002/ijc.11117. [DOI] [PubMed] [Google Scholar]
- 29.Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900–902. [PubMed] [Google Scholar]
- 30.Leung WK, To KF, Man EP, Chan MW, Bai AH, Hui AJ, Chan FK, Sung JJ. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am J Gastroenterol. 2005;100:2274–2279. doi: 10.1111/j.1572-0241.2005.50412.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama H, Hibi K, Taguchi M, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A. Molecular detection of p16 promoter methylation in the serum of colorectal cancer patients. Cancer Lett. 2002;188:115–119. doi: 10.1016/s0304-3835(01)00839-4. [DOI] [PubMed] [Google Scholar]
- 32.Zou HZ, Yu BM, Wang ZW, Sun JY, Cang H, Gao F, Li DH, Zhao R, Feng GG, Yi J. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin Cancer Res. 2002;8:188–191. [PubMed] [Google Scholar]
- 33.Hiraki M, Kitajima Y, Sato S, Nakamura J, Hashiguchi K, Noshiro H, Miyazaki K. Aberrant gene methylation in the peritoneal fluid is a risk factor predicting peritoneal recurrence in gastric cancer. World J Gastroenterol. 2010;16:330–338. doi: 10.3748/wjg.v16.i3.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamiyama H, Noda H, Takata O, Suzuki K, Kawamura Y, Konishi F. Promoter hypermethylation of tumor-related genes in peritoneal lavage and the prognosis of patients with colorectal cancer. J Surg Oncol. 2009;100:69–74. doi: 10.1002/jso.21291. [DOI] [PubMed] [Google Scholar]
- 35.Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2007;16:1935–1953. doi: 10.1158/1055-9965.EPI-06-0994. [DOI] [PubMed] [Google Scholar]
- 36.Katsumata K, Sumi T, Mori Y, Hisada M, Tsuchida A, Aoki T. Detection and evaluation of epithelial cells in the blood of colon cancer patients using RT-PCR. Int J Clin Oncol. 2006;11:385–389. doi: 10.1007/s10147-006-0590-5. [DOI] [PubMed] [Google Scholar]
- 37.Guller U, Zajac P, Schnider A, Bösch B, Vorburger S, Zuber M, Spagnoli GC, Oertli D, Maurer R, Metzger U, et al. Disseminated single tumor cells as detected by real-time quantitative polymerase chain reaction represent a prognostic factor in patients undergoing surgery for colorectal cancer. Ann Surg. 2002;236:768–775; discussion 775-776. doi: 10.1097/00000658-200212000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giribaldi G, Procida S, Ulliers D, Mannu F, Volpatto R, Mandili G, Fanchini L, Bertetto O, Fronda G, Simula L, et al. Specific detection of cytokeratin 20-positive cells in blood of colorectal and breast cancer patients by a high sensitivity real-time reverse transcriptase-polymerase chain reaction method. J Mol Diagn. 2006;8:105–112. doi: 10.2353/jmoldx.2006.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen JE, El-Deiry WS. Circulating Tumor Cells and Colorectal Cancer. Curr Colorectal Cancer Rep. 2010;6:212–220. doi: 10.1007/s11888-010-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katsuno H, Zacharakis E, Aziz O, Rao C, Deeba S, Paraskeva P, Ziprin P, Athanasiou T, Darzi A. Does the presence of circulating tumor cells in the venous drainage of curative colorectal cancer resections determine prognosis? A meta-analysis. Ann Surg Oncol. 2008;15:3083–3091. doi: 10.1245/s10434-008-0131-8. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi T, Makino M, Suzuki K, Kaibara N. Prognostic significance of reverse transcriptase-polymerase chain reaction measurement of carcinoembryonic antigen mRNA levels in tumor drainage blood and peripheral blood of patients with colorectal carcinoma. Cancer. 2000;89:970–976. [PubMed] [Google Scholar]
