Abstract
Dengue viruses (DENV) cause significantly more human disease than any other arbovirus, with hundreds of thousands of cases leading to severe disease in thousands annually. Antibodies and T cells induced by primary infection with DENV have the potential for both positive (protective) and negative (pathological) effects during subsequent DENV infections. In this review, we summarize studies that have examined T-cell responses in humans following natural infection and vaccination. We discuss studies that support a role for T cells in protection against and those that support a role for the involvement of T cells in the pathogenesis of severe disease. The mechanisms that lead to severe disease are complex, and T-cell responses are an important component that needs to be further evaluated for the development of safe and efficacious DENV vaccines.
Keywords: dengue, immune response, immunopathology, nonstructural proteins, primary infection, secondary infection, T lymphocyte, vaccine
Dengue viruses (DENV) have four closely-related serotypes that co-circulate in endemic regions. Infection results in a spectrum of clinical manifestations. Plasma leakage, a hallmark of severe DENV disease, occurs after several days of infection and coincides with viral clearance. However, pathological studies indicate that vascular endothelial cells are relatively structurally intact in patients with severe disease [1,2]. Since these patients typically experience a rapid recovery, it is thought that plasma leakage occurs due to endothelial cell malfunction, rather than lysis of infected endothelial cells [2]. This, together with evidence that increased disease severity is associated with secondary heterologous DENV infections, suggests a role for pre-existing adaptive immune responses in contributing to an immunopathologically mediated clinical outcome in severe dengue infections [3].
The pathogenesis of DENV disease is multifactorial
The primary human target cells for DENV infection in vitro include monocytes, macrophages and mature dendritic cells (Figure 1). The precise targets for DENV infection in vivo are less clear and have been challenging to identify [4], but include alveolar macrophages, phagocytes and other hematopoietic cells [5]. DENV have also been shown to replicate in B cells, a central source of antibodies [6,7]; however, some recent data suggest that B cells are not natural targets for DENV infection [8]. Antibodies to DENV can mediate a number of activities in vitro [9]. Some antibodies are able to neutralize the virus but enhance virus uptake at higher dilutions, while other antibodies do not neutralize the virus but are also able to bind to the virus and Fcγ I and II receptors, and mediate more efficient entry into the host cell [10,11]. These non-neutralizing antibodies can result in higher production of infectious particles through a process known as antibody-dependent enhancement (ADE) [12]. DENV-specific antibodies of the appropriate subclasses bound to dengue antigens on the infected cell membrane can bind to complement proteins and promote complement-dependent lysis (CDL) of infected cells and contribute to antibody-dependent cellular cytotoxicity (ADCC) of infected cells [13,14]. Much less is known about the role of γδ T cells, Th17 and NK cells in anti-DENV defense mechanisms (Figure 1). HLA restricted CD4+ and CD8+ T cells are activated upon viral infection and several epitopes have been identified in humans after natural infection. T-cell-produced cytokines have the ability to influence vascular permeability leading to plasma leakage, a hallmark of severe disease [15–17]. The outcome of DENV infection likely depends on the balance between favorable and unfavorable immune responses, host genetics, viral factors, the sequence of DENV infections and factors specific to the individual patient [3,18]. In this review, we will discuss efforts to evaluate T-cell responses to DENV infection in humans and mice and assess the contribution of T lymphocytes to protection against or pathogenesis of severe DENV disease.
Figure 1. Interactions between multiple components of the immune system during dengue virus infection.
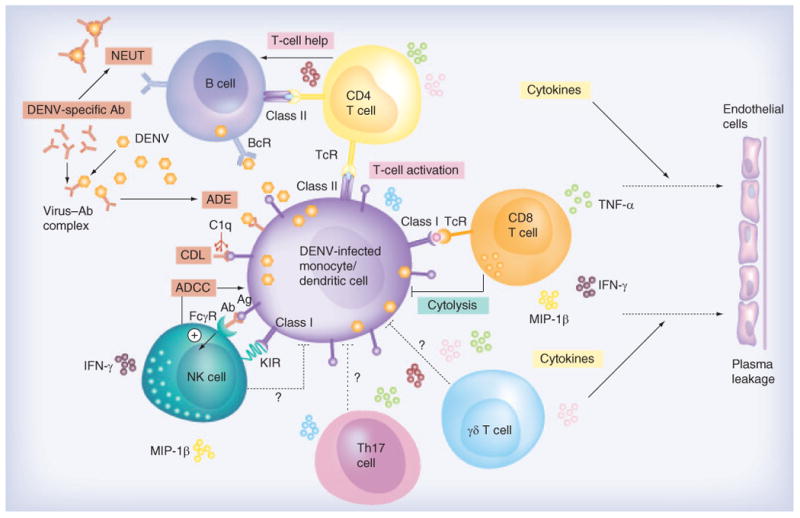
The primary targets of DENV replication are monocytes, macrophages and dendritic cells, but B cells may also be infected with DENV. Antibodies secreted by B cells can mediate a wide range of functions including neutralization, ADE, ADCC and CDL. Virus-infected target cells secrete cytokines and chemokines and attract T cells. Viral peptides are presented on MHC class I and class II presentation pathways to CD8+ and CD4+ T cells, respectively. CD4+ T cells predominantly produce cytokines but are capable of lysing virus-infected cells, and CD8+ T cells lyse virus-infected cells and produce cytokines. The role of γδ T cells, Th17 and NK cell participation in the antiviral immune defense mechanisms requires further investigation. Question marks in the figure indicate that the evidence is not clear. The result of the cascade of immune activation leads to endothelial cell permeability and plasma leakage.
Ab: Antibody; ADCC: Antibody-dependent cell-mediated cytotoxicity; ADE: Antibody-dependent enhancement; Ag: Dengue antigen; BcR: B-cell receptor; C1q: Subcomponent of complement pathway; CDL: Complement-dependent lysis; DENV: Dengue viruses; FcγR: Fc gamma receptor; KIR: Killer-like immunoglobulin receptor; NEUT: Neutralization; NK: Natural killer; TcR: T-cell receptor; Th17: T helper 17.
T-cell responses to DENV after natural infection
In order to begin to understand the contribution of DENV-specific T cells in protection or enhanced immunopathology, significant effort has been spent over the last two decades to define T-cell epitopes to DENV (Table 1). CD4+ and CD8+ T-cell epitopes have been identified on multiple proteins of DENV [19–38]. MHC class I and II restricted minimal T-cell epitopes were characterized in a subset of T cells. While T-cell epitopes have been identified on the structural proteins, the vast majority of T-cell epitopes have been found on nonstructural proteins. Our early studies, using samples from donors who received experimental live-attenuated monovalent DENV vaccines and a smaller set of samples from donors with natural infection in Thailand, demonstrated that the NS3 protein is an immunodominant protein with multiple epitopes throughout the protein [21–24,39–43]. More recently, three studies have used overlapping peptide pools or strong binding peptides to the most common HLA alleles to identify several additional T-cell epitopes in different populations around the world. Duangchinda et al. set out to study T-cell responses across the entire DENV proteome in a cohort of DENV-infected children from Khon Kaen and Songkhla hospitals in Thailand [29]. While T-cell responses to NS3 were dominant, responses to multiple proteins were observed in most infected individuals. Rivino et al. assessed CD4+ and CD8+ T cell reactivity using an overlapping 15mer peptide library spanning the DENV 2 proteome using the peripheral blood mononuclear cells(PBMCs) of adult patients from Singapore experiencing secondary DENV infection [26]. They observed that CD8+ T-cell epitopes preferentially targeted nonstructural proteins (NS3 and NS5), but CD4+ T-cell epitopes were skewed toward recognition of viral components that were also targeted by B lymphocytes (envelope, capsid and NS1). Weiskopf et al. performed a comprehensive analysis of CD8+ T-cell responses in the general population from a DENV hyperendemic area in Sri Lanka, measuring ex vivo IFNγ responses. NS3, NS4B and NS5 were the most vigorously and frequently recognized proteins and accounted for more than two-thirds of the total T-cell response [27].
