SUMMARY
During B lymphocyte development, immunoglobulin heavy chain variable (VH), diversity (DH) and joining (JH) segments assemble to generate a diverse antigen receptor repertoire. Here we have marked the distal VH and DH-JH-Eμ regions with Tet-operator binding sites and traced their 3D-trajectories in pro-B cells transduced with a retrovirus encoding Tet-repressor-EGFP. We found that these elements displayed fractional Langevin motion (fLm) due to the viscoelastic hindrance from the surrounding network of proteins and chromatin fibers. Using fractional Langevin dynamics modeling, we found that, with high probability, DHJH elements reach a VH element within minutes. Spatial confinement emerged as the dominant parameter that determined the frequency of such encounters. We propose that the viscoelastic nature of the nuclear environment causes coding elements and regulatory elements to bounce back and forth in a spring-like fashion until specific genomic interactions are established and that spatial confinement of topological domains largely controls first-passage times for genomic interactions.
INTRODUCTION
It is now established that chromosomes fold into territories that rarely intermingle (Rabl, 1885; Boveri, 1909; Bolzer et al., 2005). Chromosome territories display a confined geometry revealed by the presence of chromosome arms and bands (Sedat and Manuelidis, 1977). By comparing experimentally obtained spatial distance distributions to spatial distances derived from computer simulations of various chromatin configurations, it was predicted that the mammalian genome is organized as bundles of loops that fold into distinct domains (Münkel and Langowski, 1998; Jhunjhunwala et al., 2008). Chromosome-conformation-capture approaches have validated these findings at a global scale, revealing that mammalian genomes fold into clusters of loops that assemble into topological domains (Lieberman-Aiden et al., 2009; Dixon et al., 2012; Lin et al., 2012). On average, these domains span genomic distances comparable to the size of the Igh locus (1–3Mbp) (Dixon et al., 2012; Jhunjhunwala et al., 2008).
Antigen receptor assembly in lymphocytes occurs by the random rearrangement of variable (VH), diversity (DH), joining (JH), and constant (CH) coding elements of the immunoglobulin heavy chain locus (Igh) (Jung et al., 2006). Antigen receptor genes in pro-B cells undergo ordered rearrangement with DHJH joining preceding VH to DHJH rearrangement (Alt et al., 1984). Recombinase activating genes 1 and 2 (Rag-1/2) regulate this process. Collectively, the VH regions span a genomic distance of approximately 2.5 Mbp and fall into two distinct domains, distal and proximal VH clusters. The DH and JH elements then span a genomic distance of 50 kb and are positioned immediately upstream (~1kb) of the intronic enhancer (Eμ). Finally, constant regions downstream of the JH elements encode the Igh isotypes (Shimizu et al., 1982; Retter et al., 2007). Given the configuration of the Igh locus and distance between VH, DH, and JH genomic segments, contraction brings otherwise distant genomic neighbors into close proximity for efficient gene rearrangement.
Productive V(D)J gene rearrangement in developing B cells permits surface expression of a pre-B cell receptor (pre-BCR). Signaling through the pre-BCR suppresses RAG1/2 activity, halting continued rearrangement (Nussenzweig et al., 1988; Manz et al., 1988; Grawunder et al., 1995). Various mechanisms, including DNA methylation, chromatin remodeling, histone acetylation, germ-line transcription and transcription elongation, control the temporal and lineage specificity of V(D)J rearrangement (Jung et al., 2006; Cedar and Bergman, 2011).
During developmental progression, the Igh locus undergoes large-scale topological changes (Kosak et al., 2002; Fuxa et al., 2004; Hewitt et al., 2010; Guo et al., 2011a). In progenitor cells, the Igh alleles are sequestered at the transcriptionally repressive nuclear lamina (Kosak et al., 2002). As progenitors enter the pre-pro-B cell stage, the locus is released from the lamina to associate with recombination and/or transcription factories. Committed pro-B cells undergo large-scale conformational changes in which distinct classes of anchors merge the distal and proximal VH regions into one cloud of VH regions (Jhunjhunwala et al., 2008; Medvedovic et al., 2013). The majority of the proximal VH regions are located within close genomic proximity to the chromatin anchor CCCTC-binding factor (CTCF) sites (Degner et al., 2009). It has been proposed that CTCF positions the proximal VH regions at the base of loops that orbit the DHJH elements (Lucas et al., 2011). In addition, an insulator element that regulates Igh locus rearrangement has been identified (Guo et al., 2011b; Degner et al., 2011). The insulator element contains two CTCF binding sites, together named CBE (CTCF-binding elements). Deletion of the CBE leads to loss of ordered and lineage-specific Igh locus rearrangement involving the most proximal but not distally located VH regions (Guo et al., 2011b). The exact mechanism through which the CBE regulates proximal VH-DHJH joining remains to be elucidated.
Here we describe the 3D-trajectories adopted by the Igh locus. We found that the trajectories adopted by VH and DHJH elements displayed fractional Langevin motion due to the viscoelastic hindrance from the surrounding network of proteins and chromatin fibers, including the neighboring segments of the chromatin fiber. Using fractional Langevin dynamics modeling, we determined first-encounter times, known as first-passage times, for VH and DHJH elements and found that VH and DHJH elements have a high probability of reaching each other within minutes. We found that spatial confinement imposed by chromosomal domains is the dominant factor that determines the encounter times for coding and regulatory DNA elements. Based on these observations we propose that fLm motion causes coding (VH, DH and JH), regulatory elements (enhancer and promoters) as well as double-stranded DNA breaks to bounce back and forth in a spring-like fashion until specific genomic interactions are established. Finally, we suggest that spatial confinement of chromosomal domains largely controls the encounter times of long-range genomic interactions, including those involved in VHDHJH rearrangement, locus contraction, class switch recombination, DNA repair as well as chromosomal deletions in cancers.
RESULTS
Inserting Tet-Operator Binding Sites Adjacent to VH and DHJH Gene Segments
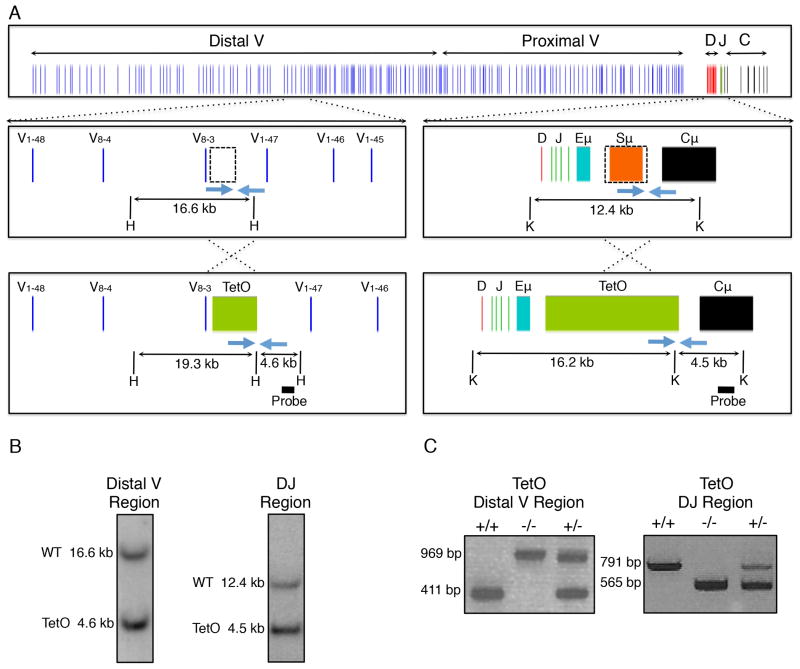
The immunoglobulin locus is structured into distinct domains comprised of distal VH, proximal VH and the DHJH regions. The VH regions are associated with promoters whereas the DHJH elements are located within close genomic proximity of the Eμ enhancer. To address how VH regions differ from DHJH elements in terms of chromatin motion, we inserted arrays of Tet-operator binding sites into the VH domains. Specifically, an array was inserted in the center of the distal VH region cluster, immediately down-stream of V8-3 (Figure 1A). A second array of either Tet-operator binding sites or Lac-operator binding sites was inserted between the DHJH elements and the Eμ enhancer (Figure 1A; Figure S1A). This region consists of a series of simple repeats involved in class switch recombination. Deletion of this genomic region diminishes class switch recombination (CSR) but does not interfere with V(D)J recombination (Luby et al., 2001).
