Summary
Elevated hepatic synthesis of fatty acids and triglycerides, driven by hyperactivation of the SREBP-1c transcription factor, has been implicated as a causal feature of the metabolic syndrome. SREBP-1c activation requires the proteolytic maturation of the endoplasmic reticulum-bound precursor to the active, nuclear transcription factor, which is stimulated by feeding and insulin signaling. Here we show that feeding and insulin stimulate the hepatic expression of PASK. We also demonstrate, using genetic and pharmacologic approaches, that PASK is required for the proteolytic maturation of SREBP-1c in cultured cells and in mouse and rat liver. Inhibition of PASK improves lipid and glucose metabolism in dietary animal models of obesity and dyslipidemia. Administration of a PASK inhibitor decreases hepatic expression of lipogenic SREBP-1c target genes, decreases serum triglycerides and partially reverses insulin resistance. While the signaling network that controls SREBP-1c activation is complex, we propose that PASK is an important component with therapeutic potential.
INTRODUCTION
The excessive synthesis and storage of lipid are prominent features of the current epidemic of metabolic disorders, including obesity, diabetes and their comorbidities. Upon feeding, fatty acids and triglycerides are synthesized primarily in the liver and adipose tissue in response to insulin signaling, and then either stored locally or exported to other tissues for use in ATP production. The Sterol Regulatory Element Binding Protein (SREBP-1c) transcription factor is a principal regulator of lipogenesis in these two tissues (Horton et al., 2002; Rosen et al., 2000). Upon activation, SREBP-1c stimulates the expression of the entire enzymatic pathway that converts acetate to fatty acids and their esterification to triacylglycerol (TAG) (Horton et al., 2003). Hyperactivation of SREBP-1c has been implicated in promoting pathologic fat synthesis and driving features of the metabolic syndrome, including hepatic lipid accumulation (or steatosis), dyslipidemia and insulin resistance (Brown and Goldstein, 2008).
While often pathological in modern humans, SREBP-1c activation in response to feeding is a normal physiological response that enables the storage of excess energy in the stable and compact TAG form. SREBP-1c activation by feeding occurs predominantly in response to insulin, which acts at multiple regulatory steps. The transcription of the SREBP-1c mRNA is strongly induced by insulin via a mechanism involving the LXR transcription factor (Chen et al., 2004; DeBose-Boyd et al., 2001; Repa et al., 2000; Schultz et al., 2000; Yoshikawa et al., 2001) and SREBP-1c autoregulation (Amemiya-Kudo et al., 2000; Chen et al., 2004). Insulin also acts, through GSK-3β inhibition and potentially through Lipin1 phosphorylation, to extend the otherwise very short half-life of active SREBP-1c (Harris et al., 2007; Peterfy et al., 2010; Peterson et al., 2011; Sundqvist et al., 2005). One of the most unique mechanisms underlying SREBP-1c activation by insulin signaling, however, is the proteolytic maturation of the ER membrane-embedded SREBP-1c precursor into the active and nuclear mature SREBP-1c transcription factor (Hegarty et al., 2005).
Analogous to its better-characterized paralog SREBP-2, SREBP-1c maturation is thought to occur through the regulated translocation of the precursor to the Golgi, where it is cleaved sequentially by two proteases, liberating the mature form from its two transmembrane segments (Horton et al., 2002; Raghow et al., 2008). The regulatory pathway linking insulin and SREBP-1c maturation is incompletely understood, but has been shown to require the canonical PI3K/Akt pathway (Krycer et al., 2010; Yellaturu et al., 2009a). More recently, evidence has accumulated that insulin-responsive SREBP-1c activation also requires the mechanistic Target of Rapamcyin Complex 1 (mTORC1) (Duvel et al., 2010; Li et al., 2010; Porstmann et al., 2008). While a portion of the insulin/Akt effect on SREBP-1c maturation appears to be dependent upon the regulation of INSIG2 gene expression (Yecies et al., 2011; Yellaturu et al., 2009b), the mechanism(s) underlying the remaining Akt/mTORC1 effect on SREBP-1c proteolytic maturation have not been identified.
PAS kinase (PASK) is an evolutionarily conserved serine/threonine kinase, which we have proposed to play an important role as a nutrient-responsive metabolic regulator (Hao and Rutter, 2008). Mice lacking the PASK gene (PASK−/−) exhibit a number of tissue-specific metabolic abnormalities, among which the most profound phenotype observed was decreased susceptibility to hepatic lipid infiltration in animals challenged with a high-fat diet (Hao et al., 2007). Hepatic steatosis is now recognized to be a common and devastating component of the metabolic syndrome (Cohen et al., 2011). However, the molecular mechanisms underlying hepatic steatosis are unknown and no therapies are currently approved for its treatment. Given the profound PASK−/− phenotype, the importance of this process in human disease, and the potential to discover new therapeutic targets, we sought to identify the mechanism whereby PASK regulates hepatic lipid metabolism.
RESULTS
PASK stimulates hepatic lipogenesis by activating SREBP-1c
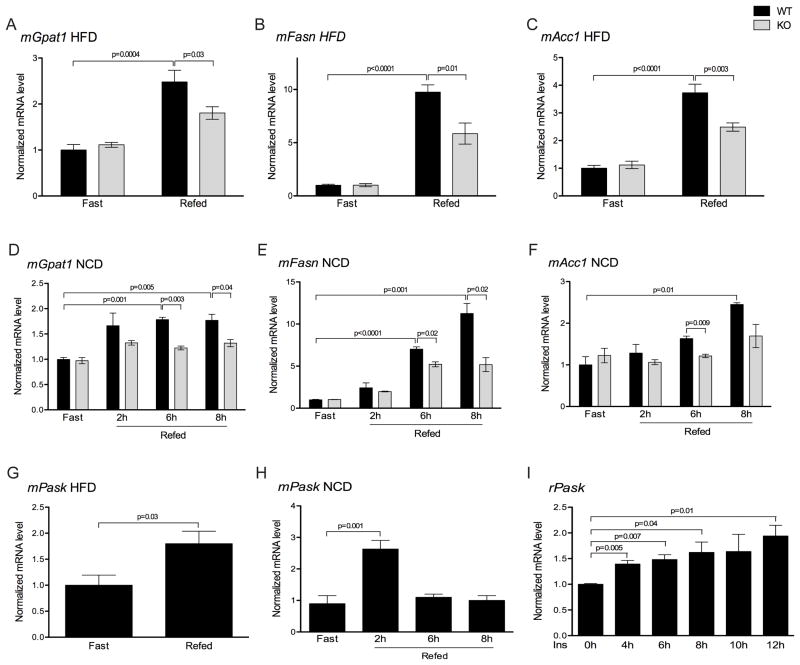
Since PASK−/− mice are protected from high-fat diet induced hepatic steatosis, we initially compared the hepatic transcriptional profiles of lipogenic genes from PASK−/− and wild-type (WT) mice in both the fasted and fed state. The expression of the genes encoding glycerol-3-phosphate acyltransferase (Gpat1), fatty acid synthase (Fasn), and acetyl-coA carboxylase (Acc1) were all similarly low in the fasted state in WT and PASK−/− liver (Figure 1A–C). While their expression increased substantially in WT mice upon feeding, this induction was blunted in PASK−/− mice (Figure 1A–C). This feeding-dependent effect led us to test the expression of these same genes in fasted and fed mice that were maintained on a normal chow diet. As with the high-fat diet, expression of lipogenic genes was similar in fasted WT and PASK−/− mice in the fasted state. Refeeding for 6 or 8 h, however, significantly increased the expression of Gpat1, Fasn and Acc1 in WT liver and this increase was absent or blunted in PASK−/− mice (Figure 1D–F).