Table 1.
T-cell epitopes recognized by virus-specific T cells.
| Protein | Amino acids† | Sequence‡ | CD4/CD8 | MHC§ | Ref. |
|---|---|---|---|---|---|
| C | 22–31 | RVSTVQQLTK | CD8 | A03/11 | [31] |
| 40–60 | LFMALVAFLRFLTIP | CD4 | [26] | ||
| 71–85 | TIKSKAINVLRGFR | CD4 | [26] | ||
| 47–55 | VLAFITFLR | CD4 | DPw4 | [19] | |
| 62–81 | TAGILKRWGTIKKSKAINVL | CD4 | [28] | ||
| 83–92 | GFRKEIGRML | CD4 | DR1,DPw4 | [19,28] | |
| 107–115 | CLIPTAMAF | CD8 | B15 | [27] | |
| 107–115 | MLIPTAMAF | CD8 | B35 | [27] | |
|
| |||||
| prM | 41– 60 | LGELCEDTITYKCPLLRQNE | CD4 | [28] | |
| 121–135 | QRIETWILRHPGFTM | CD4 | [26] | ||
| 133–141 | FTILAFLAH | CD8 | B35 | [18] | |
|
| |||||
| E | 46–55 | LKTEVTNPAV | [33] | ||
| 41–55 | LDFELIKTEAKQPAT | CD4 | [26] | ||
| 206–220 | WLVHRQWFLDLPLPW | CD4 | [26] | ||
| 236–250 | TLVTFKNPHAKKQDV | CD4 | [26] | ||
| 241–255 | KNPHAKKQDVVVLGS | CD4 | [26] | ||
| 246–160 | KKQDVVVLGSQEGAM | CD4 | [26] | ||
| 211–219 | FFDLPLPWT | CD8 | A02 | [73] | |
| 297–306 | MSYSMCTGKF | CD8 | B35 | [27] | |
| 340–359 | RDVNKEKVVGRVISSTPLAE | CD4 | [33] | ||
| 414–422 | ILGDTAWDF | CD8 | B07 | [28] | |
|
| |||||
| NS1 | 26–34 | HTWTYQEF | CD8 | B57 | [61] |
| 88–99 | IMTGDIKGIMQA | CD4 | [33] | ||
| 111–125 | LKYSWKTWGKAKMLS | CD4 | [26] | ||
| 326–350 | EDGCWYGMEIRPLKEKEENLNSLV | CD4 | [26] | ||
| 206–220 | LNDTWKIEKASFIEV | CD4 | [26] | ||
|
| |||||
| NS2a | 99–118 | RENLLLGVGLAMATTLQLPE | CD4 | [33] | |
| 108–127 | AMTTTLSIPHDLMELIDGIS | CD4 | [33] | ||
| 109–133 | SLVASVELPNSLEELGDGLAMGIMI | CD4 | [33] | ||
| 135–148 | IVTQFDNTQVGTLA | CD4 | [33] | ||
| 184–203 | SSQQKTDWIPLALTIKGLNP | CD4 | [33] | ||
| 196–216 | GSLGCKPLTMFLIAENKIWG | CD4 | [33] | ||
| 198–206 | ATGPILTLW | CD8 | B58 | [27] | |
|
| |||||
| NS2b | 52–60 | ELERAADVK | CD8 | A03/11 | [31] |
| 63–82 | DQAEISGSSPILSITISEDG | [33] | |||
| 83–102 | TMRIKDDETENILTVLLKTA | [33] | |||
| 97–106 | ILIRTGLLVI | CD8 | A0201/24 | [31] | |
|
| |||||
| NS3 | 25–32 | RIKQKGIL | CD8 | B08 | [31] |
| 64–74 | RIEPSWADVK | CD8 | A03/11 | [31] | |
| 71–79 | SVKKDLISY | CD8 | B62 | [37] | |
| 112–120 | AIKRGLRTL | CD8 | A02/24 | [31] | |
| 130–144 | GTSGSPIIDKK | CD8 | A11.1 | [25] | |
| 146–154 | VIGLYGNGV | CD4 | D R15 | [22] | |
| 157–173 | TSGTYVSAIAQAKASQE | [33] | |||
| 176–184 | NPEIEDDIF | CD8 | B35 | [27] | |
| 186–200 | RKLTIMDLHPGSGKT | CD4 | ND | [38] | |
| 194–203 | HPGAGKTKRY | CD8 | B35 | [27] | |
| 202–211 | RKYLPAIVRE | CD4 | DR15 | [36] | |
| 222–230 | APTRVVAAE | CD8 | B07 | [24] | |
| 224–234 | TRVVAAEMEEA | CD4 | DR15 | [41] | |
| 235–243 | AMKGLPIRY | CD8 | B62 | [37] | |
| 241–249 | IRYQTTATK | CD4 | DR15 | [36] | |
| 255–264 | EIVDLMCHAT | CD4 | DPw2 | [34] | |
| 276–290 | PNYNLIIMDEAHFTD | CD4 | [26] | ||
| 291–300 | DPASIAARGY | CD8 | B35 | [27] | |
| 352–362 | WITDFVGKTVW | CD4 | DR15 | [36] | |
| 351–365 | VTDFKGKTVWFVPSI | CD4 | [26] | ||
| 422–431 | RVIDPRRCMK | CD8 | A03/11 | [31] | |
| 500–508 | TPEGIIPTL | CD8 | B35 | [23] | |
| 521–530 | GEFRLRGEQR | CD8 | B40 | [27] | |
| 526–540 | LRGEARKTFVELMRR | [32] | |||
| 528–537 | GEARKTFVEL | CD8 | B40 | [27] | |
| 555–564 | INYADRRWCF | CD8 | A24 | [28] | |
| 584–598 | KEGERKKLRPRWLDA | CD4 | ND | [38] | |
| 606–614 | MALKDFKEF | CD8 | B35 | [27] | |
|
| |||||
| NS4a | 2–21 | LTLNLITEMGRLPTFMTQKA | [33] | ||
| 56–64 | LLLGLMILL | CD8 | A2 | [73] | |
| 55–64 | LLLLTLLATV | CD8 | A02/24 | [31] | |
| 6–13 | LEKTKKDL | CD8 | B08 | [31] | |
|
| |||||
| NS4b | 23–32 | TETTILDVDL | CD8 | B53 | [27] |
| 40–48 | TLYAVATTI | CD8 | A02 | [35] | |
| 49–58 | TPMLRHTIEN | CD8 | B07 | [27] | |
| 69–77 | IANQATVLM | CD8 | B35 | [27] | |
| 92–100 | VPLLAIGCY | CD8 | B35 | [27] | |
| 111–119 | VLLLVTHYA | CD8 | A2 | [73] | |
| 119–128 | AIIGPGLQAK | CD8 | A03/11 | [31] | |
| 181–189 | LLLMRTSWA | CD8 | A02 | [73] | |
| 198–206 | ATGPILTLW | CD8 | B58 | [27] | |
| 15–27 | SRLNALGKSEFQI | CD4 | [33] | ||
|
| |||||
| NS5 | 182–190 | VLNPYMPSV | CD8 | A02/24 | [31] |
| 263–282 | HVNAEPETPNMDVIGERIKR | CD4 | [33] | ||
| 291–299 | WHYDQDHPY | CD8 | B35 | [27] | |
| 291–310 | WHYDEDNPYKTWAYHGSYEV | [32] | |||
| 301–315 | KTWAYHGSYETKQTG | CD4 | [26] | ||
| 329–337 | KPWDVIPMVT | CD8 | B55 | [32] | |
| 343–351 | DTTPFGQQR | CD8 | A68 | [27] | |
| 373–382 | VMGITAEWLW | CD8 | B53 | [27] | |
| 375–383 | KITAEWLWK | CD8 | A03/11 | [31] | |
| 389–398 | KPRICTREEF | CD8 | B07 | [27] | |
| 393–402 | TPRMCTREEF | CD8 | B07/35 | [27] | |
| 563–571 | KLAEAIFKL | CD8 | A02/24 | [31] | |
Sequence positions vary slightly between strains.