Figure 1. Generation of Igh-TetO Labeled Mice.
(A) Schematic of the Igh locus showing VH, DH, JH, and CH segments, the intronic enhancer (Eμ), and switch region repeats (Sμ). Dashed boxes show genomic regions that were replaced by arrays containing 240 copies of the Tet operator. Southern blot screening strategies are indicated using HindIII (H) or KpnI (K) restriction enzymes. Blue arrows show positions of genotyping primers.
(B) Southern blot of embryonic stem cell clones positive for integration of the TetO arrays positioned adjacent to either the VH or DHJH region of the Igh locus.
(C) PCR-based genotyping results of mice harboring TetO arrays on both (+/+), one (+/−), or neither (−/−) alleles of the Igh locus.
Each construct was inserted into the embryonic stem (ES) cell genome. Briefly, ES cells were transfected with a vector in which the Tet-operator binding sites were inserted adjacent to the VH or DHJH regions. Genomic DNA was isolated from transfected ES cells and clones were examined for insertion into the Igh locus (Figure 1B). ES cells carrying the insertions adjacent to the VH and DHJH regions were used to generate chimeric mice. Mice carrying insertions in the distal VH and DHJH regions, readily went germ-line, establishing two lines of mice that either carry Tet-operator binding sites inserted into the VH, or alternatively, the DHJH regions (Figure 1C). However, mice carrying arrays of Lac-operator binding sites failed to go germ-line (Figure S1B and S1C). Upon further inspection we found that ES cells carrying Lac-operator binding sites display a large degree of chromosomal instability, possibly reflecting the inability of mice that carry Lac-operator binding sites in the genome to undergo germ-line transmission (Figure S1D). In sum, we generated mice that carry tandem arrays of Tet-operator sites either within the VH cluster or in close genomic proximity to the DHJH region.
Characterization of Mice Carrying Arrays of TET-Operator Binding Sites Inserted into the Igh Locus
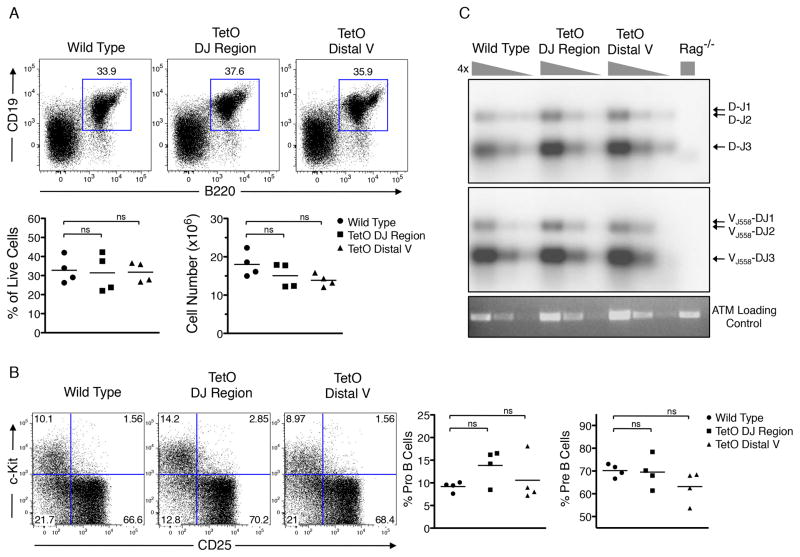
To ensure that the insertion of arrays of TetO binding sites into the Igh locus did not interfere with B cell development, we analyzed the B-lineage compartment in wild-type and mutant TetO mice. Bone marrow cells derived from 6-week old mice were isolated, analyzed for cellularity and stained for the expression of CD19 and B220 using flow cytometry. Mice containing Tet-operator binding sites inserted adjacent to either the DHJH segments or the distal VH segments displayed similar numbers of CD19 and B220 double-positive cells as compared to wild-type cells (Figure 2A). Furthermore, the proportions of pro-B versus pre-B cells were similar in the TetO mice as compared to wild-type mice (Figure 2B). These data indicate that insertion of arrays of Tet-operator binding sites at these locations did not interfere with normal production of developing B cells.
Figure 2. Characterization of Igh-TetO Labeled Mice.
(A) Femoral bone marrow cells from wild type, DHJH-TetO, and VH-TetO mice stained with B cell markers CD19 and B220. Bottom panel shows CD19+B220+ cell numbers as a fraction of live cells.
(B) Bone marrow cells stained for CD25 and C-kit. The lineage negative (CD11b, Gr1, Ter119) and B220+CD19+ population is shown.
(C) DH-JH and VHJ558-DHJH rearrangements in B220+ cells isolated from wild type, DHJH-TetO, VH-TetO and Rag−/− mice analyzed by Southern blotting using probes corresponding to VH and DHJH gene segments
See also Table S3.
To determine whether B-lineage cells carrying insertions of arrays of TetO binding sites in the Igh locus undergo efficient V(D)J recombination, DNA was isolated from pro-B cells derived from wild-type, DHJH-TetO and VH-TetO region mice as well as RAG-deficient mice. DHJH joints were readily detectable at equal levels in wild-type, DHJH-TetO and VH-TetO but not in control RAG-deficient pro-B cells (Figure 2C). Similarly, the abundance of VHDHJH joints involving the distal VH J558 cluster was not affected by the insertion of TetO sites (Figure 2C). In sum, these data indicate that insertion of arrays of Tet-operator binding sites does not affect the developmental progression of early B cell progenitors and Igh locus rearrangements.
Generation of TetR-EGFP Expression Vectors
Previous studies have utilized TetR-EYFP fusion proteins to track DNA motion in bacteria (Lau et al., 2003). Our initial attempts to detect TetR-EYFP mediated fluorescence in pro-B cells failed, plausibly caused by differences in codon usage in bacterial versus mammalian cells. However, a Tet-repressor optimized for mammalian codon usage has been utilized in studies to allow for tightly regulated control of gene expression (Urlinger et al., 2000). Hence, we fused the DNA binding domain of the modified Tet-repressor to EGFP (TetR-EGFP) along with a nuclear localization signal (PKKKRKV).