- 42.Yamaguchi K, Takagi Y, Aoki S, Futamura M, Saji S. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Ann Surg. 2000;232:58–65. doi: 10.1097/00000658-200007000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito S, Nakanishi H, Hirai T, Kato T, Kodera Y, Feng Z, Kasai Y, Ito K, Akiyama S, Nakao A, et al. Quantitative detection of CEA expressing free tumor cells in the peripheral blood of colorectal cancer patients during surgery with real-time RT-PCR on a LightCycler. Cancer Lett. 2002;183:195–203. doi: 10.1016/s0304-3835(02)00157-x. [DOI] [PubMed] [Google Scholar]
- 44.Bessa X, Piñol V, Castellví-Bel S, Piazuelo E, Lacy AM, Elizalde JI, Piqué JM, Castells A. Prognostic value of postoperative detection of blood circulating tumor cells in patients with colorectal cancer operated on for cure. Ann Surg. 2003;237:368–375. doi: 10.1097/01.SLA.0000055223.27623.F3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen WS, Chung MY, Liu JH, Liu JM, Lin JK. Impact of circulating free tumor cells in the peripheral blood of colorectal cancer patients during laparoscopic surgery. World J Surg. 2004;28:552–557. doi: 10.1007/s00268-004-7276-9. [DOI] [PubMed] [Google Scholar]
- 46.Sadahiro S, Suzuki T, Ishikawa K, Saguchi T, Maeda Y, Yasuda S, Makuuchi H, Yurimoto S, Murayama C. Detection of carcinoembryonic antigen messenger RNA-expressing cells in portal and peripheral blood during surgery does not influence relapse in colorectal cancer. Ann Surg Oncol. 2005;12:988–994. doi: 10.1245/ASO.2005.03.565. [DOI] [PubMed] [Google Scholar]
- 47.Sadahiro S, Suzuki T, Maeda Y, Yurimoto S, Yasuda S, Makuuchi H, Kamijo A, Murayama C. Detection of carcinoembryonic antigen messenger RNA-expressing cells in peripheral blood 7 days after curative surgery is a novel prognostic factor in colorectal cancer. Ann Surg Oncol. 2007;14:1092–1098. doi: 10.1245/s10434-006-9289-0. [DOI] [PubMed] [Google Scholar]
- 48.Koch M, Kienle P, Kastrati D, Antolovic D, Schmidt J, Herfarth C, von Knebel Doeberitz M, Weitz J. Prognostic impact of hematogenous tumor cell dissemination in patients with stage II colorectal cancer. Int J Cancer. 2006;118:3072–3077. doi: 10.1002/ijc.21784. [DOI] [PubMed] [Google Scholar]
- 49.Lloyd JM, McIver CM, Stephenson SA, Hewett PJ, Rieger N, Hardingham JE. Identification of early-stage colorectal cancer patients at risk of relapse post-resection by immunobead reverse transcription-PCR analysis of peritoneal lavage fluid for malignant cells. Clin Cancer Res. 2006;12:417–423. doi: 10.1158/1078-0432.CCR-05-1473. [DOI] [PubMed] [Google Scholar]
- 50.Allen-Mersh TG, McCullough TK, Patel H, Wharton RQ, Glover C, Jonas SK. Role of circulating tumour cells in predicting recurrence after excision of primary colorectal carcinoma. Br J Surg. 2007;94:96–105. doi: 10.1002/bjs.5526. [DOI] [PubMed] [Google Scholar]
- 51.Uen YH, Lin SR, Wu DC, Su YC, Wu JY, Cheng TL, Chi CW, Wang JY. Prognostic significance of multiple molecular markers for patients with stage II colorectal cancer undergoing curative resection. Ann Surg. 2007;246:1040–1046. doi: 10.1097/SLA.0b013e318142d918. [DOI] [PubMed] [Google Scholar]
- 52.Uen YH, Lu CY, Tsai HL, Yu FJ, Huang MY, Cheng TL, Lin SR, Wang JY. Persistent presence of postoperative circulating tumor cells is a poor prognostic factor for patients with stage I-III colorectal cancer after curative resection. Ann Surg Oncol. 2008;15:2120–2128. doi: 10.1245/s10434-008-9961-7. [DOI] [PubMed] [Google Scholar]
- 53.Wang JY, Lin SR, Wu DC, Lu CY, Yu FJ, Hsieh JS, Cheng TL, Koay LB, Uen YH. Multiple molecular markers as predictors of colorectal cancer in patients with normal perioperative serum carcinoembryonic antigen levels. Clin Cancer Res. 2007;13:2406–2413. doi: 10.1158/1078-0432.CCR-06-2054. [DOI] [PubMed] [Google Scholar]