Figure 1. PASK is feeding-induced and is required for normal feeding-dependent induction of lipogenic gene expression.
Wild-type (WT) and PASK−/− (KO) mice on the C57/BL6J background were maintained on a 60% high-fat diet (HFD) (A–C, G) for 8 weeks or a normal chow diet (NCD) (D–F, H) for 12 weeks. Before harvesting, mice were fasted for 24 h, or fasted for 24 h and refed either a HFD for 12 h (A–C, G) or a NCD for the indicated times (D–F, H). Livers were harvested and mRNA levels of indicated genes were measured by qRT-PCR and normalized to Cyclophilin A mRNA. Data shown are the average of n>4 ± SEM, with the “WT fasted” value set as 1. (I) Rat primary hepatocytes were serum starved overnight and incubated with 25nM insulin for the indicated times. PASK mRNA was measured by qRT-PCR and normalized to r36B4. Data shown are the average of n=3 ± SEM. The value of 0 h group was set as 1. See also Table S1 and S2.
We previously observed that PASK activity was nutrient responsive in cultured cells. This led us to hypothesize that PASK expression or activity might also be stimulated in the liver upon feeding, which might then be required for normal feeding-induced lipogenic gene expression. We therefore measured the hepatic PASK mRNA levels in fasted and fed mice. High-fat diet fed WT mice showed an increase in PASK expression upon feeding (Figure 1G). An even larger increase was observed in normal chow-fed animals upon 2 h of feeding following a fast (Figure 1H). Interestingly, by 6 h of feeding, the PASK mRNA abundance had returned to the fasted level. This acute hepatic induction of PASK by feeding is likely to be a cell-autonomous response to insulin as we observed induction of PASK mRNA by insulin in WT primary hepatocytes (Figure 1I).
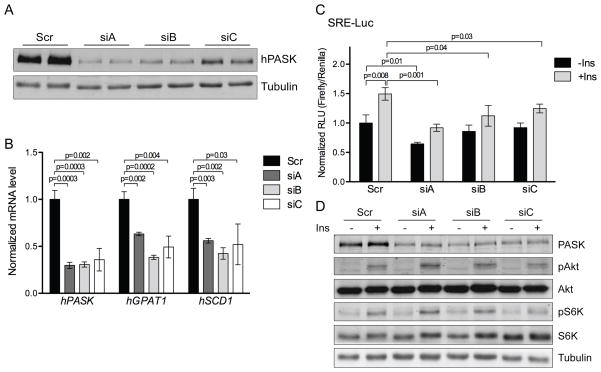
To further investigate the mechanisms whereby PASK deficiency led to reduced expression of lipogenic genes, we employed HepG2 human hepatoma cells treated with insulin, which is a major hormonal mediator of the feeding response on lipogenic gene expression. PASK knockdown by three different siRNAs in HepG2 cells (Figure 2A) caused decreased expression of GPAT1 and SCD1 upon insulin treatment (Figure 2B). Unlike the liver, however, HepG2 cells seem to have a basal level of lipogenic gene expression that is PASK-dependent. In the absence of insulin, PASK knockdown caused decreased expression of GPAT1 and SCD1 in HepG2 cells (Figure S1A). The fact that acute PASK knockdown in HepG2 cells recapitulates the regulation of gene expression observed in vivo in PASK−/− liver suggests a direct role for PASK in regulating lipogenesis.
Figure 2. PASK is required for full SREBP-1 activity.
(A) HepG2 cells were treated with scrambled or PASK-specific siRNA (siA, siB or siC), and subjected to immunoblot for PASK. (B) PASK silenced HepG2 cells were serum starved overnight and stimulated with 100nM insulin for 6 h. mRNA levels of indicated genes were measured by qRT-PCR and normalized to Tubulin mRNA levels. Data shown are the average of n=3 ± SEM, with the Scr value set as 1. (C) PASK Silenced HepG2 cells were transfected with SRE-Luc reporter, serum starved overnight and stimulated with 100nM insulin for 6 h, as indicated. Firefly and Renilla luciferase were assayed using the Dual-Reporter Luciferase Assay System. Data shown are the average of n=3 ± SD, with the “scr -ins” value set as 1. (D) Following luciferase assay, cell lysates from (C) were subjected to immunoblot for the phosphorylation state and abundance of the indicated proteins. See also Figure S1.
To determine whether this change in mRNA abundance is due to transcriptional control, we examined the effects of PASK knockdown on luciferase activity from a reporter gene driven by the promoter of either GPAT1 (Yoshida et al., 2009) or SCD1 (Bene et al., 2001). As shown in Figure S1B and S1C, PASK knockdown decreased the activity of both the GPAT1 and SCD1 promoters. As SREBP-1c is one of the major feeding and insulin-responsive regulators of GPAT1, SCD1 and the other components of the fatty acid and TAG biosynthetic pathway, we analyzed the effects of PASK knockdown on a luciferase reporter gene containing isolated SREBP binding sites (Dooley et al., 1998). PASK knockdown caused a modest reduction in SREBP activity in serum-starved conditions, which became more pronounced upon insulin treatment (Figure 2C). One potential explanation for the loss of basal and insulin-stimulated SREBP-1 activation upon PASK knockdown is that the Akt/mTORC1 pathway, which is required for SREBP-1 activation, is impaired by PASK knockdown. However, the phosphorylation state of Akt and S6K, indicative of Akt and mTORC1 activity, respectively, were both unchanged by PASK knockdown (Figure 2D), demonstrating that the effects of PASK on SREBP-1 activation occur without effects on Akt and mTORC1 activity. Over many independent experiments, PASK knockdown with each of these three different siRNAs caused a significant impairment in the fold-induction of SREBP activity by insulin (siA: n=15, p=0.003; siB: n=16, p<0.0001; siC: n=9, p=0.01). Taken together, these data support a model wherein SREBP-1c is a major mediator of the transcriptional effects of PASK on the regulation of lipid biosynthesis and that PASK is required for the full effect of insulin on SREBP-1c induction.