Sequence as reported by the cited reference. These sequences do not necessarily reflect the minimal epitope. As sequences vary between serotypes and strains these epitopes may not represent the sequence found in prevalent circulating strains.
HLA restriction was not confirmed in all studies and some were based on peptide-binding predictions.
Data taken from [18].
Studies on immune responses to a related flavivirus, yellow fever virus (YFV), also found many T-cell epitopes on NS3 [44]. In the murine system, NS3 is the major target for DENV-specific H-2k cytotoxic lymphocytes (CTL) [45], as well as Murray Valley encephalitis (MVE), Kunjun and West Nile virus-immune CTLs [46–48].
NS3 is a dominant target of CD8 T-cell epitopes
The predominant protein recognized by CD8+ CTLs in the PBMC of DENV immune individuals from different parts of the world is the nonstructural protein NS3. Although there is a predominance of recognition of determinants on the NS3 protein, there is no single immunodominant epitope. NS3 is the second largest viral protein and is one of the most highly conserved proteins among flaviviruses [49]. The crossreactive nature of CTLs against NS3 may be due to the high level of amino acid conservation among the NS3 proteins of the different serotypes. NS3 has both a protease (N terminus) and a nucleotide triphosphatase helicase (C terminus) activity. Unlike other viruses where different antigens are produced early and late during the virus lifecycle, the DENV genome encodes for a single polyprotein that gets co- and post-translationally cleaved to the individual proteins by host- and virus-specific proteases that include NS3 [50,51]. This processing means that the individual proteins are produced in equimolar concentrations. The structural proteins (C, prM and E), NSI and the N terminus of NS2a are translocated into the lumen of the endoplasmic reticulum via a series of signal and stop-transfer sequences. On the other hand, the NS3 and NS5 proteins, which are the largest proteins encoded by the DENV genome, have a cytoplasmic localization. Although speculative, possible differences in intracellular targeting and stability may lead to preferential processing and presentation of NS3 peptides.
HLA associations with disease
A number of studies have linked specific HLA class I, II, and III alleles with different DENV disease manifestations. HLA-A*0203 correlated with mild DENV disease (DF) as compared with HLA-A*0207, which correlated with severe disease (DHF) [52]. This report and others identified additional HLA class I and II correlations with likelihood for decreased (HLA-A29, A31, A33, B13, B14, B44, B52, B62, B76, B77, DRB1*04, DRB1*07, DRB1*09) or increased (HLA-A1, A24, A31, B15, B46, B51, DQ1) risk for severe DENV [53,54].
Separate studies reported that a single nucleotide polymorphism (SNP) at position 308 in the gene for tumor necrosis factor (TNF-α) is associated with DHF, and particular SNP alleles of the gene for IL-10 are associated with low levels of IL-10 protein production, and correlated with DHF [55,56]. These groups suggested that high TNF-α/low IL-10 production helped mediate severe DENV disease. Another study reported a strong genetic linkage between the SNP allele -238A in the TNF-α gene and the lymphotoxin-α (LTA)-3 haplotype, which is associated with high TNF-α and LTA-α production during acute viremia in DENV-infected patients [57,58]. Patients with TNF-238A and LTA-3 were at greater risk for developing DHF compared with DF, and DHF patients with these SNP profiles were nearly all shown to have HLA-B48 and/or HLA-B57 [58], in the Thai population [52]. Most studies have been performed in small cohorts of individuals. Such analyses underscore the need to assess extended haplotypes in larger prospective genetic studies of DENV infections from multiple ethnic populations in order to better understand potential mechanisms for the immunopathology observed in DENV infection.
T-cell immune responses in primary versus secondary DENV infections
Individuals who have been infected with a DENV serotype for the first time (primary DENV infection) have long-term protective immunity against re-infection with the same serotype. Individuals can be infected for a second time with another DENV serotype and are experiencing a secondary DENV infection. In endemic countries, most individuals have been exposed very early in life to DENV and it is challenging to obtain samples to evaluate immune responses to primary infections. Memory T cells that are re-activated during a second infection may not have optimal avidity for the corresponding epitopes of the new infecting virus because of sequence diversity between the DENV serotypes [3,59]. Therefore, it would be reasonable to predict skewed secondary T-cell responses since peptide variants found in different DENV serotypes could act as altered peptide ligands impacting the responsiveness of the T cell. Higher frequencies of epitope-specific T cells in donors with natural secondary infections might be expected since memory T cells from the first infection would be more readily activated compared with naïve T cells. We have sought to assess the frequencies of epitope-specific T cells during and after primary and secondary DENV infection to validate these predictions. For A11-NS3133–142-specific T cells, we found similar frequencies of serotype-cross-reactive T cells ex vivo in naturally infected patients with primary and secondary DENV infections [60]. For B57-NS126–34-specific T cells, since the sequence in a secondary DENV infection was identical to the sequence from an earlier primary DENV infection, we predicted that PBMC from donors with secondary infection would have particularly strong responses to the B57-NS126–34 epitope [61]. However, our findings in all but one individual with a secondary infection (n = 8/9) indicated that frequencies were similar to individuals with primary infections (n = 2), to frequencies of A11-NS3133–142 and A2-E213–221-specific T cells in the same subjects, and to the frequencies of A11-NS3133–142 T cells reported elsewhere [25,60]. In our hands, the magnitude of A11 and B57 epitope-specific T cells were not significantly different between primary and secondary infections. Owing to the limitations of sample size we did not extensively test effector functions of T cells in PBMC from these children. It is possible that the function of T cells might differ in individuals with primary versus secondary infection. Furthermore, during acute infection, trafficking of antigen-specific T cells to tissues may not allow an accurate assessment in the peripheral blood. Ideally, frequencies and functional responses need to be assessed against multiple epitopes in large prospective cohort studies to conclusively demonstrate that T-cell responses are skewed during a second infection with DENV. It has also been challenging to assess the extent to which the order of the infecting serotype affects subsequent T-cell responses.
