Abstract
The complexity of an organism’s proteome is in part due to the diversity of post-translational modifications present that can direct a protein location and function. To address the growing interest in characterizing these modifications, mass spectrometric-based proteomics has emerged as one of the most essential experimental platforms for their discovery. In searching for post-translational modifications within a target set of proteins to global surveys of particularly modified proteins within a given proteome, various experimental mass spectrometry and allied techniques have been developed. Out of twenty naturally encoded amino acids, lysine is essentially the most highly post-translationally modified residue. This chapter provides a succinct overview of such methods for the characterization of protein lysine modifications as broadly classified such as methylation and ubiquitination.
Introduction
Proteomics initially emerged with the aim of identifying and quantifying as many peptides as possible from a complex protein digest to grasp the functional significance of an organism’s genome. It became evident from the onset that such technologies could also be applied toward identifying and quantifying protein post-translational modifications (PTMs) in a similarly global manner. Growing interest in applying proteomics to characterize PTMs continues to be driven by increasing awareness of the diverse roles PTMs possess in normal and disease physiology, ranging from the addition of ubiquitin by E3 ubiquitin ligase that promotes substrate degradation to the addition of acetyl groups by histone acetyltransferases that serve as binding sites for transcriptional regulators and activate gene expression. This chapter details recent advancements in mass spectrometry (MS) and in other related disciplines that have enabled the high throughput analysis of various lysine PTMs.
Mass spectrometry overview
Mass spectrometric techniques in brief
Mass spectrometry is currently the most versatile and vital experimental platform for proteomics. Before discussing how mass spectrometry may be employed for studying lysine post-translational modifications, it is illuminating to discuss the general properties shared among mass spectrometers. First, all mass spectrometers measure the mass-dependent behavior of gas-phase ions in an electromagnetic field. To do this, all mass spectrometers store and isolate ions within a particular mass/charge ratio range (m/z) as they enter the instrument. During this process, the mass spectrum (MS) is collected, and the width of the scan range determines the complexity of the ion population and precursor ion peptides being collected. By isolating ion(s) within a set m/z value, either pre-determined by the experimenter or systematically determined by the instrument based on the ion abundances during the particular scan time, the mass spectrometer will fragment the ions and collect a second or tandem mass spectrum (MS/MS or MS2) of the fragments. As discussed later for peptides, these fragments yield invaluable information concerning the primary amino acid sequence and the modified residues of the peptides. In principle, the mass spectrometer can repeatedly isolate particular fragment ion(s), fragment again, and scan those fragments in n number of cycles for MSn acquisition.
All mass spectrometers yield information on the mass-to-charge (m/z) and relative abundance of all the ions within a give scan. Yet the mechanism of how the instrument measures m/z varies fundamentally and dramatically among different mass spectrometers. For instance, a time of flight (TOF) mass analyzer measures the time required for ions to reach the mass detector, where time is proportional to the square root of the ions’ m/z [1]. In contrast, an Orbitrap mass detector measures the frequency of axial oscillations of orbiting ions around a curved electrode, where frequency is inversely proportional to the square root of the ions’ m/z [2]. Finally, the mass analyzers accompanying a linear quadrupole ion trap detect ions axially ejected from the quadrupole with increasing radiofrequency voltage, where the resonance voltage at which ions are scanned out of the ion trap is proportional to the ions’ m/z[3].
The second characteristic shared by all mass spectrometers utilized in proteomics is actually not intrinsic to the mass spectrometer itself, but rather its interface with some form of chromatography. As most biochemical experiments occur in solution, liquid chromatography is typically used to resolve peptides from a complex sample before being introduced into the mass spectrometer, commonly with electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI). Liquid chromatography provides separation of peptide ions in a dimension orthogonal to the m/z dimension, which can already be achieved with the ion selection filters in the mass spectrometer. Common modes of separation include hydrophobicity as in reversed phase (RP) liquid chromatography (most commonly C18 based), hydrophilicity as in hydrophilic interaction liquid chromatography (HILIC), and pKa as in weak cation exchange (WCX) liquid chromatography. The careful application of chromatography vastly improves the dynamic range of peptides that the mass spectrometer can analyze per scan and can provide additional important evidence to assist in the identification of post-translational modifications.