PASK promotes SREBP-1 maturation
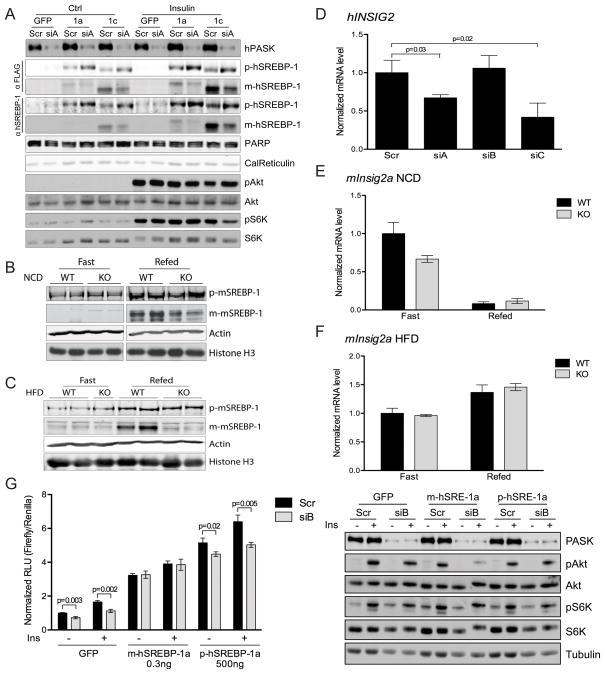
SREBP-1c is profoundly regulated by insulin at multiple levels as described previously. Given the requirement of PASK for the normal regulation of SREBP-1c, we first assessed whether it is also required for its proteolytic maturation. We examined this directly by monitoring the levels of the precursor and mature forms of SREBP-1c. We also examined the maturation of SREBP-1a, which is derived from the same gene as SREBP-1c, but is the product of an alternative promoter (Hua et al., 1995). Treatment of cells with either of two PASK-specific siRNAs (siA or siB), but not the control siRNA, blunted the insulin-responsive maturation of SREBP-1a and SREBP-1c (Figure 3A and S2A). As before, PASK knockdown impaired SREBP-1 processing while having no effect on Akt or S6K phosphorylation (Figure 3A and S2A). The insulin-responsive proteolytic maturation of SREBP-1c is also observed upon feeding in rodent liver. In WT mice maintained on a normal chow diet, we observed a significant increase in the abundance of mature SREBP-1c in the fed state relative to the fasted state (Figure 3B). This induction was severely blunted in PASK−/− mice (Figure 3B). Feeding-induced maturation of SREBP-1c was also observed in WT mice maintained on a high-fat diet, but was essentially absent in PASK−/− mice (Figure 3C).
Figure 3. PASK promotes SREBP-1 maturation.
(A) PASK silenced HepG2 cells expressing GFP, 3xFlag-tagged precursor SREBP-1a or SREBP-1c were serum starved overnight and stimulated with 100nM insulin for 6 h, as indicated. Whole cell lysate and nuclear extract were subjected to immunoblot for the phosphorylation state and abundance of the indicated proteins. (B and C) Livers were harvested from wild-type (WT) or PASK−/− (KO) mice maintained on normal chow (B) or high fat (C) diet as described in Figure 1. Whole liver lysate and nuclear extract were subjected to immunoblot for the abundance of the indicated proteins. (D) HepG2 cells were treated and human INSIG2 mRNA level was analyzed as in Figure 2B. (E and F) Mouse Insig2a mRNA level in normal chow (E) or high fat (F) diet-fed mouse liver was measured as in Figure 1. (G) Left: SRE-Luc activity was measured in PASK silenced HepG2 cells expressing GFP, mature or precursor SREBP-1a along with SRE-Luc. Right: Following luciferase assay, cell lysates were subjected to immunoblot for the phosphorylation state and abundance of the indicated proteins. See also Figure S2.
The INSIG proteins are negative regulators of SREBP-1 maturation and are regulated at many levels, including transcription and mRNA stability (Gong et al., 2006; Yabe et al., 2002; Yabe et al., 2003; Yang et al., 2002; Yellaturu et al., 2009b). Alterations in the expression of INSIG2A have been specifically implicated in mediating the effects of insulin on SREBP-1c processing in hepatic tissue (Yabe et al., 2003; Yecies et al., 2011; Yellaturu et al., 2009b). The effects of PASK knockdown on SREBP-1c maturation appear to not be through altered INSIG2 expression, however, as this gene is not induced in HepG2 cells upon PASK knockdown (Figure 3D). In fact, INSIG2 expression actually is decreased by two out of three PASK-targeted siRNAs, perhaps as a compensatory mechanism that the cell employs to attempt to restore SREBP-1c maturation. Similarly, we measured the Insig1, Insig2a and Insig2b mRNAs in WT and PASK−/− liver under fed and fasted conditions in mice maintained on either normal chow or high-fat diet. Again, under no condition did we observe an increase in Insig2a expression (Figure 3E, 3F). We also did not observe an increase in expression of the other conclude that PASK promotes SREBP-1 maturation, but does so independent of effects on INSIG gene expression.
We also addressed whether PASK might regulate the stability or transcriptional activation potential of mature, nuclear SREBP-1, other known mechanisms of SREBP-1 control. We expressed a truncated form of SREBP-1a (or GFP as a control) and asked whether PASK knockdown had any effect on SRE-driven luciferase activity. As shown in Figure 3G, PASK knockdown with siB in GFP-control cells led to decreased luciferase activity both in the presence and absence of insulin. Expression of the mature SREBP-1a, however, caused luciferase activity to become completely insensitive to PASK knockdown (Figure 3G). We were concerned that expression of this ectopic transcription factor might have overwhelmed the PASK regulatory system and the PASK insensitivity was due to an overexpression artifact. We, therefore, expressed the full-length, precursor SREBP-1a and found that PASK knockdown retained the ability to suppress luciferase activity (Figure 3G). To achieve a similar transcriptional response, we expressed much more full-length SREBP-1a, which requires processing for activation. In spite of this, full-length SREBP-1a was regulated by PASK, while the mature form of SREBP-1a was not. We, therefore, conclude that the activity of the nuclear, mature form is resistant to PASK knockdown. PASK knockdown with siA, which is more effective at PASK silencing and decreasing SREBP-1 activity (Figure 2), follows the same pattern, although PASK knockdown still causes a small but significant decrease in SREBP activity even when the mature SREBP-1a is expressed (Figure S2D). This is likely due to endogenous SREBP-1, whose activity remains PASK-dependent. As before, the activity of overexpressed full-length, precursor SREBP-1a was PASK-dependent. The loss of SREBP-1 activity upon PASK knockdown again occurred in the absence of any effect on insulin or mTORC1 signaling (Figure 3G and S2D). We initially attempted the above experiment with SREBP-1c, which has much weaker transcriptional activation potential than SREBP-1a (Shimano et al., 1997a), but we were unable to express enough full-length precursor SREBP-1c to achieve comparable luciferase activity and, therefore, technical issues have prevented rigorous interpretation of the analogous experiment with SREBP-1c.
Finally, we sought to address whether the transcriptional control of SREBP-1c might depend upon PASK. This is a difficult question to answer, however, because SREBP-1c stimulates the expression of its own promoter (Amemiya-Kudo et al., 2000; Chen et al., 2004). Therefore, post-transcriptional effects on SREBP-1c activity will indirectly result in alterations to the SREBP-1c mRNA level. As expected, we observed that the SREBP-1c mRNA was decreased in PASK−/− liver (Table S1) in parallel with the decrease in SREBP-1c target genes described previously (Figure 1). Inhibition of PASK caused a modest reduction in the activity of the mSREBP-1c promoter as determined using a luciferase reporter assay in rat primary hepatocytes stimulated with insulin (Figure S2E). This effect of PASK on promoter activity was dependent on SREBP-1 autoregulation, however, as a promoter wherein the SREBP binding site has been mutated showed no response to PASK inhibition (Figure S2E). We also observed a modest decrease in the level of the SREBP-1a mRNA (Table S1), but this is unlikely to be a major contributor to the expression of lipogenic genes in the liver as it is expressed at a much lower level than SREBP-1c (Shimomura et al., 1997). We observed no consistent alteration in the expression of SREBP-2 or its target genes in PASK−/− mice maintained on either a normal chow or high-fat diet (Table S1 and S2). Taken together, these experiments suggest that, in the absence of transcriptional control, PASK regulates the post-translational maturation of SREBP-1 in response to insulin. We can’t exclude the possibility that PASK might also regulate SREBP-1c transcriptionally, however.
