Abstract
Live cell compound screening with genetically encoded fluorescence or bioluminescence-based biosensors offers a potentially powerful approach to identify novel regulators of a signaling event of interest. In particular, compound screening in living cells has the added benefit that the entire signaling network remains intact, and thus the screen is not just against a single molecule of interest but against any molecule within the signaling network that may modulate the distinct signaling event reported by the biosensor in use. Furthermore, only molecules that are cell permeable or act at cell surface receptors will be identified as “hits,” thus reducing further optimization of the compound in terms of cell penetration. Here we discuss a detailed protocol for using genetically encoded biosensors in living cells in a 96-well format for the execution of high throughput compound screens and the identification of small molecules which modulate a signaling event of interest.
Keywords: Live cell compound screening, Genetically encoded biosensor, Cell signaling, Fluorescence resonance energy transfer, Bioluminescence
1 Introduction
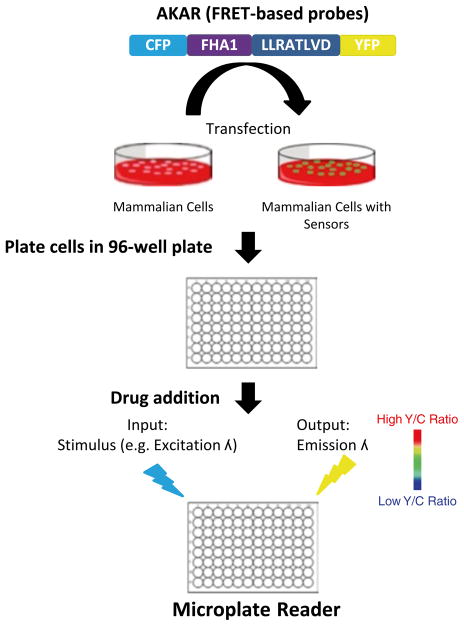
High throughput compound screening, the process in which thousands to millions of compounds are assayed against a particular molecule of interest, is a common method to identify novel small molecule modulators of a given cellular target. After further testing and optimization, ultimately such compounds can be used as chemical tools to probe the function of a target protein within a cell in a dynamic fashion, a feature that is not provided through traditional genetic manipulation [1]. Moreover, when implemented in a particular fashion, a high throughput compound screen can serve as the initial phase of the drug discovery process. In this case, a compound library is screened against a particular disease target with the intention of identifying novel therapeutic agents or new uses for existing drugs [2]. Prior to initiating a high throughput screen against a target of interest, however, it is critical to develop an assay which can rapidly and accurately report the effect of a small molecule on a target protein [1]. Specifically, in order for an assay to be used in a high throughput compound screen, it must be easily adapted to a 96-well format, or with the increasing availability and size of compound libraries, to 384- or 1,536-well format. Moreover, to be efficient in a high throughput format, the assay must be sensitive and reproducible (i.e., high signal-to-noise and low variability) and be able to be employed within a reasonable timeframe [3]. When compared to in vitro biochemical assays, achieving suitable sensitivity and reproducibility for a high throughput screen, often measured by the Z factor [4], is more challenging in cell-based assays. However, as genetically encoded biosensors can sensitively monitor signaling dynamics in living cells, when adapted accordingly, they have been proven to be a suitable assay format for live cell compound screens (Fig. 1) [5].
Fig. 1.