Localization of PTMs
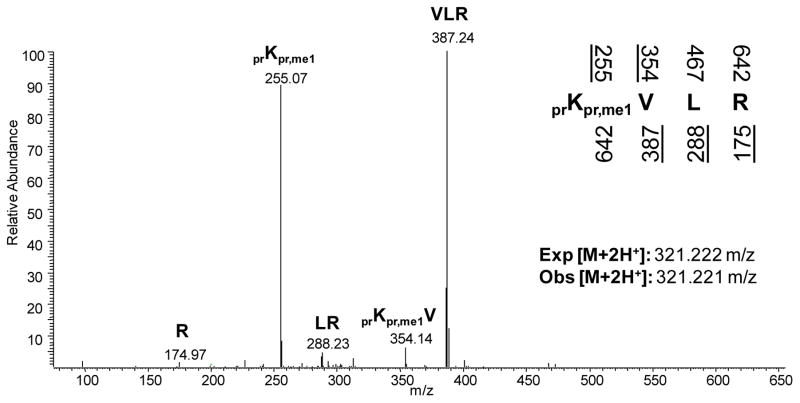
From the MS/MS, one obtains information concerning the peptide sequence that may be used to identify and localize particular post-translational modifications to specific residues. Analogous to dideoxy sequencing of oligonucleotides where one sequences from both the 5′ and 3′ direction in separate reactions using separate primers, MS sequencing operates by generating overlapping smaller peptides sharing a common N- or C-terminus. Common fragmentation methods include collisional induced dissociation (CID), electron transfer dissociation (ETD), and higher-energy C trap dissociation (HCD), each uniquely suitable for different goals of sequencing peptides or intact proteins. For example, MS sequencing of the peptide KVLR yields the fragment ions K, KV, KVL, R, LR, and VLR with the former three peptides sharing a common N-terminus and the latter three peptides sharing a common C-terminus (Figure 1). The mass difference between peptides sharing either a common N- or C-terminus corresponds to the next adjacent amino acid in the peptide sequence from either the N- or C-terminus. When that mass difference does not match any of the masses for the 20 canonical amino acids, the possibility of the residue being post-translationally modified should be considered.
Figure 1.
Tandem mass spectrum of the 20–23 histone H4 peptide (KVLR) monomethylated at K20. The underlined predicted fragments above and below the peptide sequence represent ions observed in the MS/MS. In addition to noting the fragment ions, the accurate mass of the doubly charged parent ion mass should also be considered with respect to the expected mass. Abbreviations: pr = propionyl group from the propionic anhydride derivatization, me1 = monomethylation.
High mass accuracy determination of the precursor and fragment peptide ions often facilitates the assignment of a specific PTM to a specific residue (Figure 2). For instance, the nearly isobaric trimethyl (+42.046 Da) and acetyl (+42.010 Da) modifications on most tryptic peptides can be generally resolved with a mass tolerance of < 50 ppm root mean square error. However, even when the mass difference matches perfectly with the expected mass of a modified lysine compared to an unmodified lysine, more evidence is required to prove the existence and localization of the suspected modification. Perhaps the most stringent validation test is the in vivo specific metabolic labeling of the modification itself. Analogous to the classic and arguably most rigorous experiment of 32P-radiolabeling to confirm phosphorylation, one can culture cells with a heavy isotope of the relevant metabolite that provides the source for the specific modification, for instance methionine which is metabolized into S-adenosyl methionine, the sole precursor for protein methylationby lysine and arginine methyltransferases in eukaryotic cells [4]. In general, most forms of liquid chromatography do not distinguish between 12C and 13C or 14N and 15N and thus the putative modified peptide should not only incorporate the heavy isotope, but also co-elute with the equivalently modified peptide with the light isotopes. Additionally, when examining the fragment ions in the MS/MS, the modified residue should similarly be shifted with the heavy isotope mass difference [4, 5].