Studies that have examined a role for DENV-specific T cells with pathogenesis of DENV infections
T-cell associated cytokines have been found to be elevated in the sera of patients with mild disease, and some were increased to higher levels in severe DENV disease [18,57,62–67]. While the precise cellular sources of various cytokines and chemokines found in the circulation of dengue patients are currently unknown, several studies have found that DENV-specific T cells stimulated with homologous and heterologous variant peptides in vitro also secrete these cytokines [32,38,68]. To have a better understanding of how T-cell responses relate to the onset of clinical symptoms, recent studies have used peptide MHC tetramer technology to investigate the kinetics of expansion and activation of DENV-specific T cells during acute infection and convalescence. The magnitude of A11-NS3133–142-specific T cell expansion did not correlate with disease severity in a study by Friberg et al. [60]. We compared PBMC from patients with mild and severe disease (DF versus DHF) as well as other clinical measures of disease severity such as pleural effusion index, hemoconcentration or platelet counts. A similar lack of association between the frequency of A11-NS3133–142-specific T cells and disease severity was reported in two studies in Vietnam [69,70]. A strength of these studies is the information on clinical profile, viral isolation and HLA typing in individuals with primary and secondary DENV infection [60,69,70]. Furthermore, samples were obtained during and up to 3 years following the critical phase of illness with mild and severe disease from all four serotypes [60]. Simmons et al. found T-cell IFN-γ ELISPOT responses were weakly correlated with the extent of hemoconcentration in individual patients, but not with overall disease severity [28]. Other studies had reported higher frequencies of DENV-specific T cells in patients with DHF, but these associations were found at 2 weeks [29,71] or 6 months [72] post-infection (Table 2). Differences in timing or differences in infection history (e.g., serotype of primary and secondary infection) may explain the differences in results between these studies. However, the lack of a correlation with disease severity, and the timing of peak tetramer-positive T-cell frequencies in early convalescence rather than at the time of plasma leakage, suggest that the frequency of A11-NS3133–142 tetramer-positive T cells may not be the principal determinant of disease. On the other hand, the number of samples tested during capillary leakage is very small and it is likely that highly activated T cells may be localized at the sites of infection, lost in processing or by apoptosis, and that the later memory T-cell progeny reflects the importance of those T cells in the acute phase. T-cell responses to other DENV epitopes are also likely to contribute to disease, as is suggested by our limited data on B7-NS3222–230-specific T cells in HLA-A11−B7+ subjects during the acute phase. Alternatively, characteristics of the DENV-specific T-cell response other than the quantity detected in samples of acute PBMC, for example effector responses [73], may also be more important.
Table 2.
Studies that examined a role for T cells in dengue virus pathogenesis.
| Study (year) | Target | Correlates | Time point | Ref. |
|---|---|---|---|---|
| Studies that support a role for T cells | ||||
| Green et al. (1999) | Total CD8+ | CD69+ vs disease outcome | Acute | [62] |
| Zivna et al. (2002) | B7-NS3222–231 | IFN-γ vs disease outcome | ≥6 months | [72] |
| Mongkolsapaya et al. (2003) | A11- N S 3133–142 | TET+ vs disease outcome | 2 weeks | [25] |
| Simmons et al. (2005) | C,E,M,NS3 peptides | IFN-γ vs hemoconcentration | 2 weeks | [28] |
| Mongkolsapaya et al. (2006) | A24-NS3556–564 | TET+ vs disease outcome | 2 weeks | [71] |
| Chau et al. (2008) | Total CD8+ | CD69+ vs disease outcome | Acute | [70] |
| Duangchinda et al. (2010) | All prot, A11-NS3133–142 | TNF-α, IFN-γ vs disease outcome | 2 weeks | [29] |
| Dung et al. (2010) | Total CD8+ | CD38+ HLA-DR+, HLA-DR+Ki-67+, CD38+Ki-67+ | Acute | [69] |
| Malagive et al. (2012) | NS3 specific T cells | Serum cytokines vs disease outcome | Acute | [33] |
| Mangada et al. (2002) | Total CD4 | TNF-α, IFN-γ to those who subsequently were hospitalized | Pre-secondary infection PBMC | [76] |
| Hatch et al. (2011) | Total CD4+ and CD8+ | TNF-α, IFN-γ to those who subsequently were hospitalized | Pre-secondary infection PBMC | [77] |
| Studies that do not support a role for T cells | ||||
| Simmons et al. (2005) | C,E,M,NS3 peptides | IFN-γ vs disease outcome | 2 weeks | [28] |
| Chau et al. (2008) | A11- NS3 133–142 | TET+ vs time | Acute | [70] |
| Dung et al. (2010) | A11-NS3133–142 | TET+ vs time | Acute | [69] |
| Friberg et al. (2011) | A11- NS3133–142 | TET+ vs disease outcome | Acute and 2 weeks | [60] |
PBMC: Peripheral blood mononuclear cells.
Phenotypic markers on T cells have been used to characterize effector and memory T cells in acute human viral infections [74,75]. We have used CD38, CD69 and CD71 to phenotype DENV-specific T cells in our clinical cohort. There were no significant correlations between the expression of CD38 (a marker of activation) and disease severity on A11-NS3133–142-specific T cells. This stands in contrast to the results of other studies focused on CD69 (an early activation marker) on total CD8+ T cells [62,70]. We recently assessed CD71 (transferrin receptor) expression on HLAB57-NS126–34, on A11-NS3133–142 and A2-E213–221-specific CD8+ T cells over the course of DENV infection. We observed upregulation of CD71 predominantly on DENV-specific CD8+ T cells and not on total CD8+ T cells [61]. However, CD69 and CD38 expression was similar between epitope-specific T cells and total CD8+ T cells during acute DENV infection. The finding of a novel and distinct phenotype (CD71+) in these epitope-specific T cells suggests differential activation that merits further investigation. Current data are limited and have been obtained from a small number of subjects; however, they suggest that the frequency of select epitope-specific T cells may not be the principal determinant in the association between T lymphocyte responses and disease.
Studies that have examined a role for DENV-specific T cells with protection against DENV infections
Human studies
A number of published T-cell studies have focused on the role of T cells in the pathogenesis of DENV infections. Very few studies have examined the role of T cells in protection from disease in humans. Collection of PBMC from individuals prior to infection in prospective clinical studies is key to measuring correlations between T-cell function in pre-infection PBMC and disease outcome. Therefore, whether T cells contribute to protection against DENV infection in humans remains unknown. Several prospective studies have assessed these associations. Mangada et al. compared the T-cell responses of the pre-secondary infection PBMC of patients who were hospitalized during their subsequent DENV infection to those of patients who were not hospitalized [76]. IFN-γ production in response to the infecting serotypes was significantly more common among patients who were not hospitalized. In a study performed by Hatch et al., the level of T-cell activity in preillness PBMC was compared between subjects who subsequently developed a subclinical secondary DENV infection or had a symptomatic secondary infection [77]. They found higher frequencies of cytokine-producing (TNF-α, IFN-γ and IL-2) CD4+ T cells in patients who did not develop symptomatic infection. Gunther et al. studied cellular immune responses in recipients who received a candidate tetravalent vaccine and were subsequently challenged with infectious DENV. They found that in vitro IFN-γ responses mediated by DENV-specific T cells in the peripheral blood were associated with protection against fever or viremia [78]. Lindow et al. found a trend of more multifunctional CD4+ T cells in nonviremic vaccinees relative to viremic vaccinees after administration of a low-dose DENV-1 vaccine [79]. These observations suggest that multifunctional CD4+ T cells may be indicators of individuals who are more able to control DENV infection and therefore may have less severe clinical disease.
Murine studies
DENV does not cause severe infections in immunocompetent mice. Since DENV does not block IFN signaling in murine cells, type 1 IFN knockout mice are highly susceptible to infection with laboratory strains of DENV, with paralysis commonly seen in infected mice [80]. Type 1 IFN knockout mice expressing HLA transgenes have been utilized to identify T-cell epitopes to dengue [81]. Under certain experimental conditions, when mice lacking type 1 IFNs are infected with select passaged strains of DENV, they have increased vascular leakage and TNF-α levels showing some characteristics of human DENV disease [82]. IFNα/β receptor knockout mice infected with a mouse-adapted DENV strain S221 have higher viral loads upon depletion of CD8+ T cells [83], but not after depletion of CD4+ T cells [84]. Additionally, immunization of the mice with CD8+ or CD4+ epitopes enhanced viral clearance upon subsequent DENV challenge, supporting a protective role for T cells against DENV infections.