Schematic representation of high throughput screen using genetically encodable FRET and BRET-based biosensors (e.g., AKAR)
1.1 The cAMP/PKA Signaling Pathway
Throughout this protocol, biosensors that monitor activities in the cAMP-mediated signaling pathway will be used as an illustrative example of how to use a genetically encoded biosensor in a high throughput screen. The classical route of the production of cAMP, a canonical second messenger, is initiated when an extracellular stimuli binds to a Gs-coupled G protein coupled receptor (GPCR) on the cell surface, inducing the activation of adenylyl cyclase (AC), the enzyme which catalyzes the conversion of intracellular ATP into cAMP [7]. Once produced, cAMP can activate its effector molecules, which include the cAMP-activated protein kinase (PKA) and Exchange Protein activated by cAMP (Epac), before being degraded by phosphodiesterase (PDE). Collectively, the molecular players involved in the cAMP-mediated signaling pathway operate in a highly regulated fashion to coordinate numerous cellular functions, including insulin secretion and long term memory formation [8–10]. Furthermore, pharmacological manipulation of the molecular players involved in the cAMP-mediated signaling pathway is implicated in the treatment of various diseased states. As such, there is great effort to probe the regulation of the cAMP-mediated signaling in living cells.
1.2 Genetically Encoded Biosensors for Live Cell Compound Screens
This article focuses on the application of live cell compound screens as a tool for identifying various modulators of particular signaling pathway of interest. Compared to in vitro compound screens, live cell compound screens have two additional benefits. First, since the entire signaling network remains intact, the screen will identify any compound that modulates any molecule within the signaling pathway, not just the target molecule under study. For instance, when screening for molecules that regulate intracellular levels of cAMP, a compound that acts at the levels of the GPCR, G protein, AC, or PDE could be identified. Second, the screen will only identify compounds which are cell permeable or work at the level of the cell surface, reducing the need for further optimization in terms of cell permeability [5].
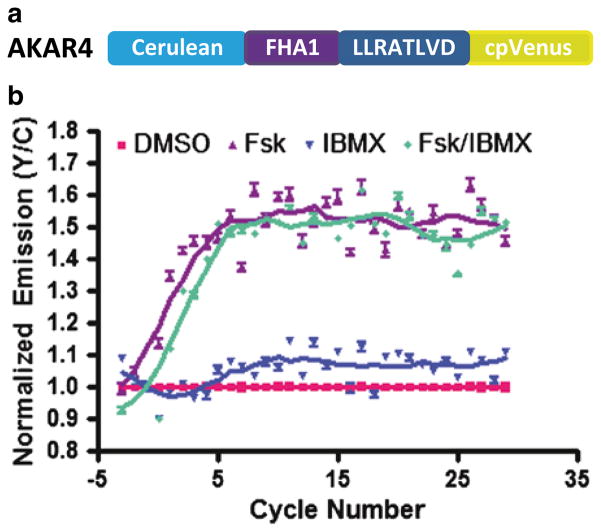
Genetically encoded biosensors have been used in live cell compound screens. A class of such biosensors is the fluorescence resonance energy transfer (FRET)-based reporters which are able to detect complex spatiotemporal dynamics of second messengers, enzyme activationctivities and protein-protein interactions in living cells (Fig. 2a) [6]. As a proof of concept, FRET-based biosensors designed to report cAMP dynamics and PKA activity were expressed in a population of cells in a multi-well format, where a microplate reader was used to detect changes in fluorescence signal over time (Fig. 2b). In doing so, these biosensors accurately identified molecules which modulated cAMP levels and the activity of PKA within the cell [5]. Similarly, FRET-based biosensors which report the activity or activation state of other promising drug targets, such as proteases and GPCRs, serve as promising tools for live cell high throughput compound screens [1, 11, 12]. An alternative design is based on bioluminescence resonance energy transfer (BRET) where a bioluminescent protein and an appropriate fluorescent protein (FP) typically serve as the donor and acceptor, respectively [12, 13]. Because BRET-based assays do not require exogenous illumination, they are not susceptible to autofluorescence and can achieve higher sensitivity than fluorescence-based assays [14, 18]. This frequently makes BRET-based assays more desirable than FRET-based methods in high throughput compound screens [15, 16]. For example, genetically encoded biosensors based on BRET have been used in a high throughput manner to detect signaling events such as second messenger dynamics and GPCR activation [12, 13]. It is also important to note that both FRET- and BRET-based assays are susceptible to compound interference. Specifically, in FRET-based screens, the inherent fluorescence of small molecules present in the compound libraries may interfere with the assay readout. Similarly, several small molecules have been shown to be direct modulators of luciferase activity, and thus would affect the readout from a BRET-based screen [19, 20]. As both FRET- and BRET-based screens have their respective strengths and limitations, care should be taken when choosing an assay for a high throughput screen [1].