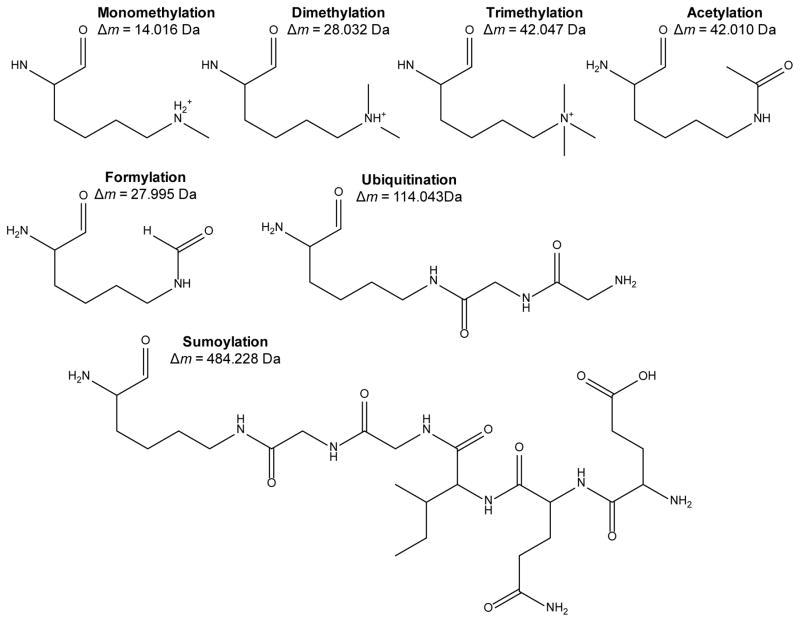
Figure 2.
Common lysine post-translational modifications, with the mass shift from the lysine residue mass provided below. Note that both the ubiquitin and SUMO moieties depicted are the tryptic fragments of the respective modification remaining on the modified lysine.
Proteomics for analyzing protein PTMs
Targeted investigations of protein lysine post-translational modifications
Among the most well documented proteins to be highly post-translationally modified are the histone proteins H1, H2A, H2B, H3, and H4, which are highly conserved among all eukaryotic cells. Histones are principally involved in the coiling of DNA into the nucleosome and intimately regulate gene expression. Much of the transcriptional and epigenetic regulation mediated by histones depends on both the identity and localization of specific post-translational modifications occurring generally on the N-terminal tails, notably lysine methylation and acetylation [6]. The chemical diversity and combinatorial occurrence of histone PTMs renders traditional assays such as Western blotting and enzyme linked immunosorbent assays with PTM-specific antibodies unreliable to interpret and quantify, yet proteomics offers the promise of high throughput and unambiguous identification of known and novel histone PTMs.
Yet MS analysis of histone modifications is not as straightforward as one may initially expect. The standard workflow for a proteomics experiment is to first reduce disulfide bonds, alkylate the free cysteines, digest with trypsin, desalt and analyze by mass spectrometry (also known as Bottom Up mass spectrometry). Reduction and alkylation of disulfide bonds is usually not necessary for histone PTM analysis as all the cysteines occur in the C-terminal portion of the protein, well removed from the majority of modifications found on the N-terminal tails. A more critical problem though is the preponderance of lysine and arginine residues in the N-terminal tail. For instance, there are eight lysine residues and seven arginine residues in the first 50 amino acids of histone H3 alone. Digestion with trypsin would result in peptides less than 4 amino acids in length, which would retain poorly under most forms of liquid chromatography. Furthermore, the adjacency of the lysine and arginine residues means that trypsin digestion would not reproducibly yield the same fragments. Finally, while trypsin can in most situations digest unmodified lysine residues, the protease is unable to digest acetylated lysines due to both steric effects and the loss of positive charge on the ε-amine group, and thereby not being stabilized by the aspartate residue of the trypsin catalytic center. Similar miscleavage events occur for mono-, di-, and trimethylated lysines in order of increasing likelihood of failed digestion. Overall, histone PTMs lead to miscleaved histone fragments depending on the modification status, a general problem for other lysine modifications.