CD8+ T-cell depletion also negated the vaccine immunized protection against a very high dose challenge with a lethal strain of DENV-infected BALB/c mice [85,86]. However, a different mouse model (HepG2-grafted SCID) suggested that DENV-specific CD8+ T cells have both protective and pathogenic roles [87]. Specifically, mice inoculated with DENV-specific CD8+ T cells and subsequently challenged with a lethal dose of DENV showed slightly reduced mortality compared with uninoculated mice (80% versus 100%); however, the mice that died did so much more quickly (day 12.8 versus day 17.4). The studies above examined mechanisms of protective immunity in mouse models during primary DENV infection or upon homologous virus re-challenge. A different study used adoptive transfer experiments to demonstrate that serotype-cross-reactive antibody was more protective against homologous and heterologous challenge with DENV-2 compared with cross-reactive cell-mediated immune responses [88].
Improvement of existing animal models
There are a number of animal models each with significant limitations [89,90]. The lack of an animal model for DHF severely limits the use of animal models to study the pathogenesis of disease. The generation of novel humanized mouse and non-human primate models presents opportunities to overcome deficiencies of other mouse models [91]. A major advantage of humanized models is the presence of human cells in a physiological setting. However, both humoral and cellular responses in humanized models need further improvement to match responses detected in humans and mechanistic studies may be challenging to perform in these models [92,93]. Infection of non-human primates results in viral replication accompanied by neutralizing antibodies and T-cell responses; however, there is only limited evidence of disease or hematologic abnormalities [94–96]. In addition, large-scale vaccine testing in non-human primate models involves significant cost and accessibility. Animal models need to be improved to reproduce the immunological response in humans in order to be reliably used to test vaccine strategies.
Induction of optimal T-cell responses
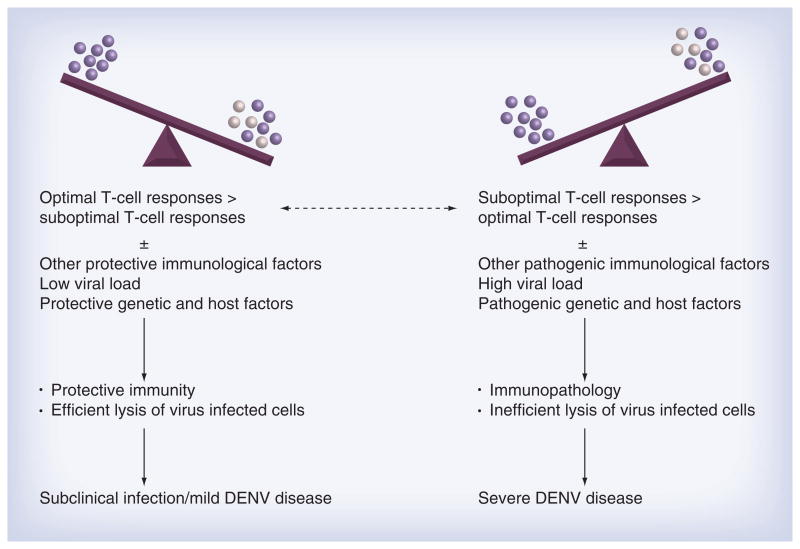
The specific definition of ‘optimal’ or ‘sub-optimal’ T-cell immune responses is unknown and there is no single metric to identify a protective T-cell response. The generation of multifunctional T cells with high-quality responses may be protective, while the generation of lesser-quality T cells is considered suboptimal [97]. Cross-reactive DENV-specific T cells have quantitative and qualitative differences in degranulation and cytokine responses to variant peptides [71, 73]. Peptide variants that differ even by a single amino acid are able to elicit strikingly different cytokine and cytolytic responses in T-cell lines [68]. We believe individuals experiencing secondary DENV infections have the potential to generate both ‘optimal’ and ‘suboptimal’ T-cell responses to multiple epitopes (Figure 2). If ‘optimal’ T-cell responses outweigh ‘suboptimal’ responses and these responses occur in the context of other ‘protective’ viral, immunological and genetic factors, the clinical outcome is positive, as is seen in >95% of secondary infections, and individuals are protected. If ‘suboptimal’ T-cell responses outweigh ‘optimal’ T-cell responses in individuals and the responses occur in the context of other associated ‘pathogenic’ risk factors, the balance is altered from a protective to pathogenic outcome.
Figure 2. Disease outcome is multifactorial.
Every person can generate ‘optimal’ or ‘suboptimal’ responses to individual DENV-specific T-cell epitopes. The total T-cell response to DENV in an individual is the cumulative response to multiple epitopes. A favorable clinical outcome occurs when overall ‘optimal’ T-cell responses occur in the context of protective host, genetic and other immunological factors. An unfavorable clinical outcome occurs when ‘suboptimal’ T-cell responses occur in the context of pathogenic host, genetic and other immunological factors. DENV: Dengue viruses.
T cell responses to multivalent DENV vaccination
Multiple DENV vaccines developed on diverse platforms are currently in clinical trials [9,98,99]. Owing to the immunopathology seen in natural secondary heterologous infections, a key consideration for vaccine manufacturers is to create a DENV vaccine that induces robust immunity to all four serotypes. The Sanofi Pasteur candidate vaccine contains four chimeric live YFV with the prM and E of each of the four DENV serotypes (DENV 1–4). Both DENV and YFV17D-204-specific CD4+ and CD8+ cellular responses induced by tetravalent chimeric DENV vaccines (CYD-TDV) were analyzed in flavivirus-naive or flavivirus-immune patients in Phase I clinical trials. Significant YFV 17D NS3-specific CD8+ responses and DENV serotype-specific T helper responses were detected in PBMC of vaccinated subjects [100]. An IFN-γ/TNF-α ratio dominated by IFN-γ, for both CD4+ and CD8+ T-cell responses, was detected with an absence of a detectable Th2 response. Responses were impacted by the YFV and DENV immune status of an individual, and a booster vaccination broadened serotype-specific responses.
In a Phase II trial in Singapore, T-cell responses were assessed before and 28 days after a first and third injection of CYD-TDV and 1 year after the third injection in a subset of 80 subjects [101]. CD4+ cytokine responses (IFN-γ/TNF-α) were detected to DENV NS3 prior to vaccination. Following vaccination, CD8+ IFN-γ responses were detected to the YFV-17D-NS3 protein in addition to a Th1 cellular response in all participants to the tetravalent vaccine, characterized by IFN-γ secretion compared with TNF-α. A booster vaccination induced more balanced responses against all four serotypes. Unfortunately, early results from a Phase II trial of this vaccine showed little protection [102].
The National Institute of Allergy and Infectious Diseases (NIAID) Division of Intramural Research has developed live, attenuated vaccines to each of the four DENV serotypes (DENV-1, 2, 3 and 4). These vaccines were designed by introducing deletions into the genomes of each virus in the 3′ untranslated region. Subjects were vaccinated with a low-dose (10 PFU) of a DENV-1 vaccine and a kinetic T-cell study was performed. Using multiparametric flow cytometry, DENV-1 specific CD4+ T cells were found to secrete IFN-γ, TNF-α and IL-2 at 21 days post vaccination [79]. Little T-cell cross-reactivity was detected to the other three DENV serotypes.
It is important to assess the characteristics (serotype-crossreactivity and effector responses) of the T lymphocyte responses to monovalent and tetravalent vaccines currently in clinical trials. Many of these trials are being conducted in populations where there is pre-existing immunity to DENV and other related flaviviruses. Immunization with chimeric flaviviruses does not seem to skew the specificity of the T lymphocyte response significantly, although certain serotypes elicited stronger responses [100]. Development of immunocompetent animal models to determine whether the immunization method (dose, location, timing and sequence) affects vaccine-mediated T-lymphocyte responses will further our understanding of T-cell responses in humans. Strategies to ‘sculpt’ vaccine-induced immune response to achieve a balanced and robust response to all four serotypes must be considered.