Fig. 2.
High throughput activity assay based on an improved AKAR (AKAR4) (a). Design of AKAR4 (b). Representative time-course curves depicting changes in emission ratios (Y/C) of HEK293T cells expressing AKAR4, treated with DMSO (1 %), Forskolin (50 μM), IBMX (100 μM), and a combination of Forskolin and IBMX (50 μM, 100 μM respectively). 1 cycle = 16 s. Error bars represent standard deviation (n = 4)
1.3 Biosensors Applicable to This Protocol
Here, we present a detailed protocol for using genetically encodeable biosensors, based on either FRET or BRET, for high throughput compound screens in living cells. As an example, we use A-kinase activity reporter 4 (AKAR4), a FRET-based biosensor designed to report PKA activity [17]. We begin by discussing strategies for transfection of the biosensor cDNA before providing instructions on passing the cells into a 96-well format. Then we outline the steps for performing the compound screen and end with tips for data analysis and identification of “hits.” The details provided are specific for HEK293T cells, but also provide a detailed outline which can be applied to numerous cell lines and compound libraries.
2 Materials
2.1 Cell Culture
Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin–streptomycin.
10 cm tissue culture dish.
96 Well Assay Plate, White Plate, Clear Bottom with Lid, Polystyrene, Sterile (Costar, No. 3610).
96 Well Assay Plate, Black Plate, Clear Bottom with Lid, Polystyrene, Sterile (Costar, No. 3603).
Poly-D-lysine: 0.1 mg/mL.
2.2 Transfection Materials
Biosensor DNA.
Dulbecco’s Phosphate Buffered Saline—without Mg2+ and Ca2+ (DPBS).
DMEM supplemented with 10 % FBS and 1 % penicillin– streptomycin.
Calcium phosphate transfection: prepare 2× HBS buffer (0.28 M NaCl, 0.05 M HEPES, 0.0015 M Na2HPO4, pH=7.05), and a separate solution of 1 M CaCl2.
2.3 Harvesting and Counting
Solution of trypsin (0.25 %) and ethylenediaminetetraacetic acid (EDTA, 0.53 mM).
Hemocytomer.
Trypan blue.
2.4 Imaging Materials and Devices
Phosphate Buffer Saline (PBS; 10×): 0.385 M Na2HPO4·7H2O, 0.17 M NaH2PO4·H2O, 0.342 M NaCl, pH 7.4.
Hank’s Balanced Salt Solution (HBSS): 0.15 M NaCl, 4 mM KCl, 1.2 mM MgCl2·6H2O, 55.5 mM Glucose, 20 mM HEPES, pH 7.2, Store at 4 °C.
FLUOstar OPTIMA Fluorescence microplate reader (BMG Labtechnologies, Inc.).
Filters: one 420DF20 excitation filter, two emission filters (470DF40 for cyan and 535DF25 for yellow).
Compound library.
2.5 Analyzing Images and Data
Spreadsheet application (e.g., Microsoft Office Excel).
3 Methods
3.1 Cell Culture and Transfection
Grow two 10 cm dishes of HEK293T cells to 60–70 % confluency (see Note 1).
-
Transfect the cells via calcium phosphate-mediated transfection.
For each dish, mix 5 μg DNA with 55 μL of 2 M CaCl2 and add ddH2O to a final volume of 500 μL.
Add the DNA solution dropwise to 500 μL of 2× HBS while the latter is being vortexed. Vortex the final solution well and incubate for 30–40 min at room temperature. The solution should turn opaque.