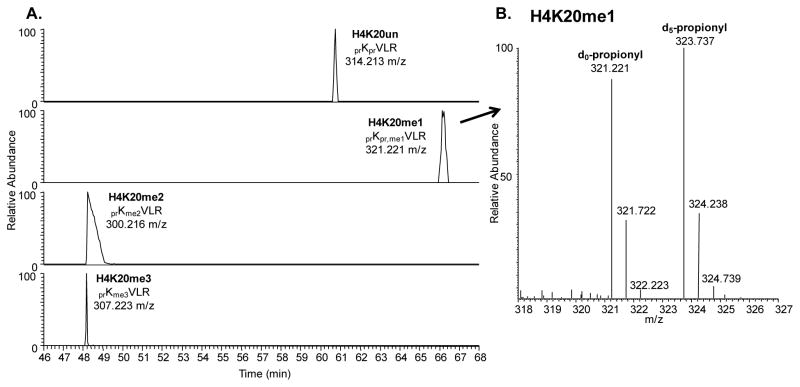
In response to this incompatibility, several derivatization methods have been developed with the general approach of blocking trypsin digestion at lysines, thereby allowing for digestion only at arginine residues [7, 8]. This chapter will focus on derivatization using proprionic anhydride, which transfers a propone moiety (+56.026 Da) to the ε-amine group of unmodified and monomethylated lysines as well as the N-terminus. After propionylation at the protein level, followed by trypsin digestion and an additional propionylation to modify the newly created N-terminus at the peptide level, the propionyl groups confer additional hydrophobicity to the histone peptides and enhance their retention in reversed phase chromatography. In addition, by allowing for trypsin digestion at only arginine residues, the same histone peptides can be reproducibly obtained across all modified lysine states. For instance, the unmodified, mono-, di-, and tri-methylated lysine 20 on histone H4 can be detected on the same 20–23 peptide (KVLR). The first advantage of this is that the ionization efficiencies of the various modified forms of the 20–23 peptide can be reasonably assumed to be comparable and thus provides additional confidence in relative quantification of the modified histone forms to each other. The second advantage is that the elution order of the various modified forms of the same propionylated histone peptide can be more readily predicted based on the nature of the modification, providing additional evidence for PTM assignment (Figure 3A). The third advantage is that one can perform the derivatization scheme as described above using an isotopically labeled propionic anhydride reagent, for instance deuterated d10-propionic anhydride, in the N-terminal capping of the histone peptides. This creates a consistent 5.029 Da offset between all light unlabeled histone peptides and the respective heavy labeled histone peptides, and allows for more rigorous quantification between histone PTM levels of different samples in a single run (Figure 3B).
Figure 3.
A. Extracted ion chromatograms of the various methylation states of the 20–23 histone H4 peptide (KVLR) under reversed phase liquid chromatography. With respect to the unmodified peptide, the monomethyated peptide should elute later due to the addition of a methyl group. Both the di- and trimethylated peptides should elute earlier than the unmodified peptide due to possessing only one propionyl group instead of two. Note the trimethylated peptide elutes earlier than the dimethylated peptide due to the charge stabilization of the ε-amine group, resulting in an overall more hydrophilic peptide. B. Mass spectrum of the monomethylated 20–23 H4 peptide, illustrating the mass shift between the light d0-propionyl derivatized peptide at 321.221 m/z and the heavy d5-propionyl derivatized peptide at 323.737 m/z. The difference in mass allows for simultaneous quantification between two different samples.
One may suspect that a simpler solution may be digestion with Arg-C which cleaves only after arginine residues and produce the identical cleavage patterns to the derivatization schemes detailed above. One key advantage of the use of Arg-C over derivatization approaches is the documented in vivo occurrence of propionyl moieties on both histone and non-histone proteins [9], and the use of Arg-C could allow for the potential quantification of these modifications. Aside from the greater cost and lower enzymatic efficiency of Arg-C, one important disadvantage of Arg-C is due to the higher charged states characteristic of the Arg-C generated histone peptides. Under the commonly used CID fragmentation method used for histone PTM sequencing, a more highly charged peptide leads to preferential non-random cleavage at certain residues throughout the peptide backboneand will be less informative toward PTM localization.