Conclusion & future perspective
With a number of DENV vaccines in preclinical, Phase I–III trials, there is an opportunity to further our understanding of immune responses to DENV vaccination. It will be important to determine whether the same correlates apply for vaccine-induced T-cell responses as for natural infection. T-cell responses elicited to different vaccines (DNA vs chimeric attenuated vs inactivated) are likely distinct and need to be evaluated. Some groups have attempted to determine if there are patterns of T-lymphocyte responses that are most strongly correlated with protection from or enhanced risk of severe DENV disease. Prospective cohorts where PBMC are collected prior to primary and secondary natural DENV infections are critical to identify the immunological predictors of protective versus pathogenic outcomes.
While substantial progress has been made in recent years, clinical and animal studies have revealed that immune responses to DENV are complicated. The presence of four closely related serotypes, interplay between multiple cellular subsets and lack of an authentic animal model has hindered progress. Gaps still remain in our understanding of DENV-specific T-lymphocyte responses and their associations with protection against or pathogenesis of severe disease. Well-designed prospective clinical studies are needed to provide the best insights in order to reduce the burden of this pathogen.
Executive Summary.
The pathogenesis of dengue viruses disease is multifactorial
Many factors including host genetics, viral factors and pre-existing immunity likely contribute to dengue virus (DENV) pathogenesis.
T-cell responses to DENV after natural infection
The majority of T-cell responses following natural infection with DENV are directed against nonstructural proteins, with responses to NS3 being dominant.
T-cell-associated cytokines are elevated in patients with severe disease.
In a limited number of studies, the frequency of epitope-specific T cells was not significantly different between primary and secondary DENV infections. Frequencies and functional responses need to be assessed against multiple epitopes in large prospective cohort studies to conclusively demonstrate that T-cell responses are skewed during a second infection with DENV.
Studies that have examined a role for T cells with pathogenesis of DENV virus infections
T-cell-derived cytokines have been found to be elevated in patients with mild and severe DENV disease.
Association between certain Class I HLA alleles and risk for disease severity supports a role for CD8 T cells in DENV pathogenesis.
The frequency and activation phenotype of epitope-specific T cells have been examined in PBMC from patients with mild and severe disease during and after the critical time period of capillary leakage.
Studies that have examined a role for T cells with protection against DENV virus infection
Prospective clinical studies are critical to clearly define a role for T cells in protection from disease in humans.
Multifunctional CD4+ T cells may be indicators of individuals who are better able to control DENV infection.
Murine studies support a protective role for T cells against DENV infections, albeit in immunocompromised animals.
An appropriate animal model for severe DENV infections is not available; however, groups are actively working to develop better models that recapitulate human disease.
T-cell responses to multivalent DENV vaccination
T-cell responses to DENV vaccines are currently being evaluated in human vaccine trials.
Multifunctional T cells may contribute to protection.
Strategies to ‘sculpt’ vaccine-induced immune response to achieve a balanced and robust response to all four serotypes must be considered.
Acknowledgments
The authors would like to thank the staff of the Queen Sirikit National Institute for Child Health, Kamphaeng Phet Provincial Hospital, the Department of Virology, Armed Forces Research Institute of Medical Sciences, Thailand, for patient recruitment, blood collection, clinical and virology information. They would like to thank their colleagues in the United States and Thailand and the volunteer subjects and parents who generously contributed towards their dengue research activities for many years. This work was financially supported by the NIH grant no P01 AI34533 and U19 AI57319.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
• of interest
- 1.Sahaphong S, Riengrojpitak S, Bhamarapravati N, Chirachariyavej T. Electron microscopic study of the vascular endothelial cell in dengue hemorrhagic fever. Southeast Asian J Trop Med Public Health. 1980;11:194–204. [PubMed] [Google Scholar]
- 2.Kurane I, Ennis FA. Immunity and immunopathology in dengue virus infections. Sem Immunol. 1992;4:121–127. [PubMed] [Google Scholar]
- 3.Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 4.Sun P, Kochel TJ. The battle between infection and host immune responses of dengue virus and its implication in dengue disease pathogenesis. Scientific World Journal. 2013;2013:843469. doi: 10.1155/2013/843469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsitis SJ, Coloma J, Castro G, et al. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80:416–424. [PubMed] [Google Scholar]
- 6.King AD, Nisalak A, Kalayanrooj S, et al. B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J Trop Med Public Health. 1999;30:718–728. [PubMed] [Google Scholar]
- 7.Lin YW, Wang KJ, Lei HY, et al. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J Virol. 2002;76:12242–12249. doi: 10.1128/JVI.76.23.12242-12249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kou Z, Quinn M, Chen H, et al. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J Med Virol. 2008;80:134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- 9.Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annu Rev Immunol. 2011;29:587–619. doi: 10.1146/annurev-immunol-031210-101315. [DOI] [PubMed] [Google Scholar]
- 10.Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flipse J, Wilschut J, Smit JM. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic. 2013;14:25–35. doi: 10.1111/tra.12012. [DOI] [PubMed] [Google Scholar]
- 13.Garcia G, Arango M, Perez AB, et al. Antibodies from patients with dengue viral infection mediate cellular cytotoxicity. J Clin Virol. 2006;37:53–57. doi: 10.1016/j.jcv.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Laoprasopwattana K, Libraty DH, Endy TP, et al. Antibody dependent cellular cytotoxicity in pre-secondary dengue virus serotype 3 (DV3) but not in DV2 infection plasma samples inversely correlated with viremia levels. J Infect Dis. 2007;195:1108–1116. doi: 10.1086/512860. [DOI] [PubMed] [Google Scholar]
- 15.Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis. 2006;19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- 16.Kurane I, Matsutani T, Suzuki R, et al. T-cell responses to dengue virus in humans. Trop Med Health. 2011;39:45–51. doi: 10.2149/tmh.2011-S09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–313. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon SJ, Zeng W, Kurane I, Ennis FA. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J Virol. 1996;70:141–147. doi: 10.1128/jvi.70.1.141-147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green S, Kurane I, Pincus S, Paoletti E, Ennis FA. Recognition of dengue virus NS1-NS2a proteins by human CD4+ cytotoxic T lymphocyte clones. Virology. 1997;234:383–386. doi: 10.1006/viro.1997.8648. [DOI] [PubMed] [Google Scholar]
- 21.Kurane I, Dai LC, Livingston PG, Reed E, Ennis FA. Defnition of an HLA-DPw2-restricted epitope on NS3, recognized by a dengue virus serotype-cross-reactive human CD4+ CD8- cytotoxic T-cell clone. J Virol. 1993;67:6285–6288. doi: 10.1128/jvi.67.10.6285-6288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurane I, Okamoto Y, Dai LC, et al. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8- cytotoxic T lymphocyte clones. J Gen Virol. 1995;76:2243–2249. doi: 10.1099/0022-1317-76-9-2243. [DOI] [PubMed] [Google Scholar]
- 23.Livingston PG, Kurane I, Dai LC, et al. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 24.Mathew A, Kurane I, Green S, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. 1998;72:3999–4004. doi: 10.1128/jvi.72.5.3999-4004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Mongkolsapaya J, Dejnirattisai W, Xu X, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003:921–927. doi: 10.1038/nm887. This study demonstrated high frequencies of dengue virus (DENV) epitope-specific T cells in patients with secondary infection, and provided evidence that these cells were originally induced by the primary DENV infection. [DOI] [PubMed] [Google Scholar]