Mix again and add the DNA solution dropwise onto the cells, being careful not to disrupt the cells.
Check the transfection efficiency after 24 h. Proceed only if there is greater than 70 % transfection efficiency (see Note 2).
3.2 Harvesting, Counting, and Plating
Wash each dish once with 5 mL of PBS.
Add 500 μL of 0.05 % trypsin to each dish. Add 9 mL of fresh DMEM to the first dish to harvest cells. Harvest the cells in the second dish using the same 9.5 mL so that the cells from both dishes are collected in the same 10 mL total volume.
Spin cells at 1,500 × g for 6–8 min.
-
Remove the media and resuspend the pellet in 1 mL of fresh media (see Note 3).
Count cells: dilute 7.5 μL of cells two fold with 7.5 μL of trypan blue, add the solution to the hemocytometer and count all of the cells in one large square. Determine the number of cells per μL according to the following equation (see Note 4): Determine the number of cells that should be plated so that 24 h later the cells form a monolayer in the well. For HEK293T cells, this is about 30,000 cells per well (see Note 1). Calculate the volume of media needed to dilute the cell suspension so that the desired number of cells can be plated in a volume of 100 μL per well (see Note 5). For example, HEK293T cells should be diluted to 300 cells per μL so that 30,000 cells will be added to each well in 100 μL. Importantly, some of the control wells should be left either without cells or with non-transfected cells.
Place the 96-well plate in the CO2 incubator and incubate overnight.
3.3 Live Cell Compound Screen
Wash the cells once with 100 μL of PBS (see Note 5).
Insert the plate into the microplate reader
Set up an imaging protocol such that both the CFP (excitation 420 ± 10 nm, emission 480 ± 10 nm) and FRET (excitation 420 ± 10 nm, emission 535 ± 10 nm) channels are acquired for each well (see Note 8).
Establish a three point baseline reading by reading all necessary channels for the entire plate three times.
Add compounds from the compound library of choice using a multichannel pipette. Add 10 μL of each compound to each well of the plate, except for the control wells (see Note 6). Mix by pipetting and be sure to change tips after each addition (see Note 7). If a robot system is available, it can be programmed to add the compounds.
Insert the plate into the plate reader and acquire data for each channel for the entire plate for a time period appropriate for the signaling events under study. For cAMP-mediated signaling, this is about 20 min.
Optional step: Perform an antagonist screen by adding a subsaturating dose of compound to each well in order to activate the signaling event under study, including half of the vehicle control wells. For example, when screening molecules which antagonize PKA activity using AKAR4, a 100 nM dose of the β2-adrenergic receptor agonist is added to each well.
3.4 Data Analysis
Using Excel, determine the average reading from the wells which had no cells or non-transfected cells for both the donor and acceptor channels. Subtract this value from the respective value for each well at each time point. These values serve as the background corrected values.
- For each time point, take the ratio of the acceptor to donor.
Normalize the calculated ratio at each time point to the value of the respective well just before drug addition.
Also, normalize the calculated ratios to the vehicle control wells.
For a time course, plot the normalized ratio over time.
Calculate the standard deviation of the vehicle control wells. A “hit” is a compound that falls 4–5 standard deviations above or below the average of the vehicle control wells.
Footnotes
The details provided are using HEK293T cells as an example. Optimization of cell culture, transfection techniques, and plating density may be needed for other cell types.
If transfection efficiency is not greater than 70 %, the signal-to- noise ratio may not be high enough to achieve high sensitivity. As an alternative to transient transfection, a cell line stably expressing the biosensor of interest could be generated and used.
Be sure not to aspirate off the pellet. It is best to turn the centrifuge tube horizontally to aspirate the media off of the side of the tube, keeping the tip away from the pellet.
The volume of one square on the hemocytometer is 0.1 mm3 or 0.1 μL, so the quick calculation is: cells/μL=#cells×Dilution Factor×10.