- 54.Castells A, Boix L, Bessa X, Gargallo L, Piqué JM. Detection of colonic cells in peripheral blood of colorectal cancer patients by means of reverse transcriptase and polymerase chain reaction. Br J Cancer. 1998;78:1368–1372. doi: 10.1038/bjc.1998.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guadagni F, Kantor J, Aloe S, Carone MD, Spila A, D’Alessandro R, Abbolito MR, Cosimelli M, Graziano F, Carboni F, et al. Detection of blood-borne cells in colorectal cancer patients by nested reverse transcription-polymerase chain reaction for carcinoembryonic antigen messenger RNA: longitudinal analyses and demonstration of its potential importance as an adjunct to multiple serum markers. Cancer Res. 2001;61:2523–2532. [PubMed] [Google Scholar]
- 56.Sadahiro S, Suzuki T, Tokunaga N, Yurimoto S, Yasuda S, Tajima T, Makuuchi H, Murayama C, Matsuda K. Detection of tumor cells in the portal and peripheral blood of patients with colorectal carcinoma using competitive reverse transcriptase-polymerase chain reaction. Cancer. 2001;92:1251–1258. doi: 10.1002/1097-0142(20010901)92:5<1251::aid-cncr1445>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 57.Silva JM, Rodriguez R, Garcia JM, Muñoz C, Silva J, Dominguez G, Provencio M, España P, Bonilla F. Detection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut. 2002;50:530–534. doi: 10.1136/gut.50.4.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douard R, Moutereau S, Serru V, Sales JP, Wind P, Cugnenc PH, Vaubourdolle M, Loric S. Immunobead multiplex RT-PCR detection of carcinoembryonic genes expressing cells in the blood of colorectal cancer patients. Clin Chem Lab Med. 2005;43:127–132. doi: 10.1515/CCLM.2005.021. [DOI] [PubMed] [Google Scholar]
- 59.Vlems FA, Diepstra JH, Cornelissen IM, Ruers TJ, Ligtenberg MJ, Punt CJ, van Krieken JH, Wobbes T, van Muijen GN. Limitations of cytokeratin 20 RT-PCR to detect disseminated tumour cells in blood and bone marrow of patients with colorectal cancer: expression in controls and downregulation in tumour tissue. Mol Pathol. 2002;55:156–163. doi: 10.1136/mp.55.3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu D, Li XF, Zheng S, Jiang WZ. Quantitative real-time RT-PCR detection for CEA, CK20 and CK19 mRNA in peripheral blood of colorectal cancer patients. J Zhejiang Univ Sci B. 2006;7:445–451. doi: 10.1631/jzus.2006.B0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JY, Wu CH, Lu CY, Hsieh JS, Wu DC, Huang SY, Lin SR. Molecular detection of circulating tumor cells in the peripheral blood of patients with colorectal cancer using RT-PCR: significance of the prediction of postoperative metastasis. World J Surg. 2006;30:1007–1013. doi: 10.1007/s00268-005-0485-z. [DOI] [PubMed] [Google Scholar]
- 62.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, Hermans J, van Krieken JH, Cornelisse CJ, Tollenaar RA. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. doi: 10.1056/NEJM199807233390403. [DOI] [PubMed] [Google Scholar]
- 63.Cagir B, Gelmann A, Park J, Fava T, Tankelevitch A, Bittner EW, Weaver EJ, Palazzo JP, Weinberg D, Fry RD, et al. Guanylyl cyclase C messenger RNA is a biomarker for recurrent stage II colorectal cancer. Ann Intern Med. 1999;131:805–812. doi: 10.7326/0003-4819-131-11-199912070-00002. [DOI] [PubMed] [Google Scholar]
- 64.Haince JF, Houde M, Beaudry G, L’espérance S, Garon G, Desaulniers M, Hafer LJ, Heald JI, Lyle S, Grossman SR, et al. Comparison of histopathology and RT-qPCR amplification of guanylyl cyclase C for detection of colon cancer metastases in lymph nodes. J Clin Pathol. 2010;63:530–537. doi: 10.1136/jcp.2009.072983. [DOI] [PubMed] [Google Scholar]
