SUMMARY
Translational readthrough, observed primarily in less complex organisms from viruses to Drosophila, expands the proteome by translating select transcripts beyond the canonical stop codon. Here we show that vascular endothelial growth factor-A (VEGFA) mRNA in mammalian endothelial cells undergoes programmed translational readthrough (PTR) generating VEGF-Ax, an isoform containing a unique 22-amino acid C-terminus extension. A cis-acting element in the VEGFA 3′UTR serves a dual function, not only encoding the appended peptide, but also directing the PTR by decoding the UGA stop codon as serine. Heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 binds this element and promotes readthrough. Remarkably, VEGF-Ax exhibits anti-angiogenic activity in contrast to the pro-angiogenic activity of VEGF-A. Pathophysiological significance of VEGF-Ax is indicated by robust expression in multiple human tissues, but depletion in colon adenocarcinoma. Furthermore, genome-wide analysis revealed AGO1 and MTCH2 as authentic readthrough targets. Overall, our studies reveal a novel protein-regulated PTR event in a vertebrate system.
INTRODUCTION
Vascular endothelial growth factor (VEGF)-A induces migration and proliferation in vascular endothelial cells (ECs), and is essential for both physiological (e.g., embryogenesis) and pathological (e.g., tumorigenesis, retinopathy, and arthritis) angiogenesis (Ferrara, 2009; Hicklin and Ellis, 2005; Nagy et al., 2007). ECs not only respond to VEGF-A, they also express and secrete it, and EC-derived VEGF-A exhibits an intracellular activity essential for vascular homeostasis (Lee et al., 2007). VEGF-A also increases vascular permeability (Senger et al., 1983) and is required for maintenance of the differentiated phenotype in ECs (Kamba et al., 2006). Deletion of VEGFA gene even at a single allele, or a 2-fold increase of VEGF-A, is embryonic lethal demonstrating a precise VEGF-A dosage requirement during development (Carmeliet et al., 1996; Ferrara et al., 1996; Miquerol et al., 2000). VEGF-A functions primarily through two tyrosine kinase receptors VEGFR1 (or flt-1) and VEGFR2 (or KDR or flk-1), and neuropilin 1 serves as an important co-receptor (Soker et al., 1998). Because VEGF-A is critical for the pathogenesis of vascularized solid tumors, agents that selectively target the protein, receptor, or signaling pathway have received substantial attention. As an example, bevacizumab, a monoclonal antibody that targets human VEGF-A, is an FDA-approved drug successfully used to treat metastatic colorectal cancer (Hurwitz et al., 2004).
The VEGFA gene on human chromosome 6 comprises 8 exons. Alternative splicing events at the 6th and 7th exons, which encode heparin-binding motifs, generate multiple isoforms all of which exhibit pro-angiogenic activity. Interestingly, an alternative splicing event within exon 8 has been reported to generate an anti-angiogenic isoform, termed VEGF-Ab (Bates et al., 2002; Harper and Bates, 2008). The splicing event in VEGFAb mRNA replaces an exon encoding CDKPRR at the C-terminus of canonical pro-angiogenic isoforms with SLTRKD (RLTRKD in B. taurus). VEGF-A expression is regulated by multiple post-transcriptional mechanisms, primarily involving the unusually long 3′ untranslated region (UTR). Heterogeneous nuclear ribonucleoprotein (hnRNP) L binds a CA-rich element, promoting VEGFA mRNA stability under hypoxia (Shih and Claffey, 1999). VEGF-A expression is also subject to exquisite translational control mechanisms. In one well-studied example, interferon-γ stimulation of myeloid cells induces binding of the heterotetrameric GAIT (interferon-γ-activated inhibitor of translation) complex to a cognate element in the VEGFA 3′UTR and inhibits its translation (Mazumder et al., 2003; Sampath et al., 2004). A protein-directed RNA switch integrates disparate signals induced by hypoxia and interferon-γ to further regulate VEGFA translation expression in myeloid cells (Ray et al., 2009). Interestingly, a mechanism that prevents complete translational silencing of VEGFA mRNA has been reported that forces a “trickle” of synthesis, underscoring the requirement for precise regulation of VEGF-A dosage in physiological conditions (Yao et al., 2012). In addition, VEGFA mRNA is subject to 5′UTR-mediated regulation in which an internal ribosomal entry site drives alternative translation-initiation from an in-frame CUG codon (Huez et al., 2001).
Translation of mRNAs normally terminates at an in-frame stop codon. However, in a few circumstances, translation can continue beyond the stop codon to a downstream stop codon, generating a C-terminal polypeptide extension. Such translational readthrough events are best understood in viruses (Li and Rice, 1989; Pelham, 1978), but also translational readthrough has been observed in bacteria (Jalajakumari et al., 1989), yeast (Namy et al., 2003), and Drosophila (Dunn et al., 2013; Robinson and Cooley, 1997; Steneberg and Samakovlis, 2001). In a few systems translational readthrough is “programmed” by cis-acting RNA elements downstream of the first stop codon, clearly distinguishing the process from readthrough resulting from errors of translation (Firth et al., 2011). Limited evidence for readthrough of vertebrate mRNAs has been shown, namely, rabbit β-globin in rabbits and rat myelin protein zero; however, the functional significance of the readthrough products, and the cis-acting RNA signals or other regulatory mechanisms have not been determined (Chittum et al., 1998; Yamaguchi et al., 2012). Moreover, to our knowledge trans-acting protein factors that bind RNA elements or otherwise regulate translational readthrough have not been observed.
Here, we demonstrate that VEGFA mRNA undergoes robust PTR in mammalian ECs. The PTR event generates a VEGF-A isoform, termed VEGF-Ax (for “extended”), with a 22-amino acid C-terminus extension. Remarkably, VEGF-Ax exhibits a function opposite to that of VEGF-A, namely, anti-angiogenic activity. Readthrough is “programmed” by a 63-nt RNA segment following the canonical stop codon that serves a dual function, encoding the peptide extension of VEGF-A and also acting as a cis-acting regulatory element. hnRNP A2/B1 binds this element and promotes readthrough. The presence of VEGF-Ax in blood and its selective expression in multiple tissues, as well as differential expression in a solid tumor indicates its pathophysiological significance. Recombinant VEGF-Ax reduced tumor progression and associated angiogenesis in a human xenograft tumor implanted in nude mice, demonstrating its anti-angiogenic property in vivo. We have expanded the generality of our findings by a genome-wide analysis that reveals additional validated readthrough targets, namely, AGO1 and MTCH2 mRNAs. Overall, our results not only demonstrate a novel PTR event in a vertebrate transcript, but also identify a new anti-angiogenic isoform of VEGF-A with a potential role as a suppressor of pathological angiogenesis.