Another example of a targeted proteomic investigation was the identification of numerous PTMs on the heterochromatin protein 1 family members HP1α, β, and γ, including lysine methylation, acetylation, and formylation [10]. As the name suggests, the HP1 members are involved in heterochromatin maintenance. For instance, HP1α recognizes and binds to trimethylated lysine 9 on histone H3 (H3K9me3), and in doing so recruits SUV39H1, which is the methyltransferase that trimethylates H3K9, and via a feedback mechanism, initiates the propagation of heterochromatin. It is perhaps not surprising then that the HP1 proteins that are critical for the modification pattern of histones would themselves be highly modified.
In order to thoroughly interrogate the PTM landscape of a few target proteins, multiple proteases with different cleavage specificities are often required for maximum sequence coverage of the target protein(s). In the case for the HP1 proteins, trypsin, chymotrypsin, and Lys-C were used in separate reactions to achieve over 90% sequence coverage combined. Sequencing grade trypsin is the most often used protease for PTM mapping due to its relative affordability, enzymatic efficiency, and strict substrate specificity for lysines and arginines. As previously discussed, though, trypsin miscleavage often occurs for modified sterically hindered lysine residues. Other proteases targeting other residues may be appropriate depending on the target protein sequence. In addition to proteases, small chemicals with unique substrate specificities may be used, such as cyanogen bromide towards unoxidized methionine residues and N-chlorosuccinimide towards tryptophan, but these chemical cleavage methods often have lower yields and unwanted side products with respect to enzymatic approaches.
The opposite approach of interrogating the modification status of intact (also known as Top Down mass spectrometry) rather than digested proteins has also been applied with histones and high mobility group (HMG) member proteins (11, 12, 13). A key advantage of not digesting the protein is that one maintains the connectivity of discrete modified residues within the same molecule. For a protein that contains a single modified site, this gain in information is trivial. Yet for highly modified proteins such as histones or HMG proteins, understanding the frequency and abundance of when a modification at one site is linked to another modification at a different site on the same protein could inform predictions on the regulation and function of those modifications.
MS analysis of intact proteins is often achieved from direct infusion of the purified protein sample into the mass spectrometer, which significantly reduces instrument sensitivity. A compromise between analyzing small tryptic peptides or the intact protein is to digest the protein into relatively large >20 residue peptides that maintain several modified sites together but can be sufficiently resolved using liquid chromatography based on the position and type of PTMs (also known as Middle Down mass spectrometry). For histones H3 and H4, this can be achieved via digestion with GluC and AspN respectively to yield the 1–50 and the 1–23 peptide respectively (14). For HMGA1a, this can be achieved via limited trypsin proteolysis to yield the 30–54 peptide (11). The larger peptides/proteins generated also exist at higher charge states as tryptic peptides, and ETD fragmentation rather than CID fragmentation for MS/MS acquisition is typically used for PTM localization.
Global Large scale surveys of protein lysine post-translational modifications
The aforementioned examples centered on proteomic investigations targeted toward highly modified proteins that can be somewhat easily isolated and where the analytical complexity originated not from the number of unique proteins, but rather by the number of unique modified forms of a few proteins. With respect to the converse, namely using proteomics to globally interrogate proteins containing particular post-translational modifications, all investigations generally start by first enriching a complex protein or peptide sample for analytes containing the particular PTM. This stems from the low abundance or stoichiometry of the modified form of the protein versus the unmodified form. For instance, Zhao and co-workers used pan-lysine acetyl antibodies to identify 388 acetylation sites from 195 proteins in mammalian cells and mouse liver mitochondria [Mol Cell 2006]. Surprisingly they discovered that a large number of these acetylated proteins derived longevity regulators and proteins involved in metabolism, implying that lysine acetylation could play a role in non-nuclear events. Other recent surveys on global protein acetylation also utilized an anti-acetylated lysine antibody to enrich tryptic digest samples for acetylated peptides, and coupled these enrichments to higher-end mass spectrometry analysis [16, 17]. Such experiments were able to detect acetylation sites of low-abundance proteins such as tumor suppressor p53, and over 700 conserved acetylated sites across 3 different cell lines [16]. Interesting insights on what role acetylation may perform can be gained from applying bioinformatic analysis on the intracellular localization and binding partners of the modified proteins. For instance, acetylation is not only found on proteins involved in roles as diverse as DNA replication to membrane trafficking [16]. Furthermore, a more targeted approach investigation acetylation of metabolic enzymes found evidence supporting a causal relation between increased metabolic activity and increased acetylation levels of the enzymes [17]. Thus, even global large scale catalogues on modified proteins can yield meaningful functional insights into a few target proteins, although not at a level of detail and resolution with more targeted approaches.