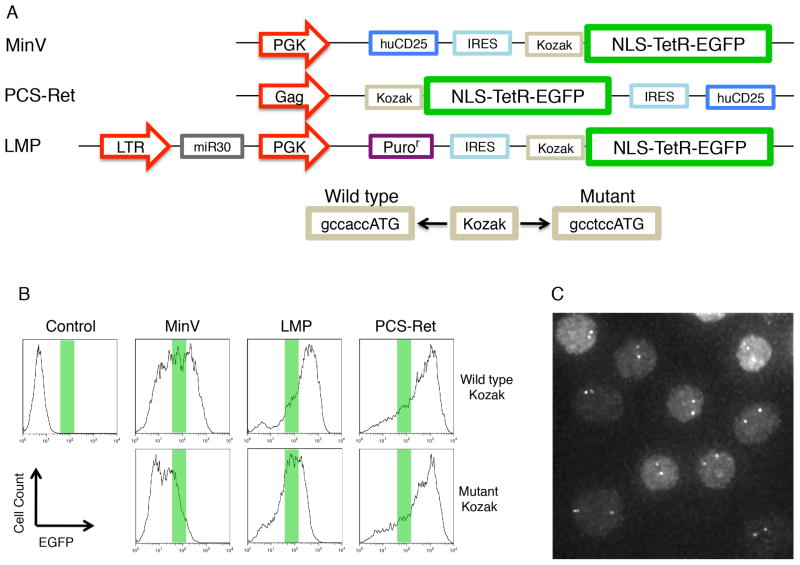
To optimize the signal-to-noise ratio of TetR-EGFP-mediated fluorescence when bound to DNA, we generated and tested multiple vectors. We modified expression levels using three distinct strategies: (1) different promoters were inserted upstream of the TetR-EGFP fusion protein; (2) TetR-EGFP expression was initiated from an internal ribosomal entry site (IRES) rather than directly from a promoter; and, (3) the Kozak sequence located upstream of the translation initiation site was mutated to dampen expression levels (Figure 3A). B-lineage cells were transduced with virus expressing TetR-EGFP driven from either the PGK promoter (MinV and LMP vectors) or the Gag promoter (PCS Ret vector), and containing either wild-type or mutated forms of the Kozak sequence (Figure 3A). Two days post transduction cells were examined for TetR-EGFP expression using flow cytometry (Figure 3B). Relatively low but detectable levels of TetR-EGFP expression was achieved using the MinV vector as well as the LMP vector carrying a mutant Kozak sequence, resulting in optimal signal-to-noise ratios of TetR-EGFP-mediated fluorescence upon binding to the Tet operator sites (Figure 3C). The LMP construct containing the mutant Kozak sequence, displayed a more uniform pattern of TetR-EGFP expression, thus we utilized this vector for subsequent monitoring of VH and DHJH motion in pro-B cells.
Figure 3. Generation and Optimization of TetR-EGFP Expression.
(A) Construction of TetR-EGFP expressing retroviral vectors. The NLS-TetR-EGFP coding sequence was cloned into three distinct retroviral vectors, MinV, PCS-Ret, and LMP. PGK or Gag promoters were used to drive EGFP expression in each of the vectors. Each construct was made using either a wild-type or mutant Kozak sequence. In the MinV and LMP vectors, the NLS-TetR-EGFP was inserted 3′ of an internal ribosomal entry site (IRES).
(B) DHJH-TetO B-lineage cells were infected with NLS-TetR-EGFP expressing retrovirus and expression levels were monitored using flow cytometry. Green bars show approximate expression levels leading to high signal to noise ratios of TetO-EGFP mediated fluorescence.
(C) DHJH-TetO B-lineage cells infected with NLS-TetR-EGFP in LMP with a mutant Kozak sequence and visualized by fluorescence microscopy. The dots in each cell show both Igh alleles.
Tracking Igh Locus Motion in B-Lineage Cells
To describe the trajectories adopted by Igh regions in physical terms, we generated strains of mice carrying TetO binding sites on both alleles. B220 positive pro-B cells from the bone marrow of VH-TetO and DHJH-TetO mice were cultured in the presence of IL7 and SCF for five days and transduced with virus expressing TetR-EGFP. Two days post transduction, TetO pro-B cells were immobilized by adherence onto poly-lysine coated optical bottom dishes and imaged (Figure 3C; Movies S1 and S2). One set of z-stacks was acquired every two seconds for 400 seconds (200 time-points) or every 40 seconds for 4000 seconds (100 time-points). The centers of mass of the TetO signals were determined for each time point. As a control to account for error due to cell motion, TetO pro-B cells were fixed with formaldehyde to prevent motion and labeled with Alexa-488 conjugated anti-GFP antibodies prior to imaging.
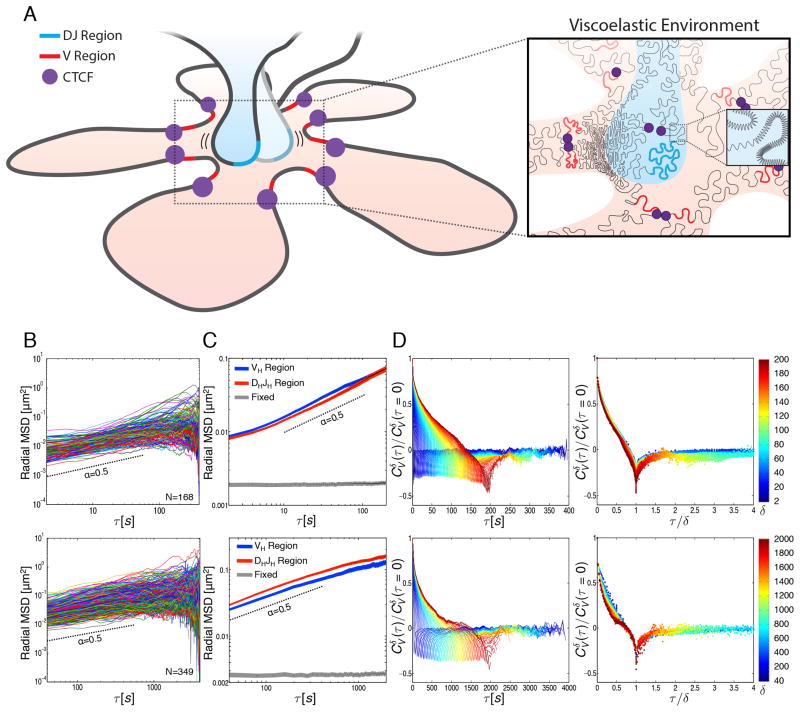
Despite attachment of cells with poly-lysine, a significant amount of cell motion, primarily cell or nuclear rotation, was observed (data not shown). To eliminate the effect of such motion in the analysis of loci trajectories, the mean squared displacement (MSD) was determined by measuring the mean squared change in distance separating the two alleles divided by two, rather than the change in the displacement of each allele. Measured this way, the radial MSD described motion in radial direction (distance between two alleles) only, thus eliminating confounding effects of cell movement and rotation (Vasquez et al., 2001; Cabal et al., 2006). As expected, the radial MSD values varied within the developing pro-B cell population, reflecting differences in cell cycle, cell size, and other intrinsic variations within the population (Figure 4B). We note that during imaging in a subset of cells we observed substantial changes in nuclear shape that resulted in large-scale movements of chromosomes and contribute to the displacement of the Igh locus gene segments. The average radial MSD of pro-B cells for both the VH-TetO and DHJH-TetO alleles were plotted as a function of the time interval τ for the values of τ between 2 and 2000 seconds (Figure 4C). As expected, Igh alleles in fixed cells did not show significant spatial displacement (Figure 4C). Anomalous diffusion coefficients (D) and scaling exponents (α) were extracted (radial MSD=2Dτα) for time intervals between 10–100 and 40–200 seconds (Figure 4C; Table S1). For wild-type TetO pro-B cells (40–200s), we found that VH-TetO motion was characterized by an anomalous diffusion coefficient (D) of 2.0 x10−3 μm2/s0.5 and a subdiffusive (i.e., less than unity) scaling exponent (α) of 0.49. For DHJH-TetO motion, we found an anomalous diffusion coefficient of 2.4 x10−3 μm2/s0.5 and subdiffusive scaling exponent of 0.52 (Table S1). These data indicate that both VH and DHJH elements in pro-B cells undergo subdiffusive motion with very similar anomalous diffusion coefficients and subdiffusive scaling exponents.