RESULTS
Endothelial Cells Express and Secrete an Anti-angiogenic Isoform of VEGF-A
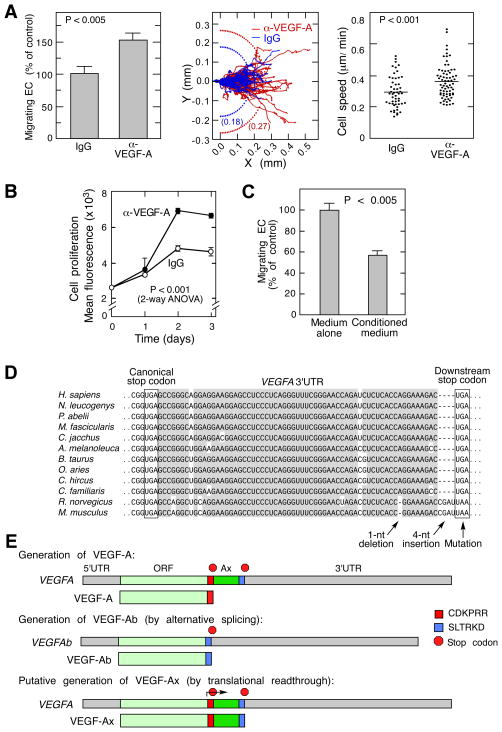
Despite recent studies of the autocrine function of EC-derived VEGF-A in blood vessel angiogenesis and homeostasis (da Silva et al., 2010; Lee et al., 2007), its paracrine function has not received much attention. We hypothesized that secreted, EC-derived VEGF-A might be an important contributor to EC migration and proliferation, which are essential for endothelial wound-healing responses and angiogenesis (Ausprunk and Folkman, 1977; Schwartz et al., 1978). Unexpectedly, a VEGF-A-neutralizing antibody enhanced migration of bovine aortic ECs as indicated by the number of migrating cells in a razor-wound assay, as well as by root-mean-square displacement and mean cell speed (Figure 1A). Likewise, the antibody increased EC proliferation determined fluorimetrically as total cell DNA (Figure 1B). Consistent with these observations, addition of EC-conditioned medium decreased EC migration (Figure 1C). These surprising results indicate that EC-derived VEGF-A inhibits their own migration and proliferation, characteristic features of anti-angiogenic VEGF-A isoforms. To test for the possible expression of VEGF-Ab isoforms we sequenced 74 VEGFA cDNA clones from bovine aortic ECs (and 6 from mouse aortic ECs), but did not find VEGF-Ab-specific mRNA, or any alternatively-spliced VEGFA mRNA that can generate anti-angiogenic isoforms. Instead, we identified only mRNAs corresponding to pro-angiogenic VEGF-A164 and -A120 (Figure S1A–C). Similarly, VEGF-Ab-specific mRNA was not detected by reverse transcriptase PCR using primers designed to differentiate between VEGF-Ab and VEGF-A isoforms (Figure S1D).
Figure 1. ECs Express and Secrete an Anti-angiogenic Isoform of VEGF-A.
(A) Neutralizing anti-VEGF-A antibody enhances EC migration. Shown are ECs migrating across razor-wound line (mean ± SE, n=3) (left), individual cell trajectories and root-mean-square displacement (center), and cell speed in the presence of anti-VEGF-A antibody (n=71) and IgG (n=52) (right).
(B) Neutralizing anti-VEGF-A antibody increases EC proliferation. Cell amount was determined fluorimetrically as DNA (mean ± SE, n=4).
(C) EC-derived conditioned medium inhibits EC migration. EC migration determined by razor-wound assay (mean ± SE, n=3).
(D) Conservation of VEGFA 3′UTR proximal region (gray); canonical and downstream, in-frame stop codons in mammals (boxed).
(E) Schematic showing generation of VEGF-A, VEGF-Ab, and VEGF-Ax isoforms.
See also Figure S1.
VEGF-Ax is a Novel Isoform Generated by Translational Readthrough of VEGFA mRNA
The paradoxical presence of anti-angiogenic VEGF-A in the absence of VEGF-Ab-specific mRNA suggested the possibility of a novel anti-angiogenic VEGF-A isoform generated from mRNA encoding the pro-angiogenic isoform. A critical clue was offered by the sequence of the VEGFA mRNA 3′UTR. In human VEGFA 3′UTR we observed an evolutionarily conserved UGA stop codon 63 nt downstream and in-frame to the canonical stop codon (Figure 1D). Importantly, a 4-nt insertion in rodent 3′UTRs is offset by a 1-nt deletion thereby preserving the in-frame nature of the stop codons. Moreover, a mutation in the downstream rodent stop codon generates the alternate UAA stop codon, providing additional evolutionary support for the significance of the in-frame stop codon. The amino acid sequence potentially encoded by the region between the stop codons is likewise conserved in mammals, but the sequence beyond the downstream stop codon exhibits weak conservation (Figure S1E,F). Based on these observations we hypothesized that VEGFA mRNA translation might extend beyond its canonical stop codon, terminating at the downstream, in-frame stop codon. This translational readthrough event in human or bovine cells would generate a VEGF-A isoform containing a 22-amino acid C-terminus extension (including an amino acid replacement of the stop codon) ending with S/RLTRKD, the same 6-amino acid terminus reported to confer anti-angiogenic activity in human VEGF-Ab (Figure 1E).
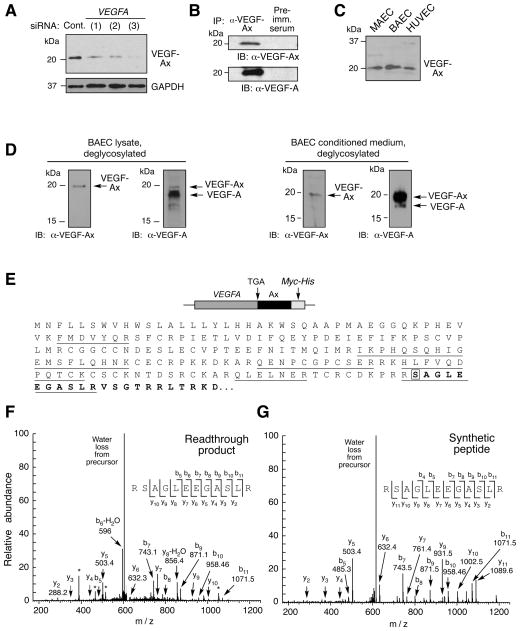
To test this hypothesis, we generated an antibody against the unique peptide, AGLEEGASLRVSGTR, generated by the proposed translational readthrough event, and not present in VEGF-Ab or any other protein (or in-silico translated mRNA). A 20-kDa protein corresponding to the predicted size of the readthrough product, designated VEGF-Ax (for extended), was identified in lysates from bovine ECs (Figure 2A). Expression was inhibited by transfection with three different VEGFA-specific siRNAs, consistent with a VEGF-A isoform. Protein immunoprecipitated with anti-VEGF-Ax antibody was recognized by an anti-VEGF-A antibody that targets the N-terminus, further verifying VEGF-Ax as a VEGF-A isoform (Figure 2B). To validate antibody specificity, HEK293 cells were transfected with myc-tagged VEGFA cDNA up to the downstream stop codon, and in which the canonical stop codon was replaced by Ala-encoding GCA to ensure efficient generation of the modified, extended isoform, denoted VEGF-AxAla. Anti-VEGF-Ax antibody recognized VEGF-AxAla but not VEGF-A, whereas anti-VEGF-A antibody recognized both isoforms (Figure S2A). As expected, anti-VEGF-Ax antibody did not recognize recombinant VEGF-Ab; however, anti-VEGF-Ab antibody did recognize VEGF-AxAla (Figure S2B). Finally, pre-adsorption of anti-VEGF-Ax antibody with AGLEEGASLRVSGTR peptide prevented recognition of the 20-kDa VEGF-Ax band, further validating antibody specificity (Figure S2C). VEGF-Ax expression also was observed in murine aortic and human umbilical vein ECs (Figure 2C), and in multiple other cell types including macrophages, hepatocytes, keratinocytes, and tumor-derived cell lines (Figure S2D). To distinguish between VEGF-Ax and VEGF-A by protein molecular weight, EC lysates were deglycosylated, subjected to 16% Tricine gel electrophoresis, and immunoblotted with anti-VEGF-A and anti-VEGF-Ax antibodies. VEGF-Ax constitutes about 10% of all VEGF-A isoforms in EC lysates; however, VEGF-Ax is the predominant isoform in conditioned medium, constituting about 85% of the total (Figure 2D).
Figure 2. VEGF-Ax is a Novel Isoform of VEGFA Generated by Translational Readthrough.
(A) VEGFA-specific siRNAs inhibit VEGF-Ax expression. Bovine ECs were transfected with three different VEGFA-specific siRNAs and VEGF-Ax in cell lysates determined by immunoblot with anti-VEGF-Ax antibody.
(B) VEGF-Ax is an authentic VEGF-A isoform. Lysates from bovine EC were immunoprecipitated with anti-VEGF-Ax antibody or pre-immune (Pre-imm.) serum and subjected to immunoblot analysis with anti-VEGF-Ax and anti-VEGF-A antibodies.
(C) VEGF-Ax is expressed by murine aortic ECs (MAEC), bovine aortic ECs (BAEC), and human umbilical vein ECs (HUVEC). Cell lysates were subjected to immunoblot analysis with anti-VEGF-Ax antibody.