- 65.Rosenberg R, Hoos A, Mueller J, Baier P, Stricker D, Werner M, Nekarda H, Siewert JR. Prognostic significance of cytokeratin-20 reverse transcriptase polymerase chain reaction in lymph nodes of node-negative colorectal cancer patients. J Clin Oncol. 2002;20:1049–1055. doi: 10.1200/JCO.2002.20.4.1049. [DOI] [PubMed] [Google Scholar]
- 66.Noura S, Yamamoto H, Ohnishi T, Masuda N, Matsumoto T, Takayama O, Fukunaga H, Miyake Y, Ikenaga M, Ikeda M, et al. Comparative detection of lymph node micrometastases of stage II colorectal cancer by reverse transcriptase polymerase chain reaction and immunohistochemistry. J Clin Oncol. 2002;20:4232–4241. doi: 10.1200/JCO.2002.10.023. [DOI] [PubMed] [Google Scholar]
- 67.Weitz J, Kienle P, Magener A, Koch M, Schrödel A, Willeke F, Autschbach F, Lacroix J, Lehnert T, Herfarth C, et al. Detection of disseminated colorectal cancer cells in lymph nodes, blood and bone marrow. Clin Cancer Res. 1999;5:1830–1836. [PubMed] [Google Scholar]
- 68.Merrie AE, van Rij AM, Dennett ER, Phillips LV, Yun K, McCall JL. Prognostic significance of occult metastases in colon cancer. Dis Colon Rectum. 2003;46:221–231. doi: 10.1007/s10350-004-6527-z. [DOI] [PubMed] [Google Scholar]
- 69.Nicastri DG, Doucette JT, Godfrey TE, Hughes SJ. Is occult lymph node disease in colorectal cancer patients clinically significant? A review of the relevant literature. J Mol Diagn. 2007;9:563–571. doi: 10.2353/jmoldx.2007.070032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soeth E, Röder C, Juhl H, Krüger U, Kremer B, Kalthoff H. The detection of disseminated tumor cells in bone marrow from colorectal-cancer patients by a cytokeratin-20-specific nested reverse-transcriptase-polymerase-chain reaction is related to the stage of disease. Int J Cancer. 1996;69:278–282. doi: 10.1002/(SICI)1097-0215(19960822)69:4<278::AID-IJC7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 71.Soeth E, Vogel I, Röder C, Juhl H, Marxsen J, Krüger U, Henne-Bruns D, Kremer B, Kalthoff H. Comparative analysis of bone marrow and venous blood isolates from gastrointestinal cancer patients for the detection of disseminated tumor cells using reverse transcription PCR. Cancer Res. 1997;57:3106–3110. [PubMed] [Google Scholar]
- 72.Weitz J, Koch M, Kienle P, Schrödel A, Willeke F, Benner A, Lehnert T, Herfarth C, von Knebel Doeberitz M. Detection of hematogenic tumor cell dissemination in patients undergoing resection of liver metastases of colorectal cancer. Ann Surg. 2000;232:66–72. doi: 10.1097/00000658-200007000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conzelmann M, Dieterle CP, Linnemann U, Berger MR. Cytokeratin 20 and guanylyl cyclase C mRNA is largely present in lymph node and liver specimens of colorectal cancer patients. Int J Cancer. 2003;107:617–628. doi: 10.1002/ijc.11425. [DOI] [PubMed] [Google Scholar]
- 74.Schlimok G, Funke I, Bock B, Schweiberer B, Witte J, Riethmüller G. Epithelial tumor cells in bone marrow of patients with colorectal cancer: immunocytochemical detection, phenotypic characterization, and prognostic significance. J Clin Oncol. 1990;8:831–837. doi: 10.1200/JCO.1990.8.5.831. [DOI] [PubMed] [Google Scholar]
- 75.Flatmark K, Borgen E, Nesland JM, Rasmussen H, Johannessen HO, Bukholm I, Rosales R, Hårklau L, Jacobsen HJ, Sandstad B, et al. Disseminated tumour cells as a prognostic biomarker in colorectal cancer. Br J Cancer. 2011;104:1434–1439. doi: 10.1038/bjc.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kienle P, Koch M, Autschbach F, Benner A, Treiber M, Wannenmacher M, von Knebel Doeberitz M, Büchler M, Herfarth C, Weitz J. Decreased detection rate of disseminated tumor cells of rectal cancer patients after preoperative chemoradiation: a first step towards a molecular surrogate marker for neoadjuvant treatment in colorectal cancer. Ann Surg. 2003;238:324–330; discussion 330-331. doi: 10.1097/01.sla.0000086547.27615.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conzelmann M, Linnemann U, Berger MR. K-ras codon 12 and 13 mutations are correlated with differential patterns of tumor cell dissemination in colorectal cancer patients. Int J Oncol. 2004;24:1537–1544. [PubMed] [Google Scholar]