Due to the availability of a PTM-specific antibody, enrichment at the peptide level is more sensible than enrichment at the protein level. Recalling that most mass spectrometric experiments analyze peptides rather than proteins, the sample complexity is far greater in the latter than in the former and while in principle the same number of acetylated peptides will be enriched with both approaches, the background of unmodified peptides is vastly greater in the latter than in the former and will complicate MS analysis. However as discussed below, there are situations when enrichment at the protein level is actually more sensible than enrichment at the peptide level.
Another analogous although unique PTM-specific enrichment has recently been developed for identifying methylated proteins, which involves performing in nucleo reactions with purified nuclei and an alkyne-containing SAM analogue [18]. The logic is that all proteins bound by endogenous methyltransferases will receive an alkyne rather than a methyl group, which can then be clicked to an azide containing epitope for pullout and enrichment. Thus, in lack of a PTM specific antibody, one could in principle add an epitope to the PTM specifically for enrichment. While PTM enrichment at the peptide level reduces sample complexity, most global surveys utilize multiple chromatographic separations to further reduce the peptide complexity. A common approach utilizes strong cation exchange (SCX) followed by RP liquid chromatography for acetylated peptide studies [17]. Other approaches rely on gel-free isoelectric focusing to generate “fractions” of peptides according to their pI values [16]. Resolving peptides in solution rather than a polyacrylamide matrix improves sample recovery, as one does not have to extract the peptides from the gel. Regardless of the specific technology used, the general principle is to apply orthogonal modes of separation to reduce sample complexity and hence increase the dynamic range of the MS analysis. These separations also have a benefit for global protein PTM characterization. Recently gel fractionation coupled to large-scale mass spectrometry based proteomics was used to interrogate human chromatin isolated through various biochemical strategies and detected over 1900 proteins from these preparations. Most interestingly, over 150 identified proteins that were lysine modified, with many of these proteins being potentially involved in transcriptional processing [19].
Once the samples are prepared for mass spectrometry, another challenge is to quantify the occurrences of modified residues. Unlike the more targeted investigations with histone acetylation, enrichment of non-derivatized modified peptides renders it difficult to normalize any detected miscleaved modified peptides with the respective shorter unmodified peptides. One approach to circumvent this issue is to apply stable isotope labeling with amino acids in cell culture (SILAC) and compare acetylation levels between samples. SILAC involves culturing one sample in standard unlabeled media and the other in media typically depleted of unlabeled lysine and arginine and supplemented with equimolar amounts of heavy isotopes of lysine and arginine [20]. The choice of both these amino acids allows for every tryptic SILAC peptide to incorporate at least one heavy amino acid. However, the challenge with developing the labeled culture is to incorporate the heavier isotopes into as high a percentage of the cellular proteins as possible. Once the SILAC tissue culture is sufficiently labeled, one mixes equal cell number of both the unlabeled and labeled samples and proceeds with MS sample preparation with the mixture with the reasonable assumption that both the unlabeled and labeled peptides will behave equivalently during the enrichment and chromatographic separation steps. Due to the isotopic mass difference, generally greater than or equal to 4 Da, one should be able to distinguish between the unlabeled and labeled peptide signal in the mass spectrum and perform relative quantification between both samples. Examples of such experiments include the already described heavy methyl SILAC experiments, where all methionine-containing and methylated peptides will contain a 4.021 Da mass shiftfor every methionine/methylgroup. [5].