(D) Separation of VEGF-Ax and VEGF-A isoforms in EC. BAEC lysates (left) and conditioned media (right) were deglycosylated and resolved on 16% Tricine gel before immunoblot analysis with anti-VEGF-Ax and anti-VEGF-A antibodies.
(E) Amino acid sequence of VEGF-Ax. Peptides identified by mass spectrometry (underline), readthrough region (bold), and recoded Ser (boxed) are highlighted.
(F) Identification of VEGFA mRNA readthrough product by MS/MS. A MS/MS spectrum was identified consistent with RSAGLEEGASLR (readthrough amino acid is underlined). The spectrum contains a total of nine C-terminal ‘y’ ions and seven N-terminal ‘b’ ions consistent with this sequence. The peptide was a low abundant component, and the spectrum contains several contaminant ions (*).
(G) Identity of readthrough peptide validated by synthetic peptide. RSAGLEEGASLR peptide was synthesized and the MS/MS spectrum determined as in (F).
See also Figure S2.
Translational readthrough in VEGFA mRNA was further established by mass spectrometric analysis of the product generated by overexpressing in HEK293 cells VEGFA cDNA containing the canonical stop codon and the Ax-specific sequence upstream of a myc-His tag to facilitate purification. In addition to detection of VEGF-A-specific peptides, selected reaction monitoring revealed a spectrum consistent with RSAGLEEGASLR (S, Ser in place of stop codon) specific for the readthrough region; the profile was confirmed by the spectrum obtained from synthetic RSAGLEEGASLR peptide (Figure 2E–G). Spectra consistent with Ser in place of the stop codon were observed in 3 out of 3 independent experiments. Targeted search for potential readthrough peptides containing the other 19 amino acids in place of the stop codon was negative, supporting Ser insertion during UGA stop codon readthrough.
Ax Element is the Necessary and Sufficient cis-acting Signal for Translational Readthrough of VEGFA mRNA
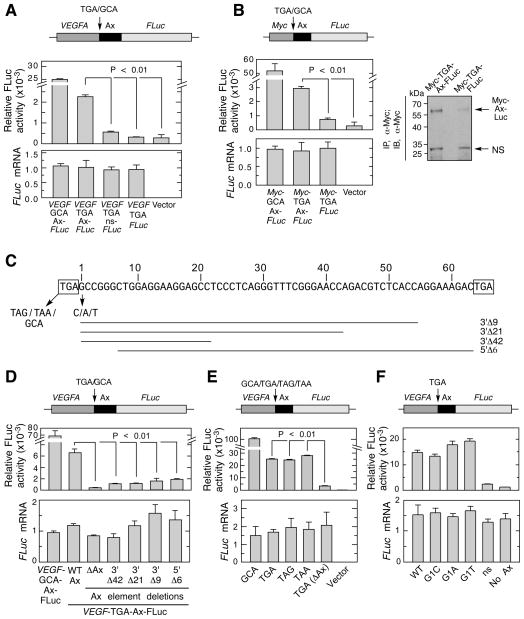
To validate translation readthrough and investigate molecular mechanisms, we established a reporter assay in which the full-length bovine VEGFA coding sequence (corresponding to VEGF-A164) up to and including the 63-nt, Ax-specific sequence (termed Ax element) was cloned upstream of and in-frame with firefly luciferase (FLuc). The downstream stop codon was excluded so that readthrough at the canonical stop codon generated a FLuc-containing chimeric protein. Transfected ECs exhibited substantially greater FLuc activity compared to vector alone or control reporters lacking the Ax element or replaced by a non-specific sequence, consistent with translation readthrough (Figure 3A). To calculate readthrough efficiency, the upstream TGA stop codon was mutated to Ala-encoding GCA to maximize FLuc expression; stop codon readthrough efficiency was about 9%. Quantitative RT-PCR analysis showed that all constructs were transcribed at similar levels, indicating that differential FLuc activity was due to differences in translation. Similar results were obtained by in vitro translation of the same reporters in rabbit reticulocyte lysates (Figure S3A). These results verify translational readthrough in VEGFA mRNA, and show the essentiality of the Ax element, consistent with a programmed translation readthrough (PTR) mechanism.
Figure 3. Ax Element is Necessary and Sufficient cis-acting Signal for VEGFA mRNA Readthrough.
(A) Ax element is sufficient for readthrough of VEGFA chimeric transcript. Plasmid containing in-frame VEGF-Ax-FLuc, and variants with TGA-to-GCA substitution, no Ax element, and Ax replaced by a non-specific sequence (ns), were transfected into ECs. FLuc activity was normalized to expression of co-transfected Renilla Luc (top), and FLuc mRNA expression determined by qRT-PCR (bottom).
(B) Ax element is sufficient for readthrough of heterologous mRNA, Chimeric plasmids containing Myc-Ax-FLuc and variants were transfected into ECs. Relative FLuc activity and mRNA expression were measured (left). Readthrough product also was determined by immunoprecipitation with rabbit anti-Myc-tag antibody followed by immunoblot with the mouse anti-Myc-tag antibody (right).
(C) Nucleotide sequence of bovine Ax element. Flanking stop codons (boxes), deletions (horizontal lines) and mutations (arrows) are shown.
(D) Plasmids containing deletions of VEGF-Ax-FLuc were transfected into ECs. FLuc activity was normalized by expression of co-transfected Renilla Luc (top); FLuc mRNA expression was determined by qRT-PCR (bottom).
(E) All three stop codons permit readthrough. ECs were transfected with VEGF-Ax-FLuc constructs containing each stop codon. Relative FLuc activity and mRNA were determined.
(F) The nucleotide immediately following the canonical stop codon (G1) does not alter readthrough efficiency. ECs were transfected with VEGF-Ax-FLuc constructs containing G-to-C, G-to-A, and G-to-T substitutions. Relative FLuc activity and mRNA were measured.
See also Figure S3.
To investigate if the Ax element drives readthrough in the context of a heterologous transcript, a construct was engineered containing Myc terminated with a TGA stop codon, upstream and in-frame to the Ax element and FLuc (Myc-Ax-FLuc). Transfection into ECs induced substantial FLuc activity compared to a control transcript without the Ax element or empty vector (Figure 3B, left). The readthrough product was detected following immunoprecipitation using anti-Myc-tag antibody by immunoblot (Figure 3B, right) and by mass spectrometry (Figure S3B). Also, Ax element-mediated readthrough of Myc-Ax-FLuc was shown by in vitro translation in rabbit reticulocyte lysate (Figure S3C). To determine the region of the Ax element necessary for function, deletions from both termini were generated (Figure 3C). Deletion of 9, 21, or 42 nt from the 3′ end or 6 nt from the 5′ end substantially reduced FLuc activity, suggesting that both termini are required, and possibly the entire element, for efficient readthrough (Figure 3D). Next, we tested the ability of Ax element to execute readthrough across the other two stop codons, UAG and UAA. Surprisingly, similar Ax element-mediated VEGFA readthrough was observed across all three stop codons (Figure 3E), thereby rejecting single stop codon-dependent mechanisms, such as UGA-dependent selenocysteine insertion (Low and Berry, 1996). Mutation of the first nucleotide after the canonical stop codon (denoted G1) did not affect the readthrough efficiency ruling out context-dependent readthrough mechanisms observed in other systems (Poole et al., 1995; Tate et al., 1999) (Figure 3F). Together, these experiments reveal the Ax element as the necessary and sufficient translational readthrough signal in VEGFA mRNA.