For cell lines which do not adhere well to plastic, such as HEK293T, it is beneficial to plate each well of the 96-well plate with poly-D-lysine beforehand. To do this, add 50 μL of 0.1 mg/mL poly-D-lysine to each well of a 96-well plate and incubate at RT for 1 h. Aspirate off the solution and wash once with PBS. Allow the plate to dry for 30 min in the laminar flow hood before plating cells.
Controls for the experiment will be a set of naïve cells, meaning no drugs or vehicle control is added to the cells. The second set of controls will be cells treated with only the vehicle control.
How much media is added to each well depends on the composition of the compound library. This example is using a library of stock concentration of 200 μM such that when 10 μL of compound is added to a well containing 190 μL of HBSS the final screening concentration is 10 μM.
If using a BRET-based biosensor, there will be some differences in the protocol for using a FRET-based biosensor. In particular, when using a BRET-based biosensor:
(a) Cells do not need to be washed with PBS.
(b) A white walled plate should be used.
(c) The desired concentration of luciferase substrate, e.g., coelenterazine-h, must be added to HBSS.
(d) Acquisition of emission wavelengths must be adjusted accordingly and no excitation light is required.
References
- 1.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High- throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3(8):466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 2.Herzberg RP. Design and implementation of high-throughput screening assay. Methods Mol Biol. 2009;565:1–32. doi: 10.1007/978-1-60327-258-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Mayr L, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9(5):580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhang JH, Chung TDY, Olderburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2) doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 5.Allen MD, DiPilato LM, Rahdar M, Ren YR, Chong C, Liu JO, Zhang J. Reading dynamic kinase activity in living cells for high- throughput screening. ACS Chem Biol. 2006;1(6):371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 6.Ni Q, Titov DV, Zhang J. Analyzing protein kinase dynamics in living cells with FRET reporters. Methods. 2006;40(3):279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Adjobo-Hermans MJW, Goedhar J, van Weeren L, Nijmeijer S, Manders EMM, Offermanns S, Gadella TWJ. Real-time visualization of heterotrimeric G protein Gq activation in living cells. BMC Biol. 2011;9(1):32. doi: 10.1186/1741-7007-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 9.Williams C. cAMP detection methods in HTS: selecting the best from the rest. Nat Rev Drug Discov. 2004;3:125–135. doi: 10.1038/nrd1306. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta- adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 11.Tian H, Ip L, Luo H, Chang DC, Luo KQ. Br J Pharm. 2007;150(3):321–324. doi: 10.1038/sj.bjp.0706988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomsen W, Frazer J, Unett D. Functional assays for screening GPCR targets. Curr Opin Biotechnol. 2005;16(6):655–665. doi: 10.1016/j.copbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Jiang LI, Collins J, Davis R, Lin KM, DeCamp D, Roach T, Hsueh R, Rebres RA, Ross EM, Taussig R, Fraser I, Sternweis PC. Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J Biol Chem. 2007;282(14):10576. doi: 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boute N, Jockers R, Issad T. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol Sci. 2002;23(8):351–354. doi: 10.1016/s0165-6147(02)02062-x. [DOI] [PubMed] [Google Scholar]
- 15.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol. 2008;3(6):346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 16.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5(1):127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 17.Depry C, Allen MD, Zhang J. Visualization of PKA activity in plasma membrane microdomains. Mol Biosyst. 2011;7:52–58. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- 18.Herbst KJ, Allen MD, Zhang J. Luminescent kinase activity biosensors based on a versatile bimolecular switch. J Am Chem Soc. 2011;133(15):5676–5679. doi: 10.1021/ja1117396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbst KJ, Allen MD, Zhang J. The cAMP-dependent protein kinase inhibitor H-89 attenuates the bioluminescence signal produced by Renilla luciferase. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorne N, Inglese J, Auld DS. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem Biol. 2010;17(6):646–657. doi: 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]