- 26•.Rivino L, Kumaran EA, Jovanovic V, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. 2013;87:2693–2706. doi: 10.1128/JVI.02675-12. This study used overlapping peptide pools and mapped T-cell responses to the DENV proteome in immune individuals from Singapore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Weiskopf D, Angelo MA, de Azeredo EL, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. This study used overlapping peptide pools and mapped T-cell responses to the DENV proteome in immune individuals from Sri Lanka. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons CP, Dong T, Chau NV, et al. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol. 2005;79:5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duangchinda T, Dejnirattisai W, Vasanawathana S, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci USA. 2010;107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Testa JS, Shetty V, Sinnathamby G, et al. Conserved MHC class I-presented dengue virus epitopes identified by immunoproteomics analysis are targets for cross-serotype reactive T-cell response. J Infect Dis. 2012;205:647–655. doi: 10.1093/infdis/jir814. This study used overlapping peptide pools and mapped T-cell responses to the DENV proteome using early convalescent peripheral blood mononuclear cells from naturally infected individuals with secondary infection from Thailand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appanna R, Huat TL, See LL, et al. Cross-reactive T-cell responses to the nonstructural regions of dengue viruses among dengue fever and dengue hemorrhagic fever patients in Malaysia. Clin Vaccine Immunol. 2007;14:969–977. doi: 10.1128/CVI.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imrie A, Meeks J, Gurary A, et al. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol. 2007;81:10081–10091. doi: 10.1128/JVI.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malavige GN, McGowan S, Atukorale V, et al. Identification of serotype-specific T cell responses to highly conserved regions of the dengue viruses. Clin Exp Immunol. 2012;168:215–223. doi: 10.1111/j.1365-2249.2012.04566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto Y, Kurane I, Leporati AM, Ennis FA. Defnition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J Gen Virol. 1998;79:697–704. doi: 10.1099/0022-1317-79-4-697. [DOI] [PubMed] [Google Scholar]
- 35.Wen J, Duan Z, Jiang L. Identification of a dengue virus-specific HLA-A*0201-restricted CD8+ T cell epitope. J Med Virol. 2010;82:642–648. doi: 10.1002/jmv.21736. [DOI] [PubMed] [Google Scholar]
- 36.Zeng L, Kurane I, Okamoto Y, Ennis FA, Brinton MA. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J Virol. 1996;70:3108–3117. doi: 10.1128/jvi.70.5.3108-3117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zivny J, DeFronzo M, Jarry W, et al. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J Immunol. 1999;163:2754–2760. [PubMed] [Google Scholar]
- 38.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 39.Kurane I, Brinton MA, Samson AL, Ennis FA. Dengue virus-specific, human CD4+ CD8- cytotoxic T-cell clones: multiple patterns of virus cross-reactivity recognized by NS3-specific T-cell clones. J Virol. 1991;65:1823–1828. doi: 10.1128/jvi.65.4.1823-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurane I, Meager A, Ennis FA. Dengue virus-specific human T cell clones. Serotype crossreactive proliferation, interferon gamma production, and cytotoxic activity. J Exp Med. 1989;170:763–775. doi: 10.1084/jem.170.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurane I, Zeng L, Brinton MA, Ennis FA. Defnition of an epitope on NS3 recognized by human CD4+ cytotoxic T lymphocyte clones cross-reactive for dengue virus types 2, 3, and 4. Virology. 1998;240:169–174. doi: 10.1006/viro.1997.8925. [DOI] [PubMed] [Google Scholar]
- 42•.Mathew A, Kurane I, Rothman AL. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J Clin Invest. 1996;98:1684–1694. doi: 10.1172/JCI118964. This was one of the early studies that identified nonstructural proteins as a dominant target of CD8 + cytotoxic T lymphocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zivny J, Kurane I, Leporati AM, et al. A single nine-amino acid peptide induces virus-specific, CD8+ human cytotoxic T lymphocyte clones of heterogeneous serotype specificities. J Exp Med. 1995;182:853–863. doi: 10.1084/jem.182.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Co MD, Terajima M, Cruz J, Ennis FA, Rothman AL. Human cytotoxic T lymphocyte responses to live attenuated 17D Yellow fever vaccine: identifcation of HLA-B35-restricted CTL epitopes on nonstructural proteins NS1, NS2b, NS3, and the structural protein E. Virology. 2002;293:151–163. doi: 10.1006/viro.2001.1255. [DOI] [PubMed] [Google Scholar]
- 45.Rothman AL, Kurane I, Lai CJ, et al. Dengue virus protein recognition by virus-specific murine CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:801–806. doi: 10.1128/jvi.67.2.801-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill AB, Mullbacher A, Parrish C, et al. Broad cross-reactivity with marked fine specificity in the cytotoxic T cell response to flaviviruses. J Gen Virol. 1992;73:1115–1123. doi: 10.1099/0022-1317-73-5-1115. [DOI] [PubMed] [Google Scholar]
- 47.Lobigs M, Arthur CE, Mullbacher A, Blanden RV. The flavivirus nonstructural protein NS3 is a dominant source of cytotoxic T cell peptide determinants. Virology. 1994;202:195–201. doi: 10.1006/viro.1994.1335. [DOI] [PubMed] [Google Scholar]
- 48.Parrish CR, Coia G, Hill A, et al. Preliminary analysis of murine cytotoxic T cell responses to the proteins of the flavivirus Kunjin using vaccinia virus expression. J Gen Virol. 1991;72:1645–1653. doi: 10.1099/0022-1317-72-7-1645. [DOI] [PubMed] [Google Scholar]
- 49.Khan AM, Miotto O, Nascimento EJ, et al. Conservation and variability of dengue virus proteins: implications for vaccine design. PLoS Negl Trop Dis. 2008;2:e272. doi: 10.1371/journal.pntd.0000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Mohan PM, Padmanabhan R. Processing and localization of dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992;66:7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Padmanabhan R. Role of protein conformation in the processing of dengue virus type 2 nonstructural polyprotein precursor. Gene. 1993;129:197–205. doi: 10.1016/0378-1119(93)90269-9. [DOI] [PubMed] [Google Scholar]
- 52.Stephens HA, Klaythong R, Sirikong M, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 53.Stephens HA. HLA and other gene associations with dengue disease severity. Curr Top Microbiol Immunol. 2010;338:99–114. doi: 10.1007/978-3-642-02215-9_8. [DOI] [PubMed] [Google Scholar]
- 54.Malavige GN, Rostron T, Rohanachandra LT, et al. HLA class I and class II associations in dengue viral infections in a Sri Lankan population. PLoS ONE. 2011;6:e20581. doi: 10.1371/journal.pone.0020581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez-Mestre MT, Gendzekhadze K, Rivas-Vetencourt P, Layrisse Z. TNF-alpha-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64:469–472. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 56.Perez AB, Sierra B, Garcia G, et al. Tumor necrosis factor-alpha, transforming growth factor-beta1, and interleukin-10 gene polymorphisms: implication in protection or susceptibility to dengue hemorrhagic fever. Hum Immunol. 2010;71:1135–1140. doi: 10.1016/j.humimm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Gagnon SJ, Mori M, Kurane I, et al. Cytokine gene expression and protein production in peripheral blood mononuclear cells of children with acute dengue virus infections. J Med Virol. 2002;67:41–46. doi: 10.1002/jmv.2190. [DOI] [PubMed] [Google Scholar]
- 58.Vejbaesya S, Luangtrakool P, Luangtrakool K, et al. TNF and LTA gene, allele, and extended HLA haplotype associations with severe dengue virus infection in ethnic Thais. J Infect Dis. 2009;199:1442–1448. doi: 10.1086/597422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr Top Microbiol Immunol. 2010;338:83–98. doi: 10.1007/978-3-642-02215-9_7. [DOI] [PubMed] [Google Scholar]