hnRNP A2/B1 is a trans-acting Enhancer of VEGFA Readthrough
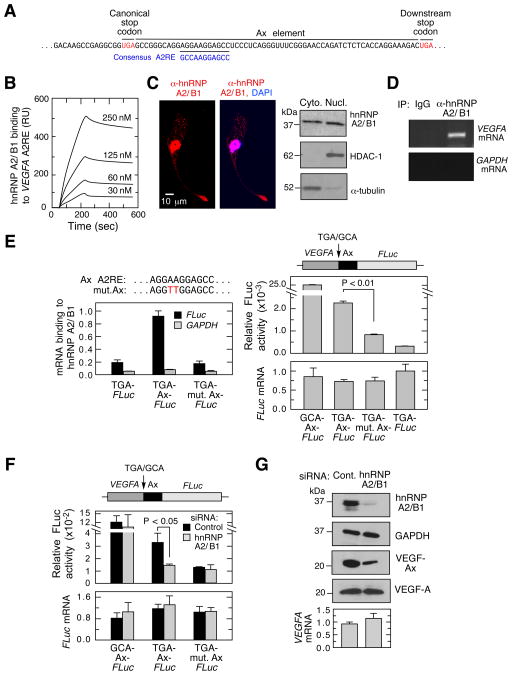
A near-consensus hnRNP A2/B1 recognition element (A2RE) was identified within the Ax element (Figure 4A). hnRNP A2/B1 is an RNA-binding protein involved in pre-mRNA processing and mRNA transport, and binds the consensus sequence 5′-GCCAAGGAGCC-3′ (He and Smith, 2009; Shan et al., 2003). Surface plasmon resonance spectroscopy revealed a high-affinity interaction between hnRNP A2/B1 and a VEGFA RNA segment containing the A2RE, with a dissociation constant (KD) of 19.2 ± 5.6 nM (Figure 4B). hnRNP A2/B1 is localized primarily in the nucleus; however, cytoplasmic localization was detected by immunofluorescence and by fractionation (Figure 4C). An RNA-binding protein immunoprecipitation (RIP) experiment confirmed the intracellular interaction between endogenous hnRNP A2/B1 and VEGFA mRNA (Figure 4D). To interrogate interaction specificity, ECs were transfected with a FLuc reporter bearing wild-type, mutant (AA-to-TT), or no Ax element. RIP followed by quantitative RT-PCR analysis showed that hnRNP A2/B1 interacted with wild-type Ax element, but not with the mutant element (Figure 4E, left) (Munro et al., 1999). Mutation of the A2RE also substantially reduced readthrough without altering mRNA expression (Figure 4E, right). The low-level readthrough in the absence of hnRNP A2/B1 binding suggests that other factors might contribute, consistent with the requirement for the entire Ax element, not just the A2RE. Similarly, siRNA-mediated knockdown of hnRNP A2/B1 in ECs reduced readthrough of the VEGFA-Ax-FLuc reporter without inhibiting FLuc activity of a construct with Ala-encoding GCA in place of the canonical stop codon or with mutant Ax element (Figure 4F). Finally, hnRNP A2/B1 knockdown reduced endogenous expression of VEGF-Ax without affecting total VEGF-A protein or mRNA in ECs (Figure 4G). These observations show the important regulatory role of hnRNP A2/B1 as a trans-acting factor promoting PTR of VEGFA mRNA.
Figure 4. hnRNP A2/B1 Facilitates VEGFA mRNA Translational Readthrough.
(A) Inter-stop codon sequence (Ax element) of human VEGFA contains a near-consensus hnRNP A2/B1 response element (A2RE).
(B) High-affinity binding of hnRNP A2/B1 to VEGFA A2RE. Surface plasmon resonance analysis of recombinant hnRNP A2/B1 binding to biotinylated RNA containing VEGFA A2RE immobilized on streptavidin sensor chip (in response units, RU).
(C) Cytoplasmic localization of hnRNP A2/B1 in ECs. Detection of hnRNP A2/B1 by immunofluorescence (left, with nuclear DAPI stain) and by immunoblot of fractionated compartments (right).
(D) hnRNP A2/B1 binds VEGFA mRNA in cells. Immunoprecipitation of hnRNP A2/B1 followed by RT-PCR using VEGFA- and GAPDH-specific primers.
(E) hnRNP A2/B1 binds the A2RE in VEGFA mRNA. VEGF-Ax-FLuc plasmid containing an Ax element with A2RE mutation (mut. Ax) was transfected into ECs. Anti-hnRNP A2/B1 immunoprecipitates were probed by qRT-PCR using FLuc- and GAPDH-specific TaqMan probes (left). Readthrough expressed as relative FLuc activity and mRNA level are shown (right).
(F) siRNA-mediated knockdown of hnRNP A2/B1 reduces readthrough of Ax element-containing plasmid. ECs were transfected with VEGF-Ax-FLuc construct and the variant containing A2RE mutation. FLuc activity was measured and FLuc mRNA determined by qRT-PCR.
(G) siRNA-mediated knockdown of hnRNP A2/B1 reduces VEGF-Ax expression. Following siRNA-mediated knockdown of hnRNP A2/B1, VEGF-Ax and VEGF-A were determined by immunoblot analysis and VEGFA mRNA by qRT-PCR.
VEGF-Ax Exhibits Anti-angiogenic Properties
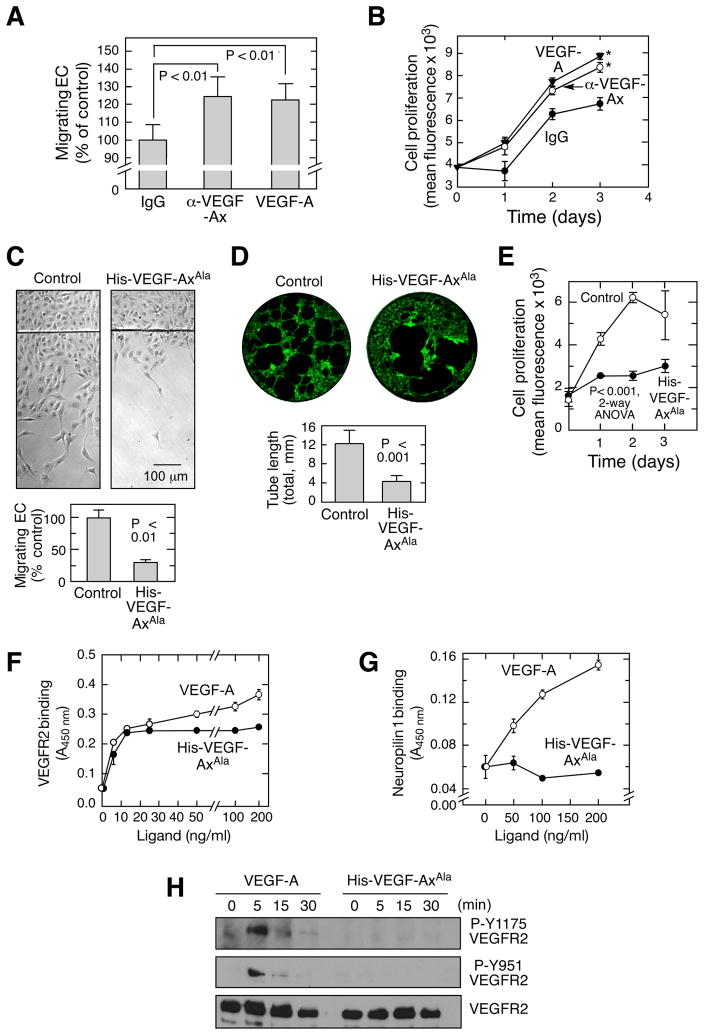
Our EC migration and proliferation experiments in the presence of VEGF-A antibody indicated secretion of an anti-angiogenic isoform of VEGF-A. Canonical VEGF-A isoforms were not detected in EC-conditioned medium immunodepleted with anti-VEGF-Ax antibody, confirming that VEGF-Ax is the predominant isoform secreted by ECs (Figure S4A). Also, the presence of RLTRKD (and absence of CDKPRR) at the C-terminus suggests that VEGF-Ax is a novel anti-angiogenic isoform. Anti-VEGF-Ax antibody increased EC migration and proliferation to an extent comparable to induction by recombinant VEGF-A, consistent with a paracrine, anti-angiogenic activity of endogenous VEGF-Ax (Figure 5A,B and S4B). Purified, recombinant His-VEGF-AxAla markedly inhibited planar EC migration, tube formation in Matrigel, and proliferation (Figure 5C–E). Similarly, siRNA-mediated knockdown of hnRNP A2/B1, which reduces VEGF-Ax production, stimulated EC migration (Figure S4C). Together, these results indicate that VEGF-Ax is the predominant isoform of VEGF-A released by ECs, exerting a potent paracrine, anti-angiogenic activity. To obtain insights into the mechanism of action of VEGF-Ax, we investigated its binding to VEGF receptors using solid phase enzyme-linked receptor-binding assays. His-VEGF-AxAla and VEGF-A bound VEGFR2, with comparable affinities, but maximal binding of the former was marginally lower at high ligand concentration (Figure 5F). Importantly, His-VEGF-AxAla did not bind neuropilin 1, a critical VEGFR2 co-receptor (Figure 5G). Likewise, His-VEGF-AxAla did not trigger canonical signaling pathways as shown by the failure to phosphorylate VEGFR2 Tyr residues Y1175 and Y951 (Figure 5H).