Pharmacological inhibition of PASK leads to decreased SREBP-1 activity
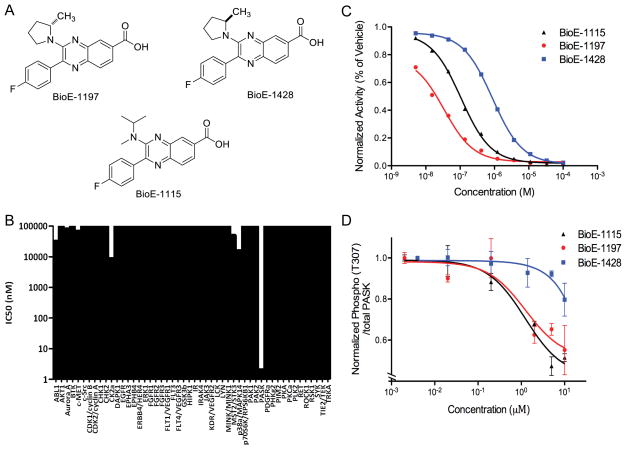
To facilitate the assessment of the importance of PASK in the regulation of SREBP-1c activity and lipid homeostasis in vivo, we developed a series of PASK inhibitors. Among them, we selected for further study two highly selective and potent inhibitors, BioE-1115 and BioE-1197 (Figure 4A). To assess their in vitro specificity, we measured the activity of PASK and 50 other protein kinases, selected to represent the breadth of the human kinase family, in the presence of BioE-1115 or BioE-1197. As shown in Figure 4B, BioE-1115 specifically inhibits PASK, with an IC50 of ~4nM. Other than PASK, casein kinase 2α was the kinase most potently inhibited by BioE-1115, having an IC50 of ~10μM. This is interesting due to the unexpected structural similarities between the kinase domains of PASK and casein kinase 2 (Kikani et al., 2010). In spite of this similarity, however, BioE-1115 was roughly 2500-fold more potent as an inhibitor of PASK than of casein kinase 2α2 (Figure 4B). BioE-1197 showed similar specificity for PASK versus the same 50 protein kinases. As shown in Figure S3A, even using an extremely high concentration of BioE-1197 (100μM), the majority of the 50 kinases were either unaffected or only modestly inhibited. PASK, however, was essentially completely inactivated and 8 other kinases were also substantially inhibited. In full dose-response inhibition curves for these nine BioE-1197-sensitive kinases, all but two were inhibited poorly, with an IC50 of greater than 50μM (Figure S3B). Similar to what we observed with BioE-1115, casein kinase 2α was inhibited by BioE-1197, but with a ~1000-fold higher IC50 compared to PASK (Figure S3B). For subsequent use as a control, we also synthesized an enantiomer of BioE-1197, which was ~100-fold less potent at inhibiting PASK in vitro (IC50= 870nM) (Figure 4C).
Figure 4. BioE-1115 and BioE-1197 are PASK-specific inhibitors.
(A) The chemical structure of BioE-1115, BioE-1197 and BioE-1428. (B) Activity of the indicated kinases was measured in the presence of either vehicle or BioE-1115 and IC50s were measured. (C) Purified PASK protein kinase activity was assayed as in (B) in the presence of the indicated concentrations of BioE-1197, BioE-1428 or BioE-1115 and the percent of vehicle-treated activity is as indicated. (D) HEK293 cells expressing a Flag-tagged PASK protein were treated with the indicated concentrations of BioE-1115, BioE-1197 or BioE-1428. PASK activity was analyzed by ELISA with both phospho-Akt substrate antibody and pan-PASK antibody. The quantitated phospho/total PASK signal is plotted ± SEM. See also Figure S3
We next examined the efficacy of BioE-1115, BioE-1197 and BioE-1428 for PASK inhibition in cultured cells. We tested this by virtue of the ability of PASK to auto-phosphorylate at Thr-307, which we have demonstrated to be solely dependent upon PASK activity (Figure S3C). In the presence of either BioE-1115 or BioE-1197, we observed a dose-dependent loss of PASK phosphorylation, with an IC50 of ~1μM (Figure 4D). As expected, the enantiomer BioE-1428 was less effective at PASK inhibition in cells, with an IC50 of >10μM (Figure 4D).
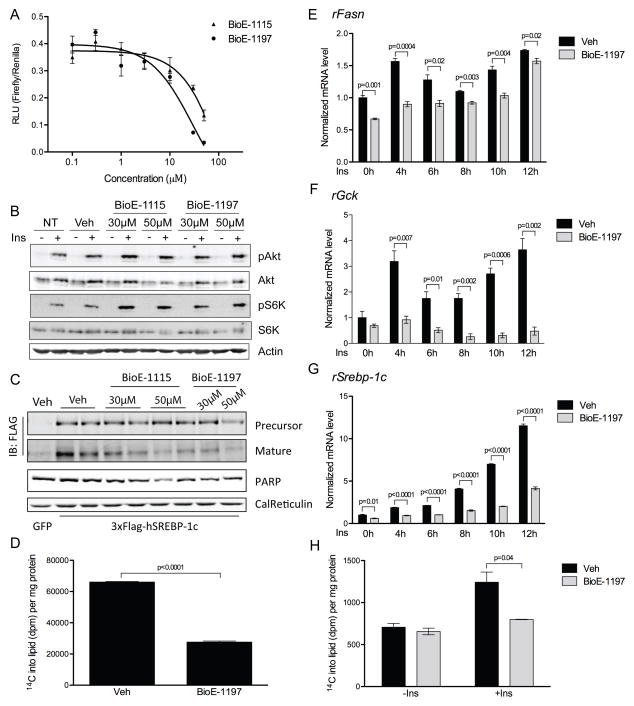
Having validated that BioE-1115 and BioE-1197 are potent and specific inhibitors of PASK, we were then able to use them to probe the acute effects of PASK inhibition. Specifically, we sought to determine whether pharmacologic PASK inhibition, like PASK knockdown and PASK deletion, leads to impaired SREBP-1 activity. We measured SRE-driven luciferase activity in the presence of varying concentrations of BioE-1115 and BioE-1197. We observed a significant reduction in SREBP activity at all concentrations above 10μM BioE-1115 and 3μM BioE-1197 (Figure 5A), without any observable effects on cell morphology or growth rate. This effect is likely to be specific for PASK inhibition, because treatment of cells with BioE-1428 caused only a modest reduction of SREBP activity even at the highest concentration tested, 50μM (Figure S3D). As with PASK knockdown, neither compound impaired the phosphorylation of Akt or S6K in response to insulin (Figure 5B and S3E) demonstrating again that the loss of SREBP-1 activity is not due to a loss of canonical insulin signaling.
Figure 5. Pharmacologic inhibition of PASK suppresses SREBP-1 activation.
(A) SRE-Luc activity was measured in HepG2 cells treated with vehicle, or the indicated doses of BioE-1115 or BioE-1197 followed by 100nM insulin for 6 h. Data are normalized to vehicle treated samples and shown as the average of n=3 ± SD. (B) HepG2 cells were treated as in (A). Cell lysates were subjected to immunoblot for the phosphorylation state and abundance of the indicated proteins. (C) HepG2 cells were infected as in Figure 3A, and treated with PASK inhibitors and insulin as in (A). Whole cell lysate and nuclear extract were subjected to immunoblot for the abundance of indicated proteins. (D) 14C-acetate incorporation into lipid was measured in BioE-1197 treated HepG2 cells and normalized to total protein in the lysate. Data shown are the average of n=3 ± SD. (E–G) Rat primary hepatocytes were treated and mRNA abundance of the indicated genes was measured as in Figure 1I. Data shown are the average of n=3 ± SEM. The value of vehicle-treated/unstimulated group was set as 1. (H) Rat primary hepatocytes were treated and measured for 14C-acetate incorporation into lipids as in (D). Data shown are the average of n=3 ± SD. See also Figure S3.