- 60.Friberg H, Bashyam H, Toyosaki-Maeda T, et al. Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci Rep. 2011;1:51. doi: 10.1038/srep00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Townsley E, Woda M, Thomas SJ, et al. Distinct activation phenotype of a highly conserved novel HLA-b57-restricted epitope during dengue virus infection. Immunology. 2013;141:27–38. doi: 10.1111/imm.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green S, Pichyangkul S, Vaughn DW, et al. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Inf Dis. 1999;180:1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 63.Green S, Vaughn DW, Kalayanarooj S, et al. Early immune activation in acute dengue is related to development of plasma leakage and disease severity. J Inf Dis. 1999;179:755–762. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- 64.Hober D, Delannoy AS, Benyoucef S, De Groote D, Wattre P. High levels of sTNFR p75 and TNF alpha in dengue-infected patients. Microbiol Immunol. 1996;40:569–573. doi: 10.1111/j.1348-0421.1996.tb01110.x. [DOI] [PubMed] [Google Scholar]
- 65.Hober D, Poli L, Roblin B, et al. Serum levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β) in dengue-infected patients. Am J Trop Med Hyg. 1993;48:324–331. doi: 10.4269/ajtmh.1993.48.324. [DOI] [PubMed] [Google Scholar]
- 66.Mustafa AS, Elbishbishi EA, Agarwal R, Chaturvedi UC. Elevated levels of interleukin-13 and IL-18 in patients with dengue hemorrhagic fever. FEMS Immunol Med Microbiol. 2001;30:229–233. doi: 10.1111/j.1574-695X.2001.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 67.Srikiatkhachorn A, Ajariyakhajorn C, Endy TP, et al. Virus-induced decline in soluble vascular endothelial growth receptor 2 is associated with plasma leakage in dengue hemorrhagic fever. J Virol. 2007;81:1592–1600. doi: 10.1128/JVI.01642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friberg H, Burns L, Woda M, et al. Memory CD8 (+) T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol Cell Biol. 2010;89(1):122–129. doi: 10.1038/icb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dung NT, Duyen HT, Thuy NT, et al. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J Immunol. 2010;184:7281–7287. doi: 10.4049/jimmunol.0903262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chau TN, Quyen NT, Thuy TT, et al. Dengue in Vietnamese infants – results of infection-enhancement assays correlate with age-related disease epidemiology, and cellular immune responses correlate with disease severity. J Infect Dis. 2008;198:516–524. doi: 10.1086/590117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 72.Zivna I, Green S, Vaughn DW, et al. T cell responses to an HLA B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J Immunol. 2002;168:5959–5965. doi: 10.4049/jimmunol.168.11.5959. [DOI] [PubMed] [Google Scholar]
- 73.Bashyam HS, Green S, Rothman AL. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J Immunol. 2006;176:2817–2824. doi: 10.4049/jimmunol.176.5.2817. [DOI] [PubMed] [Google Scholar]
- 74.Akondy RS, Monson ND, Miller JD, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183:7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller JD, van der Most RG, Akondy RS, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 76.Mangada MM, Endy TP, Nisalak A, et al. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J Inf Dis. 2002;185:1697–1703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- 77•.Hatch S, Endy TP, Thomas S, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis. 2010;203:1282–1291. doi: 10.1093/infdis/jir012. These studies have attempted to define a protective role for dengue virus-specific T cells in humans with natural infection using pre-secondary infection peripheral blood mononuclear cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gunther VJ, Putnak R, Eckels KH, et al. A human challenge model for dengue infection reveals a possible protective role for sustained interferon gamma levels during the acute phase of illness. Vaccine. 2011;29:3895–3904. doi: 10.1016/j.vaccine.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 79.Lindow JC, Borochoff-Porte N, Durbin AP, et al. Primary vaccination with low dose live dengue 1 virus generates a proinflammatory, multifunctional T cell response in humans. PLoS Negl Trop Dis. 2012;6:e1742. doi: 10.1371/journal.pntd.0001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson AJ, Roehrig JT. New mouse model for dengue virus vaccine testing. J Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiskopf D, Yauch LE, Angelo MA, et al. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol. 2011;187:4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol. 2006;80:10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yauch LE, Zellweger RM, Kotturi MF, et al. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yauch LE, Prestwood TR, May MM, et al. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. 2010;185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gil L, Lopez C, Lazo L, et al. Recombinant nucleocapsid-like particles from dengue-2 virus induce protective CD4+ and CD8+ cells against viral encephalitis in mice. Int Immunol. 2009;21:1175–1183. doi: 10.1093/intimm/dxp082. [DOI] [PubMed] [Google Scholar]
- 86.Gil L, Lopez C, Blanco A, et al. The cellular immune response plays an important role in protecting against dengue virus in the mouse encephalitis model. Viral Immunol. 2009;22:23–30. doi: 10.1089/vim.2008.0063. [DOI] [PubMed] [Google Scholar]
- 87.An J, Zhou DS, Zhang JL, et al. Dengue-specific CD8+ T cells have both protective and pathogenic roles in dengue virus infection. Immunol Lett. 2004;95:167–174. doi: 10.1016/j.imlet.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 88.Kyle JL, Balsitis SJ, Zhang L, Beatty PR, Harris E. Antibodies play a greater role than immune cells in heterologous protection against secondary dengue virus infection in a mouse model. Virology. 2008;380:296–303. doi: 10.1016/j.virol.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yauch LE, Shresta S. Mouse models of dengue virus infection and disease. Antiviral Res. 2008;80:87–93. doi: 10.1016/j.antiviral.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zompi S, Harris E. Animal models of dengue virus infection. Viruses. 2012;4:62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12:786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaiswal S, Pazoles P, Woda M, et al. Enhanced humoral and HLA-A2-restricted dengue virus-specific T-cell responses in humanized BLT NSG mice. Immunology. 2012;136:334–343. doi: 10.1111/j.1365-2567.2012.03585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaiswal S, Pearson T, Friberg H, et al. Dengue virus infection and virus-specific HLA-A2 restricted immune responses in humanized NOD-SCID IL2rgammanull mice. PLoS ONE. 2009;4:e7251. doi: 10.1371/journal.pone.0007251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koraka P, Benton S, van Amerongen G, Stittelaar KJ, Osterhaus AD. Characterization of humoral and cellular immune responses in cynomolgus macaques upon primary and subsequent heterologous infections with dengue viruses. Microbes Infect. 2007;9:940–946. doi: 10.1016/j.micinf.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 95.Mladinich KM, Piaskowski SM, Rudersdorf R, et al. Dengue virus-specific CD4+ and CD8+ T lymphocytes target NS1, NS3 and NS5 in infected Indian rhesus macaques. Immunogenetics. 2012;64:111–121. doi: 10.1007/s00251-011-0566-0. [DOI] [PubMed] [Google Scholar]
- 96.Raviprakash K, Kochel TJ, Ewing D, et al. Immunogenicity of dengue virus type 1 DNA vaccines expressing truncated and full length envelope protein. Vaccine. 2000;18:2426–2434. doi: 10.1016/s0264-410x(99)00570-8. [DOI] [PubMed] [Google Scholar]
- 97.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 98.McArthur MA, Sztein MB, Edelman R. Dengue vaccines: recent developments, ongoing challenges and current candidates. Expert Rev Vaccines. 2013;12:933–953. doi: 10.1586/14760584.2013.815412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thomas SJ, Endy TP. Critical issues in dengue vaccine development. Curr Opin Infect Dis. 2011;24:442–450. doi: 10.1097/QCO.0b013e32834a1b0b. [DOI] [PubMed] [Google Scholar]
- 100.Guy B, Nougarede N, Begue S, et al. Cell-mediated immunity induced by chimeric tetravalent dengue vaccine in naive or flavivirus-primed subjects. Vaccine. 2008;26:5712–5721. doi: 10.1016/j.vaccine.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 101.Harenberg A, Begue S, Mamessier A, et al. Persistence of Th1/Tc1 responses one year after tetravalent dengue vaccination in adults and adolescents in Singapore. Hum Vaccin Immunother. 2013 doi: 10.4161/hv.25562. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sabchareon A, Wallace D, Sirivichayakul C, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled Phase 2b trial. Lancet. 2012;380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]