Figure 5. Anti-angiogenic properties of VEGF-Ax.
(A,B) Anti-VEGF-Ax antibody inhibits EC migration and proliferation. EC migration (A) in the presence of anti-VEGF-Ax antibody or recombinant human VEGF-A (20 ng/ml) was measured by razor-wound assay. Cell proliferation (B) was measured fluorimetrically as DNA (*, P < 0.05, 2-way ANOVA).
(C–E) Anti-angiogenic properties of VEGF-Ax. Migration (C), tube formation in Matrigel (D), and proliferation (E) of bovine ECs in the presence of recombinant His-VEGF-Axala (50 ng/ml).
(F–G) Binding of VEGF-A and His-VEGF-Axala to VEGFR2 (F) and neuropilin-1 (G). Binding was measured colorimetrically by solid phase enzyme-linked receptor-binding assay.
(H) Phosphorylation status of VEGFR2. HUVECs were treated with VEGF-A and His-VEGF-AxAla for up to 30 min and immunoblotted as shown.
See also Figure S4.
Pathophysiological Significance of VEGF-Ax
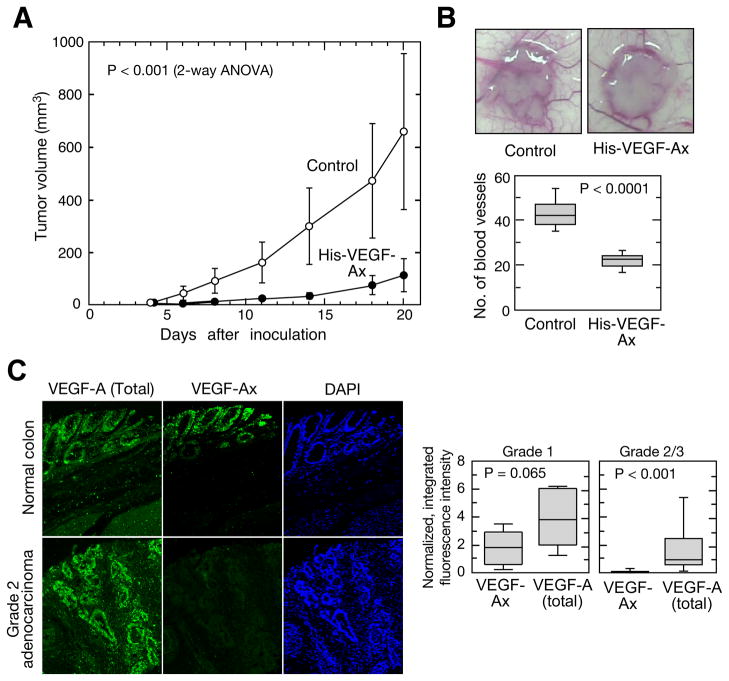
To determine the anti-angiogenic function of VEGF-Ax in vivo, we used human xeno-graft tumors in nude mice. Administration of recombinant VEGF-Ax markedly reduced the progression of HCT116 (human colon carcinoma cell)-derived tumors (Figure 6A). Furthermore, VEGF-Ax reduced tumor-associated angiogenesis as measured by the number of vessels directly feeding the tumors (Figure 6B). To further assess the physiological significance of VEGF-Ax, we determined its expression in vivo using a tissue microarray. VEGF-Ax was detected in tissue slices from human brain, colon, small intestine, spleen, and pancreas, and from serum from healthy human subjects (Figure S5A,B). Staining of human colon tissue with anti-VEGF-Ax antibody pre-adsorbed with AGLEEGASLRVSGTR peptide showed only background signal demonstrating antibody specificity in tissue immunofluorescence assays (Figure S5C). VEGF-Ax expression in tissues from pancreatic adenocarcinoma and glioblastoma was similar to that in corresponding healthy tissues (data not shown). Remarkably, VEGF-Ax expression was reduced to near-background level in cancerous tissues from grade 2 or 3 adenocarcinoma of colon, despite abundant VEGF-A expression (Figure 6C). These results not only show that VEGF-A translational readthrough occurs in vivo, but also they suggest a possible negative role of VEGF-Ax in tumor progression.
Figure 6. Anti-tumor properties of VEGF-Ax.
(A) Tumor progression in athymic nude mice. Mice were inoculated with HCT116 cells and treated sub-cutaneously with recombinant His-VEGF-Ax (10 μg/mouse; n=6 mice, 12 tumors) or buffer (n=5 mice, 10 tumors) every third day starting from 4th day after tumor cell inoculation.
(B) Tumor angiogenesis. Representative images of day 6 HCT116 tumors in athymic nude mice (top). Number of blood vessels feeding directly into tumors; whiskers show 5th and 95th percentiles (bottom; n=6 mice, 12 tumors in each group).
(C) Immunofluorescence of normal colon and colon adenocarcinoma. VEGF-Ax and total VEGF-A were quantified as integrated, background-subtracted fluorescence intensity (left); expression in Grade 1 (n=6) and Grade 2/3 colon adenocarcinoma (n=22) were normalized to healthy colon (n=5); whiskers show 5th and 95th percentiles (right).
See also Figure S5.
Genome-wide Analysis of Translational Readthrough
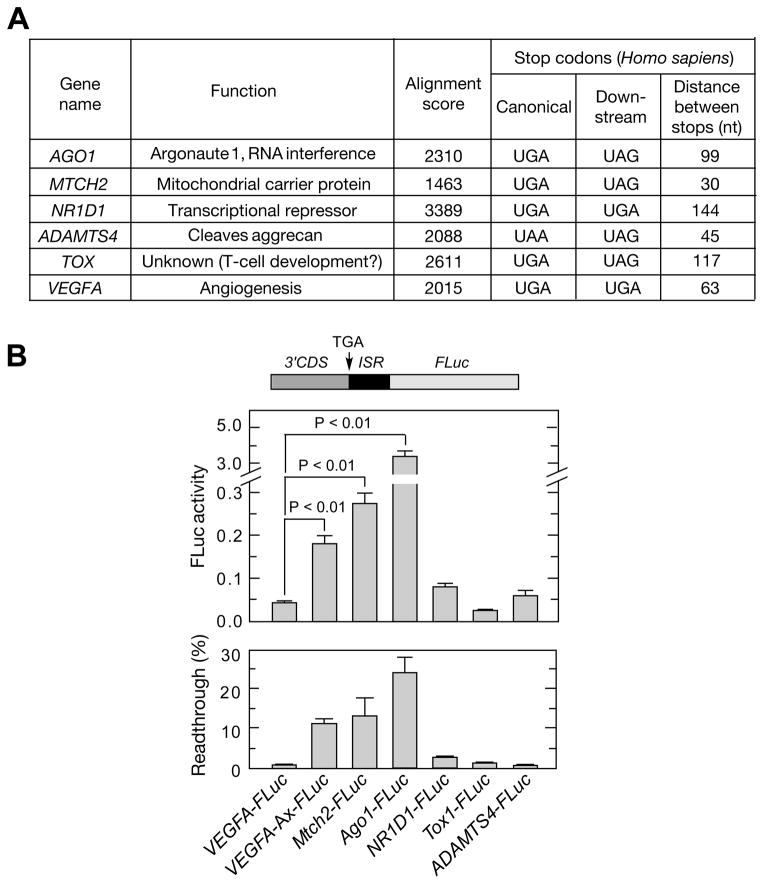
To identify additional translational readthrough candidates in mammals, we performed a genome-wide bioinformatic analysis. We interrogated a UTR database (Grillo et al., 2010) for mammalian transcripts with a stop codon in the 3′UTR in-frame to the canonical stop codon, and with amino acid conservation in the inter-stop codon region across five mammalian species (Figure S6). This screen revealed VEGFA and five high-probability readthrough candidates (Figure 7A). Candidates were tested by transfection of HEK293 cells with a plasmid containing about 700 nt of the 3′ terminus of the human coding sequence of each candidate, followed by the inter-stop codon sequence and FLuc. MTCH2 and AGO1 showed robust readthrough activity (Figure 7B). Readthrough efficiency was determined by comparison to corresponding in-frame control constructs where UGA stop codon is replaced with Ala-encoding GCA. Readthrough efficiency for VEGFA, MTCH2, and AGO1 was calculated to be 11%, 13%, and ~24%, respectively.