If BioE-1115 and BioE-1197 suppressed SREBP-1 activity through inhibition of PASK, they should act through blocking SREBP-1 proteolytic activation as was observed with PASK knockdown in cells and in liver of PASK−/− mice. Both BioE-1115 and BioE-1197 suppressed SREBP-1c maturation, as evidenced by a decrease in the mature:precursor SREBP-1c ratio, at 30 and 50μM concentrations (Figure 5C and S3F). In contrast, BioE-1428 had no effect at 30μM and the effect at 50μM was weaker than observed with BioE-1197 (Figure S3F). When quantified over five independent experiments, BioE-1197 was significantly more efficacious at suppressing SREBP-1c maturation than BioE-1428 (Figure S3G). In summary, both PASK knockdown and pharmacologic inhibition lead to impaired SREBP-1 activity, and both manipulations impact the maturation step. These complementary data support the hypothesis that PASK is required for the normal maturation and activation of SREBP-1.
We next tested whether PASK inhibition would lead to a decrease in the synthesis of fatty acids and triglycerides as would be predicted based on the impairment in SREBP-1c activity. We measured the incorporation of 14C from 14C-acetate into lipid soluble material, which is primarily triglyceride under these conditions. Treatment with 10 μM BioE-1197 caused a ~60% decrease in 14C incorporation into lipids (Figure 5D), suggesting that the impaired SREBP-1 activity is manifest at the level of lipogenesis.
Rat primary hepatocytes have been used extensively in the study of insulin-responsive SREBP-1c activation and lipogenesis. We previously observed that PASK gene expression was stimulated by insulin (Figure 1I). Therefore, we explored whether PASK inhibition would have any effect on the insulin-responsive expression of SREBP-1c target genes in these cells. Primary hepatocytes were isolated, plated, pre-treated with DMSO or BioE-1197 for 16 h and then stimulated with either insulin or vehicle over a 12 h timecourse. Insulin caused a significant increase in the mRNAs encoding fatty acid synthase and glucokinase (FASN and GCK, respectively) (Figure 5E, F). This increase was either blunted or abolished by the PASK inhibitor. In these cells, BioE-1197 caused a significant decrease in the mRNA encoding SREBP-1c both under basal and insulin-stimulated conditions (Figure 5G). Finally, we measured the incorporation of 14C from 14C-acetate into lipids. Insulin caused a significant increase in lipogenesis in vehicle-treated hepatocytes, which was completely abolished by treatment with BioE-1197 (Figure 5H).
PASK inhibition reduces SREBP-1c activity and hypertriglyceridemia in rodents
We have previously observed that PASK deficiency in mice leads to protection from many of the pathological effects of a high-fat diet (Hao et al., 2007). Most prominently, PASK−/− mice are protected from the severe diet-induced hepatic steatosis observed in wild-type mice. This lipid phenotype was accompanied by a decrease in the expression of the lipogenic program driven by SREBP-1c (Figure 1). These studies were conducted using a constitutively deleted allele of PASK, however, leading to the concern that this phenotype might be due to adaptation or a developmental abnormality. Therefore, we initiated studies to address the effects of acute inhibition of PASK in adult animal models of metabolic disease. These studies were conducted with BioE-1115 due to the more favorable in vivo pharmacologic and pharmacokinetic properties of this PASK inhibitor relative to BioE-1197 (Figure S4A).
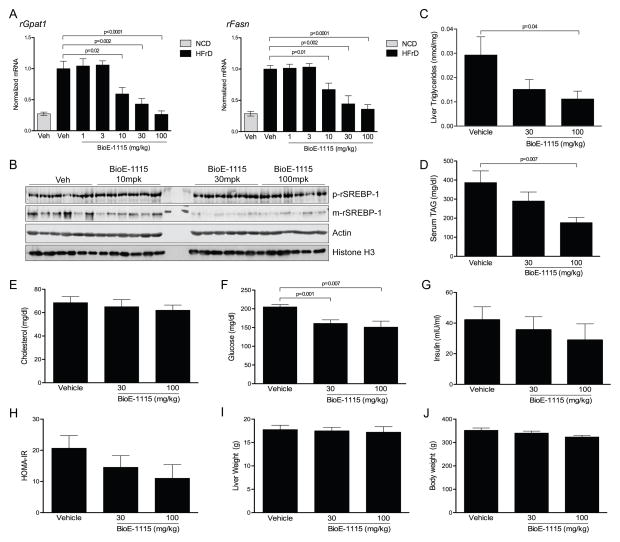
Wild-type Sprague-Dawley rats at 12 weeks of age were fed for two weeks with either a normal chow diet (NCD) or high-fructose diet (HFrD), which is known to promote dyslipidemia and insulin resistance (Hwang et al., 1987). Those fed the high-fructose diet were then orally dosed with vehicle or a range of doses of BioE-1115 once/day for one week. Following this one-week treatment, we harvested livers from these rats following refeeding and examined mRNA levels of SREBP-1c target genes. As expected, SREBP-1c target genes were upregulated in animals fed the high-fructose diet relative to control (Figure 6A and S4B–D). Among the HFrD group, rats treated with either 1 or 3 mg BioE-1115 per kg of body weight (mg/kg) showed no difference from vehicle-treated rats. Those treated with 10, 30 and 100 mg/kg, however, showed a dose-dependent suppression of the expression of Gpat1, Fasn (Figure 6A) and all other SREBP-1c target genes analyzed (Figure S4B–D). SREBP-1 maturation in liver was also suppressed in BioE-1115 treated rats at these three doses (Figure 6B), which is consistent with what we observed in PASK−/− mouse liver. SREBP-2 target genes, however, did not follow this pattern consistently (Figure S4E–I). In fact, HMG-CoA synthase 1 and SREBP-2 itself both followed the opposite pattern (Figure S4E, J), being induced in a dose-dependent manner by BioE-1115. This is particularly intriguing given that mice lacking SREBP-1 frequently die in utero, but those that survive exhibit elevated expression of SREBP-2 and its target genes, implying some sort of compensatory mechanism between the two transcription factors (Shimano et al., 1997b). One SREBP-2 target gene, that encoding HMG-CoA reductase, was significantly decreased in BioE-1197 treated animals (Figure S4F). Interestingly, this is the one SREBP-2 target gene that was also significantly underexpressed in PASK−/− mice (Table S2). Both SREBP-1c and SREBP-1a mRNA were modestly decreased at the highest doses of BioE-1115 (Figure S4K,L).
Figure 6. PASK inhibition decreases SREBP-1 activity, triglycerides and insulin resistance in animal models.
(A) Rats fed either normal chow (NCD) or high-fructose (HFrD) diet for 2 weeks were subjected to once/day treatment with vehicle or the indicated dose of BioE-1115 by oral gavage for one week. Following this regimen, rats were fasted for 24 h and then refed for 12 h. Livers were harvested and qRT-PCR was performed for the indicated genes and normalized to Cyclophilin A (n=10/group). Data shown are the average ± SEM. (B) Rats were maintained on high-fat and high-fructose diet for 18 weeks, and were treated with vehicle or indicated dose of BioE-1115 as in (A) for the last 3 weeks. Livers were harvested after rats were subjected to fast/refeeding as in (A). Whole liver lysate and nuclear extract were subjected to immunoblot for the abundance of the precursor or mature form of SREBP-1 and the indicated control proteins. (C) Triglycerides were measured in livers from the indicated treatment groups as described in (A) (n=10/group). (D–G) Serum TAG, cholesterol, glucose and insulin were measured in the animals as treated in (A). (H) Calculated HOMA-IR values. (I–J) Liver and body weight from animals in (A). See also Figure S4, S5 and S6.