Figure 4. A Viscoelastic Environment Dominates Igh DHJH and VH motion in Pro-B Cells.
(A) Model depicting fractional Langevin motion across the Igh locus. The Igh locus is organized as bundles of loops, with CTCF (purple) positioning the VH segments (red) in orbit around the DHJH region (blue). Enlarged region depicts the viscoelastic properties of the chromatin fiber caused by interactions with neighboring networks of nucleic acids and proteins as well as restoring forces within the fiber itself (shown as springs).
(B) Time-averaged radial MSD plotted as a function of time lag (τ) for the DHJH region in live B220+ B-lineage cells isolated from TetO mice. Number of cells (N) analyzed is indicated. Cells were imaged once every 2 seconds for 400 seconds (top panel) or once every 40 seconds for 4000 seconds (bottom panel).
(C) Ensemble- and time-averaged radial MSD plotted as a function of time lag (τ) for the VH and DHJH regions in live B220+ B-lineage cells as well as formaldehyde fixed pro-B cells. Radial MSD is shown for τ up to one half of total imaging time. Shaded areas represent the standard error of the mean. Dashed lines indicate a sub-diffusive scaling exponent (α) of 0.5.
(D) Velocity autocorrelation analysis of DHJH regions in live B220+ B-lineage cells (left panels). Average velocity was calculated over discretization intervals (δ) ranging from 2 to 200 seconds in 2 seconds intervals (top panel) or 40–2000 seconds in 40 seconds intervals (bottom panel). Velocity autocorrelation curves for different values of δ, plotted against a rescaled time lag (τ/δ) (right panels). The color scheme represents the values of δ from small (blue) to large (red).
See also Figures S2 and S3 and Tables S1 and S2.
VH, DHJH and Enhancer Elements Display Fractional Langevin Motion
The observations described above show that the trajectories adopted by VH- and DHJH-TetO segments are associated with an anomalous diffusive motion. Specifically, the trajectories are shown to undergo subdiffusion, a process characterized by a non-linear relationship between the mean squared displacement and the elapsed time (MSD=2Dτα with α<1), in contrast to normal diffusion in which this relationship is linear (i.e., α=1). What is the underlying mechanism of the observed subdiffusive motion? In the crowded cellular environment, several possible molecular mechanisms may give rise to subdiffusive motion of a chromosomal segment (or any microscopic particle). First, when the diffusive motion of the particle is interspersed by pauses due to the encounters with binding partners, a broad distribution of pausing times makes the particle’s motion subdiffusive, as described by the continuous time random walk (CTRW) model (Saxton, 1996; Montroll and Weiss, 1965). Second, when the particle encounters obstacles present at high concentration, the particle’s motion becomes subdiffusive, as described by the obstructed diffusion (OD) model (Saxton, 1994). Third, a particle moving through a dense network of proteins and nucleic acids experiences a response from the surroundings that is characterized by both viscous and elastic components, and hence called viscoelastic response. The viscoelastic response predisposes the particle to bounce back towards its previous position in a spring-like fashion; these reversals, or negative correlations (“memory”), in the particle’s movements lead to subdiffusive behavior and can be described by fractional Langevin motion (fLm) (Lutz, 2001). Furthermore, when the “particle” is a chromosomal segment embedded in a chromatin fiber, the rest of the fiber itself serves as the viscoelastic environment by exerting restoring forces on the segment. The discussed three mechanisms of random motion may contain contributions from nonthermal active fluctuations, which effectively modify the diffusion coefficient in the corresponding models. Despite the distinct origins of these three mechanisms, they can all lead to the subdiffusive relationship between MSD and elapsed time. Therefore, we applied additional diagnostic measures to identify the mechanism for the observed subdiffusion of VH and DHJH-TetO segments.
In order to identify the mechanism of subdiffusive motion among the discussed candidate mechanisms (CTRW, OD and fLm), we compared the time-averaged radial MSD of individual trajectories to the ensemble-averaged radial MSD. Both the time- and ensemble-averaged MSD yielded essentially the same scaling exponent, namely α ≈ 0.5, suggesting that the trajectories are ergodic (i.e., they display the same time-averaged and ensemble-averaged properties), which, in turn, supports an OD or fLm but not CTRW mechanism that underpins the subdiffusion process observed here. The observed spread of the time-averaged radial MSD for individual trajectories may be attributed to the limited time over which the trajectories were measured as well as to the inherent heterogeneity of the cells.
To obtain a more robust measure of the subdiffusive behavior, we calculated the average velocity autocorrelation function, Cδv(τ). This function indicates to what degree the average velocity over a time interval δ is correlated with the average velocity over another time interval δ that is separated by τ from the first one. We analyze the correlation properties of the average velocity, rather than the instantaneous velocity, because experimental measurements are performed at finite time intervals and thus naturally yield the velocity averaged over a time interval. Cδv(τ) was calculated for different values of δ and plotted as a function of the time lag τ (Figure 4D: left panel; Figure S2). For all δ, Cδv(τ) dipped into negative values before decaying to zero. The observed negative values of Cδv(τ), which are indicative of negative correlations, cannot be explained by CTRW mechanism, which does not lead to correlations. A negative dip in Cδv(τ) can arise in several contexts: (i) OD, (ii) fLm, as well as (iii) localization errors in noisy images or (iv) an extreme spatial confinement. To resolve the remaining ambiguity in the origin of the negative dips in the average velocity autocorrelation, we analyzed the behavior of Cδv(τ) as a function of the ratio (τ/δ) of the two temporal parameters: the time lag (τ) and the time interval (δ) over which the velocity was calculated. Remarkably, when plotted against the rescaled time lag, τ/δ, all of the Cδv(τ) curves collapsed onto a single master curve, indicating that the motion of VH and DHJH segments possesses the property of self-similarity, or similar patterns at different temporal scales (Figure 4D: right panel; Figure S2). Such a collapse is not expected to occur for negative correlations in the average velocity caused by OD, localization errors or by an extreme confinement, but rather is a signature of a fLm, or viscoelastic, mechanism (Weber et al., 2010; Weber et al., 2012). The observed negative values of Cδv(τ) thus indicate a negative correlation (a reversal) in velocity of the Igh locus segment caused by the elastic component (“push back”) of the viscoelastic response within the cellular environment. As VH and DHJH regions of the chromatin fiber collide with a dense network of nucleic acids and proteins, as well as stretch and compress the neighboring segments of the fiber, motion in one direction is likely to be followed by motion in the opposite direction, i.e. the network pushes back (Figure 4A). We conclude that the viscoelastic environment within topological domains is in large part responsible for the subdiffusive motion observed for VH and DHJH coding as well as regulatory elements such as enhancers and promoters in B-lineage cells.