Figure 7. Genome-wide Analysis to Identify Targets of Translational Readthrough.
(A) Properties of candidate genes selected for experimental validation. Candidates were selected using a bioinformatic screening protocol (Figure S6).
(B) Validation of translational readthrough targets. Chimeric plasmids containing about 700 nt of the 3′-terminus of human coding sequences upstream of the inter-stop codon region and in-frame with FLuc were transfected into HEK293 cells. Relative FLuc activities normalized to FLuc mRNA were determined by qRT-PCR (top). Readthrough efficiency is expressed as percent of FLuc activity of normal translation controls in which canonical UGA stop codon is replaced by GCA (bottom).
See also Figure S6.
DISCUSSION
Organisms have evolved a plethora of mechanisms at multiple levels to expand their proteomes relative to their surprisingly limited number of distinct genes. Among the most employed examples are alternative splicing and alternative polyadenylation at post-transcriptional level, alternative initiation at translational level, and protein cleavage and modification at the post-translational level. There are multiple examples but little mechanistic information about proteome expansion by translational readthrough that can generate proteins with extended C-termini, and potentially with alternate functions compared to the shorter, canonical form. Although some mechanistic insights have been provided by studies of translational readthrough in RNA viruses, little is known about mechanisms of translational readthrough in vertebrates and its role in proteome expansion. Here we reveal a programmed translational readthrough (PTR) event in VEGFA transcript that generates an anti-angiogenic isoform, VEGF-Ax. Readthrough events have been described in vertebrate transcripts, for example, rabbit β-globin and rats myelin protein zero (Chittum et al., 1998; Yamaguchi et al., 2012), and a recent ribosome profiling experiment provided evidence for translational readthrough of 42 mRNAs in human fibroblasts (Dunn et al., 2013). However, the functional significance of the readthrough product has not been shown in any of these cases.
Basal readthrough levels ranging from 0.02% to 1.4% have been observed in yeast and mammalian cells (Dunn et al., 2013). In viruses, where multiple readthrough events have been established, 6% efficiency in murine leukemia virus (MLV) and 2–5% in Sindbis have been reported (Houck-Loomis et al., 2011; Li and Rice, 1989). In these cases the nucleotide sequence downstream of the canonical stop codon serves as a signal directing readthrough, establishing them as authentic “programmed” events unambiguously distinguishable from translation errors (Firth et al., 2011; Rajon and Masel, 2011). For example, a 63-nt RNA element downstream of the stop codon induces translational readthrough in the MLV gag-pol gene (Houck-Loomis et al., 2011). In our studies, the readthrough efficiency of VEGFA ranges from 7 to 25%. VEGFA mRNA readthrough is “programmed” by the Ax element, a 63-nt long RNA sequence between the two stop codons that can drive readthrough in a heterologous system. We also identify hnRNP A2/B1 as a trans-factor that binds the A2RE in the Ax element and positively regulates VEGFA readthrough. This finding is particularly noteworthy in that regulation of translational readthrough by a trans-factor has not been reported.
Although context-specific regulation of readthrough is suggested by a ribosome profiling study which showed differential readthrough of nine transcripts in two Drosophila cell types (Dunn et al., 2013), molecular mechanisms underlying such regulation have not been elucidated in any system. As one possible mechanism we speculate that the Ax element in coordination with hnRNP A2/B1, interacts directly or indirectly (e.g., by inducing a pause) with the translating ribosomes and prevents eukaryotic releasing factor 1 (eRF1) binding to the stop codon. As a consequence, the UGA stop codon could instead be recognized by a suppressor tRNA or by near-cognate tRNAs resulting in readthrough. This mechanism is supported by several observations: (i) protein interactome studies suggest that hnRNP A2/B1 interacts with several ribosomal proteins, including S7, S19, S28, L27a and L39 (Havugimana et al., 2012; Kristensen et al., 2012), (ii) ribosome occupancy is asymmetric at the stop codon, occupying 6 nt downstream of the stop codon (Ingolia et al., 2009). The A2RE element starts at the 11th nt following the stop codon, possibly facilitating an interaction between hnRNP A2/B1 and a ribosome paused at the stop codon, (iii) aminoglycosides, which induce readthrough, also bind ribosomes (Howard et al., 2004; Kondo et al., 2005), and (iv) the observed readthrough of all three stop codons suggests that a single tRNA species may not be involved. A deep mechanistic understanding to the readthrough mechanism might require structural studies of the relationship of the Ax element and its binding partner with the paused ribosome.
Serine was identified as the amino acid inserted in the place of the UGA codon during VEGFA mRNA readthrough. Interestingly, the tRNA that inserts selenocysteine (Sec) in place of UGA codons, namely tRNA[Ser]Sec, specifically recognizes UGA codons, and is initially charged with Ser before it is phosphorylated and converted to Sec by Sec synthase (Turanov et al., 2011). Moreover, Ser-tRNA[Ser]Sec can suppress termination at a UGA codon in rabbit β-globin mRNA in vitro (Chittum et al., 1998; Diamond et al., 1981). Thus, Ser-tRNA[Ser]Sec is a candidate for Ser insertion during VEGFA mRNA read-through. Alternatively, a Ser-tRNASer can contribute to readthrough as a consequence of compromised ribosome fidelity. Although only Ser was identified in place of the UGA codon of VEGFA mRNA, other amino acids might be inserted; Trp, Cys, and Arg are likely candidates as they have been observed in other readthrough events involving the UGA stop codon, namely, in rabbit β-globin (Chittum et al., 1998) and gag-pol MLV (Feng et al., 1990). However, these and other amino acids likely constitute a minor fraction as they were not observed even after targeted mass-spectrometry analysis. The observed readthrough across all stop codon types was unexpected, but nonsense suppression across all three stop codons has been reported in mammalian systems (Beier and Grimm, 2001; Hatfield et al., 1990). Recognition of all three stop codons by eRF1 is also consistent with our finding of promiscuous readthrough (Kisselev et al., 2003).
Translational readthrough contributes significantly to the proteome expansion in yeast, Drosophila, and human fibroblasts (Dunn et al., 2013; Jungreis et al., 2011; von der Haar and Tuite, 2007). Using a bioinformatic, genome-wide approach based on evolutionary conservation of the intra-stop codon sequence, we have identified five additional putative readthrough targets, none of which have been previously described. Translational readthrough was experimentally validated in two mRNAs, AGO1 and MTCH2. MTCH2 induces apoptosis by recruiting tBID (truncated BH3-interacting domain death agonist) to mitochondria (Zaltsman et al., 2010); AGO1 (or EIF2C1) encodes Argonaute 1, an important participant in the microRNA and RNA interference systems (Kim et al., 2006). Although the other candidates, NR1D1, TOX1, and ADAMTS4, were inactive in our luciferase-based readthrough assay in HEK293 cells, they might be readthrough targets in other conditions or cells. The AGO1 and MTCH2 3′UTRs do not share sequence similarity with the Ax element, nor do they possess an A2RE, suggesting that their readthrough mechanisms are likely to be different from that of VEGFA mRNA.