In addition to the marked change in SREBP-1c target gene expression, we also observed a reversal of the dyslipidemic features associated with a high-fructose diet as BioE-1115 treatment caused a decrease in hepatic TAG (Figure 6C). Similarly, serum TAG was also decreased in a dose-dependent manner by BioE-1115 administration (Figure 6D), while serum cholesterol was basically unaffected (Figure 6E). Due to the normalization of triglyceride concentrations, we speculated that BioE-1115 might have salutary effects on glucose homeostasis as well. Indeed, the PASK inhibitor caused a significant decrease in serum glucose (Figure 6F). This drop in glucose levels is almost certainly a consequence of increased insulin sensitivity, rather than increased insulin secretion, as serum insulin levels are slightly lower (rather than higher) in drug-treated animals (Figure 6G). A calculated measure of insulin resistance, HOMA-IR, is decreased in a dose-dependent manner by BioE-1115 administration (Figure 6H). Neither dose of BioE-1115 caused a significant change in either liver weight or body weight (Figure 6I,J), suggesting that the effects on lipid and glucose homeostasis were not due to overt toxicity.
We next performed a study of similar design except that the HFrD-fed rats were treated for 90 days with either vehicle or BioE-1115. The effect on SREBP-1c target gene expression was enhanced with the longer treatment time. At 3 mg/kg BioE-1115, both Fasn and Acc1 expression was significantly suppressed by PASK inhibition and at 10, 30 and 100 mg/kg, expression of these genes was restored to that in normal chow fed animals (Figure S5A). The effect on SREBP-1c activity appears to be specific for the liver as there were no significant differences in the expression of SREBP-1c or its target genes in abdominal fat (Figure S6A) or in gastrocnemius muscle (Figure S6B) upon BioE-1115 treatment. After 90 days of treatment and at the interim timepoint of 45 days, we observed a significant dose-responsive decrease in serum triglycerides (Figure S5B, C). As in the shorter dosing period, BioE-1115 decreased serum glucose levels as measured by glycated hemoglobin (HbA1c), which is a measure of chronic glycemia (Figure S5D, E). After both 51 and 90 days of dosing, there was no drug-dependent difference in body weight between any of the vehicle or treatment groups (Figure S5F, G).
DISCUSSION
The regulation of SREBP-1c is complex, both in terms of the regulatory stimuli and how they impinge upon SREBP-1c activity (Raghow et al., 2008). The most widely studied and understood stimulus that promotes SREBP-1c activation is insulin and various components of its downstream signaling pathway, including Akt and mTOR (Jeon and Osborne, 2012). These related mechanisms combine to elicit a profound activation of SREBP-1c in response to feeding. We show herein that PASK is transcriptionally induced by feeding in vivo in the liver and is also induced in a cell-autonomous manner by insulin in primary hepatocytes. This induction appears to be related to SREBP-1c activation as PASK is required for normal feeding and insulin-responsive SREBP-1c activity. Inhibition of PASK also impairs insulin-responsive lipid biosynthesis, which is driven by SREBP-1c. This requirement for PASK in SREBP-1c activations has been demonstrated in four model systems and using three distinct modalities for blocking PASK activity. SREBP-1c driven transcription is impaired by: genetic deletion of PASK in mice fed either normal chow or high-fat diet, pharmacologic inhibition and siRNA-mediated knockdown in cultured HepG2 cells, and pharmacologic inhibition in both rat primary hepatocytes and high-fructose fed rats. Not only do these disparate manipulations cause the same physiological effect, they also appear to act via the same biochemical mechanism. This provides a compelling argument that PASK is an important regulator of SREBP-1c activation.
Insulin regulates SREBP-1c at almost every level of its expression and stability. Expression of the SREBF-1 gene is robustly stimulated by insulin (Raghow et al., 2008). The precursor SREBP-1c protein is synthesized in a latent, inactive form that is embedded in the endoplasmic reticulum membrane. Upon insulin stimulation, SREBP-1c translocates to the Golgi, where the protein encounters two proteases that cleave it to release an N-terminal fragment that is competent to bind DNA and activate gene expression. SREBP-1c maturation is controlled by two associated proteins, SCAP and INSIG (Raghow et al., 2008). Insulin increases the transcriptional activation potential of mature SREBP-1c (Dif et al., 2006; Kotzka et al., 1998). Finally, insulin also impedes the otherwise rapid degradation of the mature, nuclear form of SREBP-1c (Raghow et al., 2008). In total, this multiplicity of regulatory processes enables a robust induction of SREBP-1c by insulin that is both rapid and sustained.
The principal regulatory point at which PASK acts appears to be the proteolytic maturation of the precursor to mature SREBP-1c. This is best evidenced by the decrease in mature form and increase in precursor form observed in cells subjected to PASK knockdown or inhibition. A specific decrease in the mature form is also observed in PASK−/− mice in the fed state. We cannot eliminate the possibility, however, that PASK might also regulate the synthesis or stability of the SREBF-1 mRNA. We observed that PASK−/− liver, PASK knockdown HepG2 cells and primary hepatocytes with PASK inhibition all have lower SREBP-1c mRNA level than the control situation. But, this could be explained by the fact that SREBP-1c positively regulates its own gene expression (Amemiya-Kudo et al., 2000; Chen et al., 2004). If the loss of PASK impairs the proteolytic activation of SREBP-1c, this would secondarily lead to a decrease in the SREBP-1c mRNA level. In fact, we found that PASK inhibition decreased the activity of a luciferase reporter driven by the SREBP-1c promoter in response to insulin, but this effect was completely eliminated when the autoregulatory SREBP binding site within the promoter was mutated. It remains possible, however, that PASK acts at other steps to regulate SREBP-1c activity, but our data does show that normal insulin-responsive maturation requires PASK.
The mechanisms whereby insulin signaling promotes the proteolytic activation of SREBP-1 are still incompletely understood. It has been observed that Akt, in response to insulin signaling, leads to the phosphorylation of SREBP-1c in a manner that correlates with activation (Yellaturu et al., 2009a). While the functional significance of this SREBP-1c phosphorylation has not been established yet, it is possible that PASK might directly phosphorylate SREBP-1c to promote its maturation. More recent work has shown that insulin signals through both mTORC1-dependent and mTORC1-independent pathways to promote SREBP-1 activation (Wan et al., 2011; Yecies et al., 2011). The latter involves the regulation of INSIG2a, which is a negative regulator of SREBP-1 (Yecies et al., 2011; Yellaturu et al., 2009b). PASK does not appear to act through this mechanism as INSIG2 mRNA is not higher upon PASK knockdown, as would be expected if this were the explanation for impaired SREBP-1 processing. In fact, the INSIG2 mRNA is lower upon PASK knockdown perhaps as a compensatory effect in response to impaired SREBP-1 activation.
The proteins and mechanisms connecting mTORC1 activation with stimulation of SREBP-1 processing are currently unknown. Our data demonstrate that PASK is not required for Akt or mTORC1 activation in response to insulin signaling, thus leading us to conclude that PASK either acts downstream of mTORC1 or in a parallel pathway to promote SREBP-1 activation. The precise placement of PASK in this signaling network and definition of the mechanisms connecting the nodes of this network await further studies.