3D-Trajectories Adopted by the Immunoglobulin DHJH Segments in B Cell Progenitors
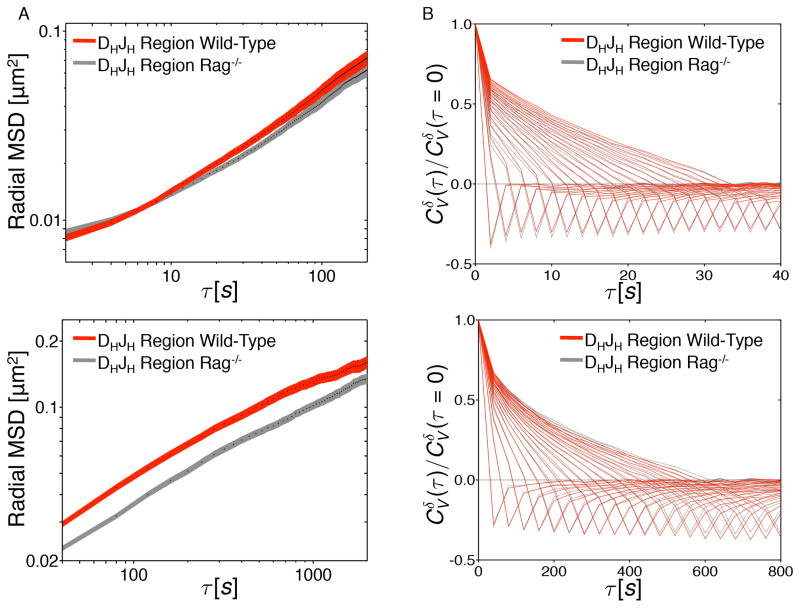
Recent studies have identified an insulator, named the CBE element, acting to suppress VHDHJH gene rearrangement involving the most proximal VH region cluster prior to the formation of a DHJH joint (Guo et al., 2011b; Degner et al., 2011). To determine whether the CBE affects DHJH motion, DHJH-TetO mice were crossed with RAG-deficient mice. Pro-B cells, derived from the bone marrow of DHJH-TetO RAG-deficient mice, were cultured and transduced with virus expressing TetR-EGFP. DHJH motion was monitored as described above (Figure 5A). As expected, the subdiffusive scaling exponents (α) for DHJH motion in RAG-deficient TetO and wild-type TetO proB cells were equivalent (α ≈ 0.5) (Table S1). Consistent with these observations, the velocity autocorrelation functions for the DHJH region were nearly identical for wild-type and RAG-deficient TetO pro-B cells (Figure 5B). The apparent diffusion coefficient associated with DHJH motion increased only slightly (2.4x10−3 versus 1.8x10−3 μm2/s0.5) in wild-type TetO pro-B cells as compared to RAG-deficient TetO pro-B cells (Figure 5A; Table S1). Taken together, these observations indicate that the CBE insulator does not substantially modulate the diffusion coefficients and subdiffusive scaling exponents associated with DHJH elements.
Figure 5. 3D-Trajectories Adopted by the Immunoglobulin Heavy Chain Locus Motion in Wild-Type and Rag-deficient pro-B Cells.
(A) Ensemble- and time-averaged radial MSD plotted as a function of time lag (τ) for the DHJH regions in live B220+ B-lineage cells derived from wild type and Rag−/− mice. Cells were imaged once every 2 seconds for 400 seconds (top panel) or once every 40 seconds for 4000 seconds (bottom panel). Radial MSD is shown for τ up to one half of total imaging time. Shaded areas represent the standard error of the mean.
(B) Velocity autocorrelation analysis of DHJH regions in wild type and Rag−/− cells. Velocity was calculated over time intervals (δ) ranging from 2 to 40 seconds in 2 seconds steps (top panel) or 40–800 seconds in 40 seconds steps (bottom panel).
Simulating VH and DHJH Encounters
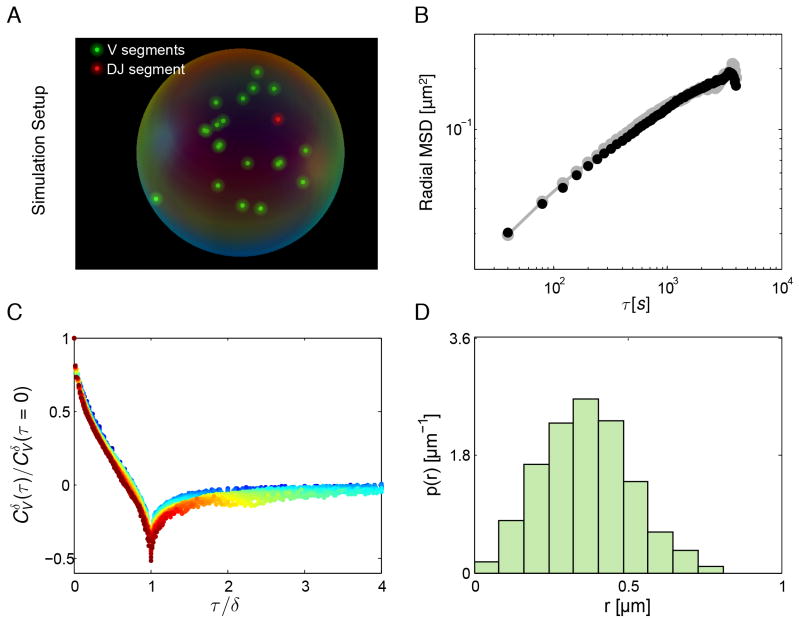
To investigate further the underlying mechanisms of motions associated with coding and regulatory DNA elements, VH and DHJH dynamics were modeled as an overdamped fractional Langevin motion in a confined sphere of radius R (Jeon and Metzler, 2010). The sphere mimicked the confinement imposed by a chromosomal domain, as the majority of genomic interactions have been demonstrated to occur within domains (Lieberman-Aiden et al., 2009; Dixon et al., 2012; Lin et al., 2012). The initial positions of the VH and DHJH segments were chosen by randomly drawing from the steady-state distribution (Figures 6D). The correlated fractional Gaussian noise was generated using the circulant embedding method as described previously (Dietrich and Newsam, 1997). The increment for each of the x-, y- and z-components of motion was calculated and applied at each time step. Reflective boundary condition was applied whenever the diffusing segments were found to be outside of the confinement volume. Parameters in the simulations were chosen such that the anomalous diffusion coefficient D=2.4 x 10−3 μm2/s0.5 and the scaling exponent α=0.5 of the simulated motion match those extracted from the radial MSD of our experimental measurements. Two values for the confinement radius were used. The value R=0.5 μm was estimated based on previous 3D-FISH measurements (Jhunjhunwala et al., 2008) of spatial distances between the DHJH and VH elements, by matching the steady-state distribution of distances between the DHJH and VH gene segments in simulations (Figure 6D) with that measured by 3D-FISH in B-lineage cells. The value R=1μm was estimated based on the best match between the radial MSD from the simulations and those obtained experimentally in the present study (Figure 6B). Once VH and DHJH segments were within interaction distance r0, the corresponding first-passage time was recorded. The interaction distance r0 was chosen to be 30nm based on the dimensions of the chromatin fiber. Simulations were repeated 1000 times (Figure 6A; Movies S3 and S4).