The concept of anti-angiogenic VEGF-A isoforms was introduced by the report of an alternative splicing event within the terminal exon of VEGFA to generate VEGFAb mRNA that replaces CDKPRR at the C-terminus with SLTRKD conferring anti-angiogenic properties in vitro (Bates et al., 2002). Several laboratories have shown that recombinant or over-expressed VEGF-Ab inhibits in vivo angiogenesis in murine tumor models, in mammary tissue of transgenic mice, and during neovascularization in mouse retina, rabbit cornea, and rat mesentery (Cebe-Suarez et al., 2008; Cebe Suarez et al., 2006; Cromer et al., 2010; Konopatskaya et al., 2006; Manetti et al., 2011; Qiu et al., 2008; Rennel et al., 2008; Rennel et al., 2009; Woolard et al., 2004). Lastly, immunological evidence for robust VEGF-Ab expression has been reported in multiple cells and tissues (Harper and Bates, 2008). Despite these promising observations, the in vivo expression of VEGF-Ab remains controversial, based in part on the observation that the RT-PCR methods used for detection and quantitation are prone to error (Bates et al., 2013; Harris et al., 2012). Similarly, we were unable to detect VEGFAb mRNA isoforms in ECs despite robust expression of anti-angiogenic activity. At present, detection by anti-VEGF-Ab antibody is considered the strongest support for expression of endogenous VEGF-Ab. However, this evidence is confounded by the presence of the same SLTRKD hexapeptide at the C-termini in both VEGF-Ax and VEGF-Ab, and anti-VEGF-Ab antibodies raised against this region do not distinguish between the two isoforms. In contrast, the antibody raised against the first 15 amino acids of the peptide extension of VEGF-Ax is highly specific for VEGF-Ax (Figure S2B). New tools and approaches are required to resolve the relative in vivo expression of the various pro-and anti-angiogenic isoforms. Development of antibodies that recognize VEGF-Ab, but not VEGF-Ax, possibly by targeting the junction of the sequence encoded by exon 7 and SLTRKD, would be extremely helpful. Also, RNA sequencing analysis could not only provide strong support for expression of VEGFAb isoforms, but also can offer insight into the relative amounts of differentially spliced VEGFA and VEGFAb isoforms in a given tissue or cell type.
VEGF-Ax exhibits potent anti-angiogenic activity as evidenced by inhibition of EC migration, proliferation, and tube formation in vitro, and tumor growth and associated angiogenesis in vivo. A report of enhanced EC sprouting in aortic rings from mice heterozygous for EC-specific VEGFA gene deletion (da Silva et al., 2010) is consistent with our finding of predominant anti-angiogenic VEGF-Ax in EC-conditioned medium. The discovery of anti-angiogenic VEGF-A isoforms compels a need for cognizance and caution in applying VEGF-A-targeted therapies for treatment of VEGF-A-dependent malignancies, as coincident diminution of VEGF-Ax may exacerbate angiogenesis and tumorigenesis (Harper and Bates, 2008; Varey et al., 2008). Moreover, usefulness of VEGF-A as a biomarker might also need to be re-considered in the view of potential presence of both pro- and anti-angiogenic isoforms. We speculate that expression of VEGF-Ax is an innate mechanism that evolved to limit EC movement and angiogenesis during tissue homeostasis and to reduce pathological angiogenesis. However, in some conditions exemplified by colon adenocarcinoma in our study, a loss of expression of VEGF-Ax might induce angiogenesis and exacerbate pathogenesis. Thus, abnormally low levels of VEGF-Ax in plasma or tissues can serve as a biomarker for prognosis or effective susceptibility to current anti-VEGF treatment. Moreover, because of its robust anti-angiogenic activity, VEGF-Ax provides a potential therapeutic agent against diseases characterized by excessive angiogenesis.
EXPERIMENTAL PROCEDURES
Cell culture
Bovine aortic endothelial cells (ECs) were cultured in DME/Ham’s F-12 medium containing 5% fetal bovine serum (FBS) and used in all experiments unless specified otherwise. Human umbilical vein ECs (HUVECs) were cultured in EC growth medium with SingleQuots supplements (Lonza). HEK293 cells were cultured in DMEM medium containing 10% FBS. Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
Luciferase-based translational readthrough assay
Plasmids containing chimera of FLuc downstream of test sequences (VEGFA, Myc, TOX, ADAMTS4, AGO1, NR1D1, or MTCH2) were transfected (500 ng/well) into bovine ECs in 24-well plates using Lipofectamine 2000. Plasmid expressing Renilla luciferase (RLuc) (50 ng/well) was co-transfected as efficiency control. After 48 h, cells were lysed and FLuc and RLuc activity were measured using Dual-Luciferase Reporter Assay System (Promega) in Victor3 1420 Multilabel Plate Counter (Perkin-Elmer). The same constructs were subjected to in vitro coupled transcription-translation (500 ng/reaction) using TNT T7 coupled rabbit reticulocyte lysates (Promega) and FLuc activity measured by Luciferase Assay System (Promega).
Animal studies
Nu/Nu athymic nude male mice were inoculated subcutaneously on both flanks with HCT116 human colon carcinoma cells. Mice were injected subcutaneously with His-VEGF-Ax (10 μg/mouse) obtained from HEK293 cells stably expressing bovine VEGF-Ax with Ser in place of the canonical stop codon. HiTrap Heparin HP Columns (GE Healthcare) were used for purification. Control mice were injected with buffer containing 25 mM Tris and 150 mM NaCl (pH 8). Injections were given every third day starting from the 4th day after tumor inoculation. Tumor progression was monitored by caliper determination of volume [(shortest diameter)2 × (longest diameter) × 0.525]. For determination of tumor angiogenesis, treatment was started on the second day after tumor inoculation, and blood vessels feeding directly into day 6 tumors were counted under a dissecting microscope at 12.5x magnification. Images were captured using PSMT5 operating microscope (World Precision) with 12.5x objective lens; every visible vessel touching the circumference of the tumor nodule was scored as a single vessel by an observer blinded to the treatment.
Supplementary Material
HIGHLIGHTS.
Discovery of VEGF-Ax, a novel, anti-angiogenic isoform of VEGF-A
VEGF-Ax is generated by programmed translational readthrough (PTR) of VEGFA mRNA
Ax is a dual function RNA element that drives PTR and encodes peptide extension
PTR is enhanced by hnRNP A2/B1, a trans-acting protein that binds Ax element
Acknowledgments
This work was supported in part by National Institutes of Health grants P01 HL029582, P01 HL076491, R01 GM086430, and R21 HL094841 (to P.L.F.). S.M.E. was supported by a fellowship from the American Heart Association, Great Rivers Affiliate. Shared instrument grant, S10 RR031537, from the National Institutes of Health (to B.W.) was used to purchase the Orbitrap mass spectrometer. Mouse studies were supported by the Athymic Animal and Xenograft Core, NCI P30 CA043703-23, of the Case Comprehensive Cancer Center (to D.L.). We are grateful to Donna Driscoll and Jodi Bubenik for helpful discussions. S.M.E. and P.L.F. are inventors on a patent that has been filed related to this work.