Not only did administration of the PASK inhibitor cause profound decreases in the hepatic expression of SREBP-1c target genes and the generation of mature SREBP-1, it also impacted hepatic and systemic metabolic parameters. Both hepatic and serum triglycerides were normalized, which might be expected based on the suppression of lipogenic gene expression in the liver. In addition, we observed a significant decrease in serum glucose in high-fructose fed rats treated with the PASK inhibitor for 7 days and in HbA1c in rats treated for either 45 or 90 days. In conjunction with the modest decrease in serum insulin, this is suggestive of enhanced insulin sensitivity in inhibitor-treated animals.
A pathological vicious cycle has been described wherein the hypoglycemic effects of insulin are blunted, but insulin maintains the ability to activate SREBP-1c and lipogenesis (Biddinger et al., 2008; Brown and Goldstein, 2008). Enhanced lipogenesis, and the increasingly dyslipidemic state, exacerbate the “selective insulin resistance” causing more insulin release. We hypothesize that PASK inhibition prevents this vicious cycle by mitigating the toxic lipogenesis that can occur under conditions of insulin resistance and hyperinsulinemia. We previously showed that PASK−/− mice exhibit improved insulin sensitivity and resistance to hepatic steatosis elicited by a high-fat diet (Hao et al., 2007). We now also show that treatment of high-fructose fed rats with a PASK inhibitor completely normalized their elevated expression of SREBP-1c target genes and hypertriglyceridemia.
Importantly, we observed no overt toxicity upon genetic depletion of PASK (PASK−/− mice) or upon extended treatment with doses of the PASK inhibitor that are 10–30 fold higher than those required to see significant effects on lipogenic gene expression and serum triglycerides. Such a treatment regimen had no effect on body weight, even after 90 days of treatment. It also had no effect on the expression of SREBP-1c or its target genes in two other tissues of metabolic importance, skeletal muscle and adipose tissue. In contrast, inhibition of mTOR Complex 1 with rapamycin caused decreased lipogenesis in adipose tissue, which was accompanied by decreased expression of SREBP-1 (Pereira et al., 2013). The selectivity of the effect of PASK inhibition is also in contrast to the phenotype observed for the majority of mice lacking SREBP-1, which die in utero (Shimano et al., 1997b), while mice lacking PASK are viable. Therefore, we conclude that PASK is not required for the basal activation of SREBP-1 or for the activation of SREBP-1 in all tissues. Our data suggests an important role for PASK in the hepatic activation of SREBP-1c in response to feeding. However, it is possible that PASK has functions in other tissue types that may contribute to the beneficial effect on metabolism we observed upon PASK inhibition. Nonetheless, this observation is of particular interest in light of the potentially causal role that hepatic SREBP-1c activation has been proposed to play in metabolic disease in humans. Others have shown that genetic or pharmacological inhibition of SREBP maturation improves hepatic and whole-body metabolism (Moon et al., 2012; Tang et al., 2011). We therefore propose that SREBP-1c activation by PASK is an important feature of the mammalian metabolic syndrome and should be explored as a therapeutic opportunity in humans.
EXPERIMENTAL PROCEDURES
Animals
PASK−/− mice were described previously (Hao et al., 2007). Age-matched male wild-type and PASK−/− mice were maintained on either a normal chow diet (NCD) for 12 weeks, or a 60% high fat diet (HFD) for 8 weeks from 8–10 weeks of age (60% fat by calories, Research Diets Inc.). For fasting-refeeding studies, mice were either fasted for 24 h, or fasted for 24 h and re-fed a NCD or a HFD for indicated time periods before euthanasia and organ harvest. All procedures were approved by the Institutional Animal Care and Use Committee of University of Utah.
Sprague-Dawley (SD) male rats (Charles River Laboratories) were maintained on a high fructose diet (60% fructose, Research Diets Inc.) or NCD for 2 weeks before experimentation. Rats were housed under standard vivarium conditions (12 h light/dark cycle) with water and chow ad libitum. All studies were approved by the Institutional Animal Care and Use Committee guidelines of the University of Utah and/or St. Louis University as appropriate.
Luciferase Assay
HepG2 cells were cotransfected with (1) pGPAT-Luc, pSCD-Luc or pSRE-Luc; (2) a construct expressing CMV-driven Renilla luciferase (Promega); (3) pQCXIN-GFP, pcDNA3.1-2xFlag-mSREBP-1a or pQCXIN-3xFlag-pSREBP-1a construct, as indicated, using Lipofectamine LTX (Invitrogen), according to the manufacture’s instructions. Rat primary hepatocytes were cotransfected with wild-type or mutant pSREBP-1c-Luc and Renilla luciferase using Lipofectamine 2000 (Invitrogen), according to the manufacture’s instructions. After transfection, cells were serum-starved overnight, with additional treatment of (1) vehicle, BioE-1197 or BioE-1428, and/or (2) 100nM insulin for 6 or 12 h before harvest, as indicated. Firefly and Renilla luciferase were assayed using the Dual-Reporter Luciferase Assay System (Promega).
Measurement of De Novo Lipogenesis
HepG2 cells or rat primary hepatocytes were treated with vehicle or 10μM BioE-1197 in serum-free media overnight. On the next day, cells were transferred to new media with vehicle or 30μM BioE-1197 and 100nM insulin for 6 h as indicated, and were labeled with 10μCi/ml [1-14C]-acetate (Perkin Elmer) for the last 4 h before harvest. Cells were washed twice with PBS and then lysed in 0.5% Triton X-100. The lipid fraction was extracted by adding chloroform and methanol (v/v 2:1) followed by dH2O, with vortexing. Samples were then centrifuged at 1500 rpm for 15 min, and the organic (bottom) phase containing lipids was used to measure 14C incorporation on a Beckman LS 6500 scintillation counter. The results were normalized to protein concentration of lysates.
ELISA
HEK293T cells were transfected with pcDNA3.1-Flag-PASK (wild-type or kinase-dead) construct using Lipofectamine 2000 (Invitrogen), according to the manufacturers instructions. After 18 h, cells were re-seeded into 96-well plates in DMEM/1% FBS. Cells were then treated with DMSO or drug for 16 h, followed by lysis in 0.2ml lysis buffer (20mM Na2HPO4, 0.5% Triton, 0.1% SDS, 0.02% azide, 1mM NaF, 1mM glycerphosphate, 1mM Na3VO4). The lysates were then applied to MaxiSorb 96 well plate (Nunc), which was previously coated with α-FLAG capture antibody (M2, Sigma) and blocked with 3% BSA (in 1X PBS). After incubation at 4°C for 1.5 h, the plates were washed with high salt washing buffer (20mM Na2HPO4, 0.5% Triton X-100, 0.1% SDS, 0.02% NaN3, 0.1% BSA and 1M NaCl) followed by low salt buffer (150mM NaCl). Then the plates were incubated for 2 h with antibody to either phospho-Akt substrate (9614, Cell Signaling) or hPASK (U2501), followed by high and low salt buffer washes. Subsequently, plates were incubated with HRP-conjugated secondary antibody for 1 h and then washed with high and low salt buffer and 1X PBS. Phospho-AKT substrate or PASK antibody dependent luminescence signal was assayed using LumiGLO chemiluminescent substrate system (KPL), according to manufacturer’s instructions. Inhibition curve and IC50 were determined using Prism software (GraphPad).