Similar global surveys have also been performed searching for lysine formylation [21]. This modification was found across a fairly large number of chromatin-associated proteins such as histone and high mobility group proteins. However, one problem with modifications such as formylation is that they may arise from the presence of formaldehyde or formic acid potentially used during sample preparation and are thus artifacts. Another problem is the similar mass shift between a formyl and a dimethyl group, and analogous to a trimethyl and an acetyl group, high mass accuracy is necessary to resolve this difference between both possible modifications.
In contrast to an acetyl, methyl, or formyl group, both ubiquitin and small ubiquitin-like modifier (SUMO) groups are extremely large post-translational modifications well over several kilodaltons in mass, making it somewhat difficult to analyze by mass spectrometry directly. The attachment of ubiquitin via an isopeptide bond between the C-terminus of ubiquitin and the ε-amine group of a lysine residue on the substrate generally leads to proteosomal, lysosomal, or vacuolar degradation of the substrate [22]. The attachment of SUMO also occurs via an isopeptide bond formation with lysine residues on the substrate, and is believed to antagonize substrate binding to other complexes and to oppose the addition of ubiquitin and thus its downstream consequences [23]. In addition to their large size, an additional difficulty in studying these modifications is their rapid turnover. With respect to ubiquitin, turnover is achieved via the activities of E3 ubiquitin ligase and deubiquitinating carboxy terminal hydrolases, and with respect to SUMO, the activities of E3 SUMO ligase and Ulp domain-containing desumoylating enzymes [22, 23]. The net consequence of rapid turnover for these modifications is their exceptionally low stoichiometry with respect to the unmodified substrate, and hence, mandating some form of enrichment for any proteomic interrogation of both classes of modified proteins.
The ease of genetic manipulation in the Saccharomyces cerevisiae model system has enabled a clever alternative to enrichment for ubiquitinated and sumoylated peptides, namely by generating strains that express ubiquitin or SUMO with an epitope-tag, for instance a histidine tag [24, 25, 26]. Since both ubiquitin and SUMO groups are cleavable by proteases, one cannot enrich at the peptide level because any N-terminal tag originally on the ubiquitin and SUMO proteins will be removed from the substrate. Thus, unlike the global proteomic investigations on acetylation, enrichment at the modified protein rather than peptide level is required for ubiquitination and sumoylation studies using this approach. Following epitope pulldown, one would proceed with protease digestion and multiple dimensional chromatographic separations prior to MS analysis. The subsequent bioinformatics search for ubiquitinated and sumoylated peptides must consider both the protease miscleavage site at the modified lysine as well as the mass shift not from the intact ubiquitin or SUMO moiety but rather from the cleaved C-terminal fragment still attached after protease digestion(27, 28)(Figure 2).
In contrast to generating laboratory strains expressing tagged-ubiquitin or –SUMO proteins, another approach is to indirectly enrich for the modified proteins using an antibody-conjugated protein domain that selectively interacts with the modification. For sumoylation, a recent study has used 32–133 RING-finger 4 fragment, which interacts specifically with polymerized branched SUMO groups [27]. Such an approach allows purification of sumoylated substarted in a wider range of cell types less amenable to molecular cloning.
Because one digests away the ubiquitin and SUMO chains to a single small fragment, one can only assay the presence and levels of total ubiquitination or sumoylation on that residue and will be unable to understand the branched pattern of the modifications. This is a similar issue with histone PTM quantification when one cannot link different modified sites to each other on the same original molecule. One solution to assay the branching pattern is to introduce in vitro generated SUMO branched fragments into the mass spec [28]. The lower sample complexity allows one to subsequently generate a reference tandem mass spectrum, parent ion charge state distribution, and retention time for each branched fragment that can be matched to the actual in vivo SUMO branched sample, thereby facilitating the bioinformatic search for sumoylated substrates.