Footnotes
None of the other authors have any financial conflict of interest with the information in this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ausprunk DH, Folkman J. Migration and proliferation of endothelial cells in preformed and newly formed blood vessels during tumor angiogenesis. Microvasc Res. 1977;14:53–65. doi: 10.1016/0026-2862(77)90141-8. [DOI] [PubMed] [Google Scholar]
- Bates DO, Cui TG, Doughty JM, Winkler M, Sugiono M, Shields JD, Peat D, Gillatt D, Harper SJ. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–4131. [PubMed] [Google Scholar]
- Bates DO, Mavrou A, Qiu Y, Carter JG, Hamdollah-Zadeh M, Barratt S, Gammons MV, Millar AB, Salmon AH, Oltean S, et al. Detection of VEGF-A(xxx)b isoforms in human tissues. PloS ONE. 2013;8:e68399. doi: 10.1371/journal.pone.0068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Cebe Suarez S, Pieren M, Cariolato L, Arn S, Hoffmann U, Bogucki A, Manlius C, Wood J, Ballmer-Hofer K. A VEGF-A splice variant defective for heparan sulfate and neuropilin-1 binding shows attenuated signaling through VEGFR-2. Cell Mol Life Sci. 2006;63:2067–2077. doi: 10.1007/s00018-006-6254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebe-Suarez S, Grunewald FS, Jaussi R, Li X, Claesson-Welsh L, Spillmann D, Mercer AA, Prota AE, Ballmer-Hofer K. Orf virus VEGF-E NZ2 promotes paracellular NRP-1/VEGFR-2 coreceptor assembly via the peptide RPPR. FASEB J. 2008;22:3078–3086. doi: 10.1096/fj.08-107219. [DOI] [PubMed] [Google Scholar]
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- Cromer W, Jennings MH, Odaka Y, Mathis JM, Alexander JS. Murine rVEGF164b, an inhibitory VEGF reduces VEGF-A-dependent endothelial proliferation and barrier dysfunction. Microcirculation. 2010;17:536–547. doi: 10.1111/j.1549-8719.2010.00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva RG, Tavora B, Robinson SD, Reynolds LE, Szekeres C, Lamar J, Batista S, Kostourou V, Germain MA, Reynolds AR, et al. Endothelial alpha3beta1-integrin represses pathological angiogenesis and sustains endothelial-VEGF. Am J Pathol. 2010;177:1534–1548. doi: 10.2353/ajpath.2010.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A, Dudock B, Hatfield D. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981;25:497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. ELife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1990;87:8860–8863. doi: 10.1073/pnas.87.22.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2009;29:789–791. doi: 10.1161/ATVBAHA.108.179663. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011;39:6679–6691. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SJ, Bates DO. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat Rev Cancer. 2008;8:880–887. doi: 10.1038/nrc2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Craze M, Newton J, Fisher M, Shima DT, Tozer GM, Kanthou C. Do anti-angiogenic VEGF (VEGFxxxb) isoforms exist? A cautionary tale. PLoS ONE. 2012;7:e35231. doi: 10.1371/journal.pone.0035231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Smith DW, Lee BJ, Worland PJ, Oroszlan S. Structure and function of suppressor tRNAs in higher eukaryotes. Crit Rev Biochem Mol Biol. 1990;25:71–96. doi: 10.3109/10409239009090606. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480:561–564. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Anderson CB, Fass U, Khatri S, Gesteland RF, Atkins JF, Flanigan KM. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol. 2004;55:422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- Huez I, Bornes S, Bresson D, Creancier L, Prats H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol Endocrinol. 2001;15:2197–2210. doi: 10.1210/mend.15.12.0738. [DOI] [PubMed] [Google Scholar]
- Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalajakumari MB, Thomas CJ, Halter R, Manning PA. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3:1685–1695. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, Kellis M. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamba T, Tam BY, Hashizume H, Haskell A, Sennino B, Mancuso MR, Norberg SM, O’Brien SM, Davis RB, Gowen LC, et al. VEGF-dependent plasticity of fenestrated capillaries in the normal adult microvasculature. Am J Physiol Heart Circ Physiol. 2006;290:H560–576. doi: 10.1152/ajpheart.00133.2005. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J, Francois B, Urzhumtsev A, Westhof E. Crystallographic studies of Homo sapiens ribosomal decoding A site complexed with aminoglycosides. Nucleic Acids Symp Ser (Oxf) 2005:253–254. doi: 10.1093/nass/49.1.253. [DOI] [PubMed] [Google Scholar]
- Konopatskaya O, Churchill AJ, Harper SJ, Bates DO, Gardiner TA. VEGF165b, an endogenous C-terminal splice variant of VEGF, inhibits retinal neovascularization in mice. Mol Vis. 2006;12:626–632. [PubMed] [Google Scholar]
- Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 2012;9:907–909. doi: 10.1038/nmeth.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GP, Rice CM. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- Manetti M, Guiducci S, Romano E, Ceccarelli C, Bellando-Randone S, Conforti ML, Ibba-Manneschi L, Matucci-Cerinic M. Overexpression of VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, leads to insufficient angiogenesis in patients with systemic sclerosis. Circ Res. 2011;109:e14–26. doi: 10.1161/CIRCRESAHA.111.242057. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- Nagy JA, Dvorak AM, Dvorak HF. VEGF-A and the induction of pathological angiogenesis. Annu Rev Pathol. 2007;2:251–275. doi: 10.1146/annurev.pathol.2.010506.134925. [DOI] [PubMed] [Google Scholar]
- Namy O, Duchateau-Nguyen G, Hatin I, Hermann-Le Denmat S, Termier M, Rousset JP. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:2289–2296. doi: 10.1093/nar/gkg330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978;272:469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Bevan H, Weeraperuma S, Wratting D, Murphy D, Neal CR, Bates DO, Harper SJ. Mammary alveolar development during lactation is inhibited by the endogenous antiangiogenic growth factor isoform, VEGF165b. FASEB J. 2008;22:1104–1112. doi: 10.1096/fj.07-9718com. [DOI] [PubMed] [Google Scholar]
- Rajon E, Masel J. Evolution of molecular error rates and the consequences for evolvability. Proc Natl Acad Sci USA. 2011;108:1082–1087. doi: 10.1073/pnas.1012918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennel ES, Hamdollah-Zadeh MA, Wheatley ER, Magnussen A, Schuler Y, Kelly SP, Finucane C, Ellison D, Cebe-Suarez S, Ballmer-Hofer K, et al. Recombinant human VEGF165b protein is an effective anti-cancer agent in mice. Eur J Cancer. 2008;44:1883–1894. doi: 10.1016/j.ejca.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennel ES, Varey AH, Churchill AJ, Wheatley ER, Stewart L, Mather S, Bates DO, Harper SJ. VEGF(121)b, a new member of the VEGF(xxx)b family of VEGF-A splice isoforms, inhibits neovascularisation and tumour growth in vivo. Br J Cancer. 2009;101:1183–1193. doi: 10.1038/sj.bjc.6605249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Examination of the function of two kelch proteins generated by stop codon suppression. Development. 1997;124:1405–1417. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Schwartz SM, Haudenschild CC, Eddy EM. Endothelial regneration. I. Quantitative analysis of initial stages of endothelial regeneration in rat aortic intima. Lab Invest. 1978;38:568–580. [PubMed] [Google Scholar]
- Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, Dvorak HF. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001;2:593–597. doi: 10.1093/embo-reports/kve128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate WP, Mansell JB, Mannering SA, Irvine JH, Major LL, Wilson DN. UGA: a dual signal for ‘stop’ and for recoding in protein synthesis. Biochemistry Mosc. 1999;64:1342–1353. [PubMed] [Google Scholar]
- Turanov AA, Xu XM, Carlson BA, Yoo MH, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv Nutr. 2011;2:122–128. doi: 10.3945/an.110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varey AH, Rennel ES, Qiu Y, Bevan HS, Perrin RM, Raffy S, Dixon AR, Paraskeva C, Zaccheo O, Hassan AB, et al. VEGF 165 b, an antiangiogenic VEGF-A isoform, binds and inhibits bevacizumab treatment in experimental colorectal carcinoma: balance of pro- and antiangiogenic VEGF-A isoforms has implications for therapy. Br J Cancer. 2008;98:1366–1379. doi: 10.1038/sj.bjc.6604308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Woolard J, Wang WY, Bevan HS, Qiu Y, Morbidelli L, Pritchard-Jones RO, Cui TG, Sugiono M, Waine E, Perrin R, et al. VEGF165b, an inhibitory vascular endothelial growth factor splice variant: mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res. 2004;64:7822–7835. doi: 10.1158/0008-5472.CAN-04-0934. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Hayashi A, Campagnoni CW, Kimura A, Inuzuka T, Baba H. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J Biol Chem. 2012;287:17765–17776. doi: 10.1074/jbc.M111.314468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH, Vaz FM, De Leonardis F, Fiermonte G, Palmieri F, et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.