Chemicals and Dosing Formulations
BioE-1197, BioE-1428 and BioE-1115 were synthesized by Pharmaron. All compounds were made up in a vehicle formulation of 0.5% methylcellulose and 0.025% Tween-80 (Sigma) in ddH20. Dosages were calculated, compounds weighed and placed in a 15ml glass homogenizer (Kimble Chase) to which vehicle was added and the compound ground to a fine suspension, transferred to a screw top tube and the homogenizer was rinsed twice with vehicle and then brought to final volume. Compounds were made up every 4–5 days based on animal weight and stored at room temperature. Animals were orally dosed once a day between 7am – 9am.
BioE-1115 in the Sprague Dawley Diet-induced Obesity Model
Male SD rats (n=8–12) were obtained in this study with an average weight of 129.4 ± 0.63 g and were maintained on a high fructose diet for 2 weeks prior to experimentation. The rats were dosed by oral gavage once a day for 7 or 90 days at doses of 1, 3, 10, 30 and 100 mg/kg of BioE-1115 or with vehicle. Body weights were taken every day for compound formulation. Animals were fasted for 24 h and re-fed for 12 h prior to the termination of the experiment. All animals received their respective compound dosage three hours prior to termination. Animals were euthanized by CO2 asphyxiation and cardiac puncture was performed for final serum analysis. Liver tissue was taken during necropsy, weighed and snap frozen in liquid N2. Blood was centrifuged at 3500 rpm for 10 min and serum was collected and analyzed for insulin, glucose, cholesterol and triglycerides using a Beckman CX 5Pro (Beckman Coulter).
Statistical Analysis
Data are presented as mean ± standard deviation unless otherwise indicated. A two-tailed equal variance t test was used to compare differences, and the null hypothesis was rejected at the 0.05 level.
Supplementary Material
Highlights.
PASK is required for feeding and insulin induced expression of lipogenic genes.
PASK activates SREBP-1 by promoting its maturation.
PASK inhibitor improves hepatic and whole body dislipidemia in obese rats.
Acknowledgments
We wish to thank members of the Rutter lab for helpful discussions; Ryan Doering and Addie Walkup for assistance with the mouse studies; John M. McCall for support of the medicinal chemistry; Phil Needleman, William J. Rutter and Brendan Manning for helpful advice; Tim Osborne for the pSRE-Luc and 2xFlag-mSREBP-1a constructs; Wesley I. Sundquist for the pQCXIN-GFP construct; Alan Diehl for a tagged-SREBP-1 plasmid. This work was supported by RO1DK071962 (J.R.) and by Synergenics.
Footnotes
AUTHOR CONTRIBUTIONS
X.W., W.I.S., I.D., C.K.K., H.S., B.S.Z and G.A.N. designed and conducted experiments; D.R. led the medicinal chemistry program; X.W., J.M., J.T.B, G.A.N. and J.R. designed experiments, analyzed data and wrote the manuscript.
DR, WIS, ID, HS, BSZ, JM, JTB, GAN, JR are employees, consultants, and/or shareholders of BioEnergenix, LLC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES and CITATIONS
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. The Journal of biological chemistry. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- Bene H, Lasky D, Ntambi JM. Cloning and characterization of the human stearoyl-CoA desaturase gene promoter: transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochemical and biophysical research communications. 2001;284:1194–1198. doi: 10.1006/bbrc.2001.5102. [DOI] [PubMed] [Google Scholar]
- Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Aleman JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell metabolism. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Selective versus total insulin resistance: a pathogenic paradox. Cell metabolism. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E. Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. The Biochemical journal. 2006;400:179–188. doi: 10.1042/BJ20060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. The Journal of biological chemistry. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell metabolism. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Hao HX, Cardon CM, Swiatek W, Cooksey RC, Smith TL, Wilde J, Boudina S, Abel ED, McClain DA, Rutter J. PAS kinase is required for normal cellular energy balance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15466–15471. doi: 10.1073/pnas.0705407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Rutter J. The role of PAS kinase in regulating energy metabolism. IUBMB life. 2008;60:204–209. doi: 10.1002/iub.32. [DOI] [PubMed] [Google Scholar]
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC., Jr Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. The Journal of biological chemistry. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- Hegarty BD, Bobard A, Hainault I, Ferre P, Bossard P, Foufelle F. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of clinical investigation. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- Hwang IS, Ho H, Hoffman BB, Reaven GM. Fructose-induced insulin resistance and hypertension in rats. Hypertension. 1987;10:512–516. doi: 10.1161/01.hyp.10.5.512. [DOI] [PubMed] [Google Scholar]
- Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikani CK, Antonysamy SA, Bonanno JB, Romero R, Zhang FF, Russell M, Gheyi T, Iizuka M, Emtage S, Sauder JM, et al. Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation. The Journal of biological chemistry. 2010;285:41034–41043. doi: 10.1074/jbc.M110.157594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzka J, Muller-Wieland D, Koponen A, Njamen D, Kremer L, Roth G, Munck M, Knebel B, Krone W. ADD1/SREBP-1c mediates insulin-induced gene expression linked to the MAP kinase pathway. Biochemical and biophysical research communications. 1998;249:375–379. doi: 10.1006/bbrc.1998.9161. [DOI] [PubMed] [Google Scholar]
- Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends in endocrinology and metabolism: TEM. 2010;21:268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YA, Liang G, Xie X, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Brown MS, Goldstein JL, Horton JD. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell metabolism. 2012;15:240–246. doi: 10.1016/j.cmet.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MJ, Palming J, Rizell M, Aureliano M, Carvalho E, Svensson MK, Eriksson JW. The immunosuppressive agents rapamycin, cyclosporin A and tacrolimus increase lipolysis, inhibit lipid storage and alter expression of genes involved in lipid metabolism in human adipose tissue. Molecular and cellular endocrinology. 2013;365:260–269. doi: 10.1016/j.mce.2012.10.030. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Harris TE, Fujita N, Reue K. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. The Journal of biological chemistry. 2010;285:3857–3864. doi: 10.1074/jbc.M109.072488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends in endocrinology and metabolism: TEM. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes & development. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes & development. 2000;14:1293–1307. [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes & development. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. The Journal of clinical investigation. 1997a;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. The Journal of clinical investigation. 1997b;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. The Journal of clinical investigation. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell metabolism. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, Song BL. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell metabolism. 2011;13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Wan M, Leavens KF, Saleh D, Easton RM, Guertin DA, Peterson TR, Kaestner KH, Sabatini DM, Birnbaum MJ. Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell metabolism. 2011;14:516–527. doi: 10.1016/j.cmet.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell metabolism. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Mansbach CM, 2nd, Siddiqi SA, Park EA, Raghow R, Elam MB. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. The Journal of biological chemistry. 2009a;284:7518–7532. doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaturu CR, Deng X, Park EA, Raghow R, Elam MB. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional down-regulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP). SREBP-1c complex. The Journal of biological chemistry. 2009b;284:31726–31734. doi: 10.1074/jbc.M109.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Harada N, Yamamoto H, Taketani Y, Nakagawa T, Yin Y, Hattori A, Zenitani T, Hara S, Yonemoto H, et al. Identification of cis-acting promoter sequences required for expression of the glycerol-3-phosphate acyltransferase 1 gene in mice. Biochimica et biophysica acta. 2009;1791:39–52. doi: 10.1016/j.bbalip.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Molecular and cellular biology. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.