Figure 6. Modeling Anomalous Diffusion of VH and DHJH Elements as Fractional Langevin Motion.
(A) VH and DHJH segments confined in a sphere of radius R and subject to fractional Langevin motion using experimentally obtained values of anomalous diffusion coefficient (D) and subdiffusive exponent (α) as physical parameters.
(B) Radial MSD obtained from simulated (D=0.0024 μm2/s0.5 and R=1 μm) and experimental measurements. Black symbols are calculated from simulated trajectories. Gray symbols are DHJH MSD calculated from measurements in wild-type pro-B cells.
(C) Velocity autocorrelation functions generated from simulations with velocity computed for different values of the discretization interval (δ) exhibit a collapse on a master curve upon rescaling of the time lag τ by δ, as do their experimentally determined counterparts (Fig. 4D).
(D) The steady-state distribution of distances between a DHJH and a VH segment generated from the simulation (D=0.0024 μm2/s0.5 and R=0.5 μm) agrees well with the experimental distribution of spatial distances previously measured in pro-B cells by 3D-FISH (Jhunjhunwala et al. 2008).
To validate the simulations beyond ensuring a good match between the experimental and simulated MSD and between the experimental and simulated distributions of distances, we calculated the velocity autocorrelation functions (with R=1μm) derived from the simulated trajectories. When calculated for different values of the discretization interval (δ) and plotted against the rescaled time lag (τ/δ), the velocity autocorrelation curves collapsed onto a master curve (Figure 6C), as did their experimentally determined counterparts (Figure 4D: right panel). Collectively, these results indicate that the simulations performed with the experimentally determined physical parameters of VH and DHJH elements reproduce, at the quantitative level, the key aspects of the anomalous diffusion observed in developing B cells.
First-Passage Times for VH and DHJH Encounters
In the murine bone marrow, pro-B cells proliferate with a turnover rate of approximately 1–2 days. Hence, VH-DHJH encounters need to occur within this time, which raises the question: how long does it take for DHJH elements to reach a VH region? This question can be addressed via fractional Langevin dynamics modeling using experimentally determined physical parameters of VH and DHJH elements.
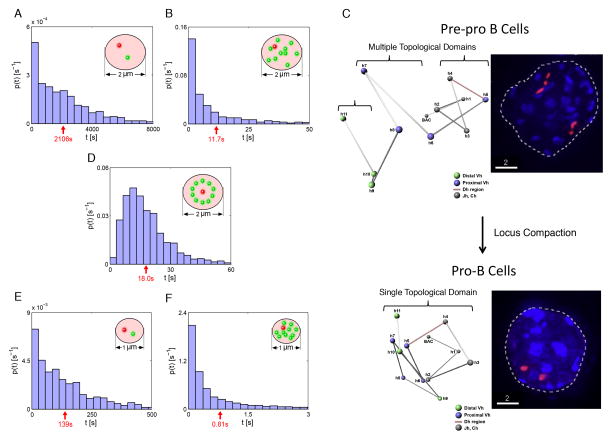
We initiated the simulation using a single pair of DHJH and VH elements and a confinement radius of 1 μm. We found that it takes approximately 30 minutes on average (Figure 7A) for a DHJH and a VH element to find each other. The mean first-passage time decreased more than 100-fold, to 12 s, when a single copy of DHJH and 100 copies of VH elements were present (Figure 7B).
Figure 7. First-Passage Time Distributions for VH and DHJH Segments Undergoing fractional Langevin Motion.
(A) Simulating the behavior of a single pair of VH and DHJH segments in a spatial confinement of radius 1.0 μm and with initial positions randomly chosen from the distribution shown in Figure 6D generated after the system is given enough time to reach the steady state. Motion of VH and DHJH segments as parts of the polymer chain and constrained by anchors is modeled as fractional Langevin motion in a confined sphere of radius R. The distribution of first passage times, the times for the DHJH segment to come within 30 nm of the VH segment, is shown. Red arrow indicates mean first passage time (MFPT).
(B) First passage time distributions for simulated motion of 100 VH and one DHJH segment in a confinement of radius 1.0 μm.
(C) Left panels show 3D-FISH studies revealing the organization and locus contraction of the Igh locus in pre-pro-B and pro-B cells. In pre-pro-B cells, VH and DHJH segments are physically separated into distinct topological domains. Upon differentiating into committed pro-B cells the proximal and distal VH regions merge. Right panel indicates the entire Igh locus labeled by overlapping fluorescently labeled BAC probes. Note distinct topological domains in pre-pro-B and pro-B cells (adapted from Jhunjhunwala et al., 2008).
(D) Simulation as performed in (B) but with VH segments distributed on a sphere with the DHJH segment in the center.
(E, F) Simulations as performed in (A) and (B) but with a confinement of radius 0.5 μm.
Prior to V(D)J recombination the VH and DHJH regions are constrained to distinct topological domains (Jhunjhunwala et al., 2008). In pro-B cells, at the onset of recombination, locus compaction results in the merging of these domains (Figure 7C). Such events bring into question how the confinement of DNA within topological domains affects first-passage times. To determine to what degree the encounter time is affected by confinement, we repeated the simulations but now using a confinement radius of 0.5 μm (Figure 7E and 7F), i.e. half the size of that used in Figures 7A and 7B. Notably, decreasing the radius of confinement two-fold decreased the mean first-passage time for VH and DHJH elements by 16-fold.
Simple dimensional analysis allows us to determine the general scaling of the encounter time with the confinement radius and the diffusion coefficient. The dimensions of MSD are [MSD]=[Length]2=[D][Time]α (we use the standard notations […] for “the dimensions of …”), and hence [Time] = [Length]2/α [D]−1/α. Because the relevant parameter with the dimensions of length in this first-passage time problem is the radius of confinement (R) (Condamin et al., 2007), we find that the interaction frequency (equal to the inverse of the mean first-passage time) scales with the confinement radius as R−2/α and with the diffusion coefficient as D1/α. Applying this scaling with the value of α extracted from the radial MSD plot (Figure 4C), we find that, upon decreasing the radius of confinement two fold, the interaction frequency is predicted to increase 24 = 16-fold. In contrast, increasing the value of the diffusion coefficient two fold leads to a merely 22 = 4-fold increase in the interaction frequency. Both of these predictions are consistent with the observations from our simulations. We conclude that the size of confinement largely determines the frequency of interactions between coding elements (such as VH and DHJH regions) and regulatory elements (enhancers and promoters).
Recent observations have suggested that the VH regions lie in an orbit around a cavity containing the DHJH elements (Lucas et al., 2011). We modified the simulations such that VH regions were distributed on the surface of an imaginary sphere with the DHJH elements positioned in the center (Figure 7D). The mean first-passage time for VH and DHJH elements was only modestly affected as compared to the initial random configuration, although the different initial conditions changed the distribution of first-passage times (Figures 7B and 7D). Taken together, this analysis indicates that confinement size is a dominant factor for first-passage times of genomic elements, and that VH and DHJH elements have a high probability to reach each other within minutes in developing B cells.