- 78.Riethdorf S, Wikman H, Pantel K. Review: Biological relevance of disseminated tumor cells in cancer patients. Int J Cancer. 2008;123:1991–2006. doi: 10.1002/ijc.23825. [DOI] [PubMed] [Google Scholar]
- 79.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18:244–252. doi: 10.1097/PPO.0b013e318258b78f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 82.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 83.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 85.Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol. 2010;11:1–13. doi: 10.1007/s11864-010-0115-3. [DOI] [PubMed] [Google Scholar]
- 86.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 87.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 88.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 89.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41:685–692. doi: 10.5483/bmbrep.2008.41.10.685. [DOI] [PubMed] [Google Scholar]
- 90.Bolocan A, Ion D, Ciocan DN, Paduraru DN. Prognostic and predictive factors in colorectal cancer. Chirurgia (Bucur) 2012;107:555–563. [PubMed] [Google Scholar]
- 91.Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001;47:624–630. [PubMed] [Google Scholar]
- 92.Duffy MJ, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39:718–727. doi: 10.1016/s0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 93.Juhl H, Stritzel M, Wroblewski A, Henne-Bruns D, Kremer B, Schmiegel W, Neumaier M, Wagener C, Schreiber HW, Kalthoff H. Immunocytological detection of micrometastatic cells: comparative evaluation of findings in the peritoneal cavity and the bone marrow of gastric, colorectal and pancreatic cancer patients. Int J Cancer. 1994;57:330–335. doi: 10.1002/ijc.2910570307. [DOI] [PubMed] [Google Scholar]
- 94.Cohen AM, Garin-Chesa P, Hanson M, Weyhrauch K, Kemeny N, Fong Y, Paty P, Welt S, Old L. In vitro detection of occult bone marrow metastases in patients with colorectal cancer hepatic metastases. Dis Colon Rectum. 1998;41:1112–1115. doi: 10.1007/BF02239432. [DOI] [PubMed] [Google Scholar]
- 95.Schott A, Vogel I, Krueger U, Kalthoff H, Schreiber HW, Schmiegel W, Henne-Bruns D, Kremer B, Juhl H. Isolated tumor cells are frequently detectable in the peritoneal cavity of gastric and colorectal cancer patients and serve as a new prognostic marker. Ann Surg. 1998;227:372–379. doi: 10.1097/00000658-199803000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schlimok G, Funke I, Holzmann B, Göttlinger G, Schmidt G, Häuser H, Swierkot S, Warnecke HH, Schneider B, Koprowski H, et al. Micrometastatic cancer cells in bone marrow: in vitro detection with anti-cytokeratin and in vivo labeling with anti-17-1A monoclonal antibodies. Proc Natl Acad Sci USA. 1987;84:8672–8676. doi: 10.1073/pnas.84.23.8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schneider BM, Schlimok G, Riethmüller G, Witte J. [Bone marrow micrometastases in colorectal cancers] Fortschr Med. 1989;107:59–63. [PubMed] [Google Scholar]
- 98.Lindemann F, Schlimok G, Dirschedl P, Witte J, Riethmüller G. Prognostic significance of micrometastatic tumour cells in bone marrow of colorectal cancer patients. Lancet. 1992;340:685–689. doi: 10.1016/0140-6736(92)92230-d. [DOI] [PubMed] [Google Scholar]