Future prospects for proteomics for PTM analysis
Recent work has demonstrated that mass spectrometry is an ideal technique for characterization and discovery of lysine modifications, as evident by even new lysine modifications being still revealed such as lysine succinylation [29]. Although much progress has been made toward developing and applying mass spectrometry for PTM investigations, there is still a demand for better chromatographic resolution of modified peptides, more rigorous bioinformatics platforms to analyze post-translational modifications effectively, and more efficient biochemical methods to enrich for modified proteins. Finally, even if these particular demands are met in the coming years and one can successfully identify and quantify all the modified proteins within a cell, proteomics as a field will always need to evolve with more targeted assays, for instance site-directed mutagenesis or knockdown experiments, and even other similarly global experiments, such as microarrays, in order to achieve a truly functional understanding of lysine post-translational modifications.
Summary
Various mass spectrometers equipped with different types of mass analyzers possess unique advantages and tradeoffs for detecting post-translationally modified peptides and proteins. A consideration of which type of mass spectrometer is more appropriate for the specific experiment planned is necessary for successful PTM quantification.
The identification of a post-translationally modified peptide or protein must be validated by accurate parent ion mass, tandem mass spectrum, elution order, metabolic labeling, isotopic abundances, and other orthogonal lines of evidence.
Multiple proteases must often be used to ensure as complete sequence coverage as possible for PTM interrogations on a few target proteins. Furthermore, chromatographic resolution of the peptide sample will dramatically improve the dynamic range of detecting modified analytes in a background of mostly unmodified analytes.
Various enrichment methods for low-level modifications such as ubiquitination and sumoylation have been developed and should be applied to further increase the dynamic range of detection.
Quantification of modified peptides can be achieved labeled-free, in which case with respect to the unmodified peptide, or using labeled approaches such as SILAC or d5-propionyl derivatization.
Acknowledgments
We thank all members of the Garcia lab for constructive discussion during the preparation of the chapter. We extend apologies to those whose original research could not be referenced due to space considerations. BMZ is supported by the NSF Graduate Research Fellowship. BAG is supported by a NJCCR SEED grant, a National Science Foundation (NSF) Early Faculty CAREER award, an NIH Innovator award (DP2OD007447) from the Office of the Director, NIH, and by NSF grant (CBET-0941143).
References
- 1.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 2.Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier transform mass spectrometry. Mol Cell Proteomics. 2011;10:M111.009431. doi: 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Londry FA, Hager JW. Mass selective axial ion ejection from a linear quadrupole ion trap. J Am Soc Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 5.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plazas-Mayorca MD, Zee BM, Young NL, Fingerman IM, LeRoy G, Briggs SD, Garcia BA. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Z, Tang Y, Chen Y, Kim S, Liu H, Li SS, Gu W, Zhao Y. Molecular characterization of propionyllysines in non-histone proteins. Mol Cell Proteomics. 2009;8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeRoy G, Weston JT, Zee BM, Young NL, Plazas-Mayorca MD, Garcia BA. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–2442. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young NL, Plazas-Mayorca MD, DiMaggio PA, Flaniken IZ, Beltran AJ, Mishra N, LeRoy G, Floudas CA, Garcia BA. Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a. J Am Soc Mass Spectrom. 2010;21:960–970. doi: 10.1016/j.jasms.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Zhang K, Zou Y, Perna A, Wang Y. A quantitative study on the in vitro and in vivo acetylation of high mobility group A1 proteins. J Am Soc Mass Spectrom. 2007;18:1569–1578. doi: 10.1016/j.jasms.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 17.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binda O, Boyce M, Rush JS, Palaniappan K, Bertozzi CR, Gozani O. A chemical method for labeling lysine methyltransferase substrates. Chembiochem. 2011;12:330–334. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torrente MP, Zee BM, Young NL, Baliban RC, LeRoy G, Floudas CA, Hake SB, Garcia BA. Proteomic interrogation of human chromatin. PLoS One. 2011 doi: 10.1371/journal.pone.0024747. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Wiœniewski JR, Zougman A, Mann M. Nε-formylation of lysine is a widespread post-translational modification of nuclear proteins occuring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 24.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 25.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 26.Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Reports. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal ACO. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivostrategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]