DISCUSSION
Although our insights into the folding patterns of the genome is still rudimentary, recent studies have suggested a highly ordered Igh locus configuration. It has been proposed that anchors such as CTCF act to spatially sequester the VH regions, positioning them “in orbit” around a cavity that contains a DHJH element (Figure 4A) (Lucas et al., 2011). Such a rosette structure would help ensure that all of the various VH segments and DHJH segments have the opportunity to find each other in a V(D)J recombination center. Here we found that the DHJH and VH elements undergo fractional Langevin motion indicative of a viscoelastic environment likely caused by a surrounding network of VH and DHJH DNA segments, neighboring DNA segments, tethers such as CTCF and other factors associated with the chromatin fiber. The value of the scaling exponent α ≈ 0.5 in B cells approached those observed for chromosomes in other species, including multiple bacterial species, yeast and human cells (Table S2) (Cabal et al., 2006; Bronstein et al., 2009; Weber et al., 2010). We suggest that fractional Langevin motion prevents rapid displacement of the DHJH elements away from the VH region cluster. Thus, fractional Langevin motion may provide a physiological advantage: DHJH regions would be bouncing back and forth against genomic elements across the Igh locus fiber until they pair with nearby VH regions in transit (Figure 4A).
The process of V(D)J gene rearrangement is tightly regulated. Recent studies have demonstrated that ordered rearrangement involving proximal VH regions is enforced by an insulator element (CBE), which separates the VH regions from the DHJH regions (Guo et al., 2011b). Our measurements revealed only modest differences in DHJH motion in B cell progenitors that are arrested prior to the onset of DHJH rearrangements versus pro-B cells that carry DHJH joints. How then is proximal VH-DHJH recombination regulated? It is conceivable that the region containing the CBE insulator element assumes a somewhat stiff conformation or a rodlike structure. Consequently the proximal VH and DHJH regions and the intervening rod-like structure may display fractional Langevin motion like any other genomic region, but since the CBE region may not be able to fold back upon itself, VH and DHJH interactions would be rare. Alternatively, as suggested previously, the CBE may act to form a loop with other CTCF binding sites located downstream of the Igh locus to sequester the DHJH regions away from the VH region cluster or by suppressing anti-sense transcription across the proximal VH domains preventing proximal VH-DH rearrangement prior to the formation of DHJH joints (Guo et al., 2011b; Degner et al., 2011). Future studies involving the tracking of DHJH motion in relation to distal VH and proximal VH motion should distinguish between these possibilities.
The simulation results indicate that spatial confinement is the dominant factor that regulates the probability of interaction between VH and DHJH elements. Simple dimensional analysis suggests that the encounter frequency scales with the confinement radius as R−2/α and with the diffusion coefficient as D1/α. For the value of α (0.5) determined from our measurements, this means that decreasing the radius of confinement two fold increases the interaction frequency 16-fold, while, in contrast, increasing the value of the diffusion coefficient two fold increases interaction frequency merely 4-fold. We conclude that the degree of confinement largely determines the frequency of interactions between coding elements (such as VH and DHJH regions) and regulatory elements (enhancers and promoters). These findings have implications relating to chromosomal organization, gene regulation, V(D)J recombination, class switch recombination, chromosomal translocations and chromosomal deletions.
Chromosome-conformation capture studies have demonstrated that the large majority of genomic interactions occur within chromatin domains (1–3 Mbp) and with much lower frequencies between domains (Lieberman-Aiden et al., 2009; Dixon et al., 2012; Lin et al., 2012). Thus, the organization of the genome into topological domains constrains the 3D-trajectories that are adopted by chromatin fiber. We suggest that the spatially confined geometry is also maintained by the viscoelastic properties of mammalian nuclei, suppressing the rapid diffusion of coding and regulatory DNA elements between chromatin domains. However, topological domains are not static during developmental progression. For example, during the transition from the pre-pro-B to the pro-B cell stage the genome undergoes large-scale changes in domain organization (Lin et al., 2012). Prominent among these are the antigen receptor loci. Specifically, the distal VH and proximal VH regions merge during the transition from the multipotent progenitor to the committed pro-B cell stage (Kosak et al., 2002; Fuxa et al., 2004; Jhunjhunwala et al., 2008). The Igκ locus also undergoes locus contraction in B cell progenitors (unpublished observations). Similarly, the TCR α and β loci contract in cells prone to undergo TCR rearrangement (Roldan et al., 2005). How does locus contraction affect DNA recombination? The simulation results indicate that topological domains merge or segregate during developmental progression to modulate the encounter times of genomic elements localized within topological domains. The merging of topological domains during developmental progression is not restricted to antigen receptor loci. An important example involves the EBF1 locus, which, during the transition from the pre-pro-B to the pro-B cell stage, dissociates from the nuclear lamina to merge with transcriptionally active domains and may allow enhancers and promoters located across this vast locus to interact with high frequencies, ultimately leading to the induction of a B-lineage specific program of gene expression (Lin et al., 2012). In sum, we suggest that the merging or segregation of topological chromatin domains affects the encounter times of VH and DHJH or promoter and enhancer elements to permit rapid changes in gene expression during developmental progression.
Somatic recombination during B cell development is not restricted to V(D)J recombination. In the peripheral lymphoid organs, class switch recombination (CSR) changes the expression of immunoglobulins from one isotype to another. In activated B cells, switch regions join after double stranded breaks (DSBs) are generated (Zarrin et al., 2007). It has been suggested that DSBs may rapidly diffuse to promote efficient synapse formation and class switch recombination and that topological confinement may play a key role in this process (Alt et al., 2013; Gostissa et al., 2014). Our simulation results provide support for such a model. In conjunction with previous spatial distance measurements across the switch regions, we predict that DSBs generated across the switch regions reach each other within minutes and that topological confinement is the dominant factor in establishing rapid first-passage times (Jhunjhunwala et al., 2008).
Our findings also have implications for chromosomal deletions that frequently are associated with cancer. Intrachromosomal interstitial deletions associated with illegitimate rearrangements involving non-antigen receptor genes and cryptic recombination signal sequences are frequently linked with lymphoid cancers (Onozawa and Aplan, 2012). Recent studies revealed that proximity of two DSBs is the critical factor in determining the preferences for DSBs translocating in cis (Zhang et al., 2012; Alt et al., 2013). These observations are consistent with the simulation results presented here indicating that spatial confinement is a dominant component in controlling the encounter times of long-range genomic interactions. Hence, we suggest that the predisposition of two DSBs to join intrachromosomally might lead to the development of T-ALL and other types of lymphoid malignancies if the chromatin topology associated with such breaks would permit rapid first-passage times (Alt et al., 2013; Bunting and Nussenzweig, 2013). Finally, we note that rapid first-passage times of paired DSBs may similarly underpin the physical mechanism by which chromosomal deletions are generated in a wide variety of malignancies (Solimini et al., 2012; Beroukhim et al., 2010).
EXPERIMENTAL PROCEDURES
TetR-EGFP Constructs and Retrovirus Production
A TetR-EGFP construct was created by fusing a TetR (tTA2S) with enhanced GFP (EGFP). tTA2S has been previously described (Urlinger et al., 2000). A nuclear localization signal (PKKKRKV) was added to the 5′ end by PCR. EGFP vectors are available from Clontech. An XbaI site was added to the 3′ end of tTA2S and the 5′ end of EGFP for fusion. Finally, either a wild-type (gccaccATG) or mutant (gcctccATG) Kozak sequence was added along with restriction sites for cloning into the MinV, PCS Ret, or LMP vectors. LMP is available from Thermo Scientific and the TetR-EGFP was inserted in place of the vector’s EGFP cassette. MinV and PCS Ret have been previously described (Hawley et al., 1996; Sayegh et al., 2003). Primers used are listed in Table S3. Complete vectors were purified by CsCl gradient and transfected into 293T cells along with retroviral packaging vectors by calcium phosphate transfection. Media was replaced the following morning and viral supernatant was harvested one day later and stored at −80°C until used.