- 99.Silly H, Samonigg H, Stöger H, Brezinschek HP, Wilders-Truschnig M. Micrometastatic tumour cells in bone marrow in colorectal cancer. Lancet. 1992;340:1288. doi: 10.1016/0140-6736(92)92987-q. [DOI] [PubMed] [Google Scholar]
- 100.Pantel K, Schlimok G, Angstwurm M, Weckermann D, Schmaus W, Gath H, Passlick B, Izbicki JR, Riethmüller G. Methodological analysis of immunocytochemical screening for disseminated epithelial tumor cells in bone marrow. J Hematother. 1994;3:165–173. doi: 10.1089/scd.1.1994.3.165. [DOI] [PubMed] [Google Scholar]
- 101.Leinung S, Würl P, Schönfelder A, Weiss CL, Röder I, Schönfelder M. Detection of cytokeratin-positive cells in bone marrow in breast cancer and colorectal carcinoma in comparison with other factors of prognosis. J Hematother Stem Cell Res. 2000;9:905–911. doi: 10.1089/152581600750062354. [DOI] [PubMed] [Google Scholar]
- 102.Tórtola S, Steinert R, Hantschick M, Peinado MA, Gastinger I, Stosiek P, Lippert H, Schlegel W, Reymond MA. Discordance between K-ras mutations in bone marrow micrometastases and the primary tumor in colorectal cancer. J Clin Oncol. 2001;19:2837–2843. doi: 10.1200/JCO.2001.19.11.2837. [DOI] [PubMed] [Google Scholar]
- 103.Werther K, Normark M, Brünner N, Nielsen HJ. Cytokeratin-positive cells in preoperative peripheral blood and bone marrow aspirates of patients with colorectal cancer. Scand J Clin Lab Invest. 2002;62:49–57. doi: 10.1080/003655102753517208. [DOI] [PubMed] [Google Scholar]
- 104.Schoppmeyer K, Frühauf N, Oldhafer K, Seeber S, Kasimir-Bauer S. Tumor cell dissemination in colon cancer does not predict extrahepatic recurrence in patients undergoing surgery for hepatic metastases. Oncol Rep. 2006;15:449–454. [PubMed] [Google Scholar]
- 105.Bosch B, Guller U, Schnider A, Maurer R, Harder F, Metzger U, Marti WR. Perioperative detection of disseminated tumour cells is an independent prognostic factor in patients with colorectal cancer. Br J Surg. 2003;90:882–888. doi: 10.1002/bjs.4129. [DOI] [PubMed] [Google Scholar]
- 106.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hayashi N, Ito I, Yanagisawa A, Kato Y, Nakamori S, Imaoka S, Watanabe H, Ogawa M, Nakamura Y. Genetic diagnosis of lymph-node metastasis in colorectal cancer. Lancet. 1995;345:1257–1259. doi: 10.1016/s0140-6736(95)90922-2. [DOI] [PubMed] [Google Scholar]
- 108.Hayashi N, Arakawa H, Nagase H, Yanagisawa A, Kato Y, Ohta H, Takano S, Ogawa M, Nakamura Y. Genetic diagnosis identifies occult lymph node metastases undetectable by the histopathological method. Cancer Res. 1994;54:3853–3856. [PubMed] [Google Scholar]
- 109.Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, Diener MK, Büchler MW, Koch M, Weitz J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714–1726. doi: 10.1053/j.gastro.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 110.Mohan HM, O’Connor DB, O’Riordan JM, Winter DC. Prognostic significance of detection of microscopic peritoneal disease in colorectal cancer: a systematic review. Surg Oncol. 2013;22:e1–e6. doi: 10.1016/j.suronc.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 111.Puglisi MA, Tesori V, Lattanzi W, Gasbarrini GB, Gasbarrini A. Colon cancer stem cells: controversies and perspectives. World J Gastroenterol. 2013;19:2997–3006. doi: 10.3748/wjg.v19.i20.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Spila A, Ferroni P, Cosimelli M, D’Alessandro R, Abbolito MR, Mariotti S, Aloe S, Carone MD, Graziano F, Tedesco M, et al. Comparative analysis of CA 242 and CA 19-9 serum tumor markers in colorectal cancer patients. A longitudinal evaluation. Anticancer Res. 2001;21:1263–1270. [PubMed] [Google Scholar]