Imaging
Imaging was performed under normal growth conditions using phenol-red free media. Cells were plated on Fluorodish poly-D-lysine coated plates (World Precision Instruments) and imaged using a 100X 1.4NA oil immersion objective on a Zeiss CSU spinning disk confocal microscope with a Yokogawa spinning disk scan head and an EM-CCD camera. 20–30 Z sections were obtained with a spacing of 0.5 μm and acquired at a rate of one stack every two seconds for 200 total time points or one stack every 40 seconds for 100 total time points. Laser intensity was 6% and exposure time was 30ms for 2 seconds intervals and 45ms for 40 seconds intervals. For fixed cell measurements, cells were fixed for 10 minutes in 4% paraformaldehyde in PBS and quenched 5 minutes in 0.1M Tris-HCl pH 7.4 and stained using a 1:2000 dilution of Alexa 488 conjugated rabbit monoclonal anti-GFP from Life Technologies (G10362). Axial elongation was corrected prior to calculating probe-to-probe distances (Figure S3).
Supplementary Material
HIGHLIGHTS.
3D-trajectories adopted by the immunoglobulin heavy chain locus
Fractional Langevin motion and VH-DHJH genomic interactions
First-passage times for coding and regulatory DNA elements
Spatial confinement largely controls encounter times for genomic interactions
Acknowledgments
We thank Peter Geiduschek, Alex Bortnick, and Roy Riblet for editing the manuscript. We thank John Sedat, Richard Flavell, Cornelia Zorca and Lena Koslover for stimulating discussions. We thank Claudia Bossen, Joe Pogliano and Ralf Metzler for advice. We thank James Fitzpatrick and Jamie Kasuboski (Waitt Advanced Biophotonics Center Core Facility) for help with imaging and image analysis. J.S.L. was supported by a training grant from the National Institutes of Health (Cellular and Molecular Genetics). The studies were supported by grants from the National Science Foundation Faculty Early Career Development Award (MCB-0845099) and NSF Center for Theoretical Biological Physics Grant PHY-0822285 to O.K.D. and the National Institutes of Health (AI00880 and AI082850) to C.M.
Footnotes
AUTHORS CONTRIBUTIONS
J.S.L. initiated the study, designed and performed the experiments, analyzed the data and contributed to the writing of the manuscript. Y.Z. analyzed the data, performed the simulations and contributed to the writing of the manuscript. O.K.D. and C.M. supervised the study and contributed to the writing of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. Embo J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibrobast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, et al. The landscape of somatic copy-number alterations across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Die Blastomerenkerne von Ascaris meglocephala und die Theorie der Chromosomenindividualitaet. Archiv für Zellforschung. 1909;3:181–268. [Google Scholar]
- Bronstein I, Israel Y, Kepten E, Mai S, Shav-Tal Y, Barkai E, Garini Y. Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys Rev Lett. 2009;103:018102. doi: 10.1103/PhysRevLett.103.018102. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nature Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- Condamin S, Bénichou O, Tejedor V, Voituriez R, Klafter J. First-passage times in complex scale-invariant media. Nature. 2007;450:77–80. doi: 10.1038/nature06201. [DOI] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich CR, Newsam GN. Fast and exact simulation of stationary Gaussian processes through circulant embedding of the covariance matrix. SIAM J Sci Comput. 1997;18:1088–1107. [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Schwer B, Chang A, Dong J, Meyers RM, Marecki GT, Choi VW, Chiarle R, Zarrin AA, Alt FW. Igh class switching exploits a general property of two DNA breaks to be joined in cis over long genomic distance. Proc Natl Acad Sci USA. 2014;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy chain locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Goldman SJ, Leonard JP, Hawley TS. Retroviral vectors for production of interleukin-12 in the bone marrow to induce a graft-versus-leukemia effect. Ann NY Acad Sci. 1996;795:341–345. doi: 10.1111/j.1749-6632.1996.tb52687.x. [DOI] [PubMed] [Google Scholar]
- Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- Jeon JH, Metzler R. Fractional Brownian motion and motion governed by the fractional Langevin equation in confined geometries. Phys Rev E. 2010;81:021103. doi: 10.1103/PhysRevE.81.021103. [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby TM, Schrader CE, Stavnezer J, Selsing E. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J Exp Med. 2001;193:159–168. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JS, Bossen C, Murre C. Transcription and recombination factories: common features? Curr. Opin Cell Biol. 2011;23:318–324. doi: 10.1016/j.ceb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz E. Fractional Langevin equation. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;64:051106. doi: 10.1103/PhysRevE.64.051106. [DOI] [PubMed] [Google Scholar]
- Manz J, Denis K, Witte O, Brinster R, Storb U. Feedback inhibition of immunoglobulin gene rearrangement by membrane mu, but not by secreted mu heavy chains. J Exp Med. 1988;168:1363–1381. doi: 10.1084/jem.168.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q, Huis, In‘t Veld PJ, Guo C, Yoon HS, Denizot Y, Holwerda SJ, de Laat W, Cogné M, Shi Y, Alt FW, Busslinger M. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 2013;39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montroll EW, Weiss GH. Random walks on lattices. II. J Math Phys. 1965;6:167–181. [Google Scholar]
- Münkel C, Langowski J. Chromosome structure described by a polymer model. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1998;57:5888–5896. [Google Scholar]
- Nussenzweig MC, Shaw AC, Sinn E, Campos-Torres J, Leder P. Allelic exclusion in transgenic mice carrying mutant human IgM genes. J Exp Med. 1988;167:1969–1974. doi: 10.1084/jem.167.6.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozawa M, Aplan PD. Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Genes, Chromosomes and Cancer. 2012;51:525–535. doi: 10.1002/gcc.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. Über Zellteilung. Morphologisches Jahrbuch. 1885;10:214–330. [Google Scholar]
- Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im TH, Ludewig M, Nordsiek G, Severitt S, Thies S, Mauhar A, Blocker H, Muller W, Riblet R. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J Immunol. 2007;179:2419–2427. doi: 10.4049/jimmunol.179.4.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok J. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys J. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous diffusion due to binding: a Monte Carlo study. Biophys J. 1996;70:1250–1262. doi: 10.1016/S0006-3495(96)79682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;6:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Sedat J, Manuelidis A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harbor Symp Quant Biol. 1977;42:331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Shimizu A, Takahashi N, Yaoita Y, Honjo T. Organization of the constant-region gene family of the mouse immunoglobulin heavy chain. Cell. 1982;28:499–506. doi: 10.1016/0092-8674(82)90204-5. [DOI] [PubMed] [Google Scholar]
- Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, Brruows AE, Anselmo AN, Bredemeyer AL, Li MZ, Berouhkhim R, Meyerson M, Elledge SJ. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez J, Belmot AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Current Biology. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Thompson MA, Moerner WE, Spakowitz AJ, Theriot JA. Analytical tools to distinguish the effects of localization error, confinement, and medium elasticity on the velocity autocorrelation function. Biophys J. 2012;102:2443–2450. doi: 10.1016/j.bpj.2012.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Del Vecchit C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.