Abstract
The centrosome is the main organizer of the microtubule cytoskeleton in animals, higher fungi and several other eukaryotic lineages. Centrosomes are usually located at the centre of cell in tight association with the nuclear envelope and duplicate at each cell cycle. Despite a great structural diversity between the different types of centrosomes, they are functionally equivalent and share at least some of their molecular components. In this paper, we explore the evolutionary origin of the different centrosomes, in an attempt to understand whether they are derived from an ancestral centrosome or evolved independently from the motile apparatus of distinct flagellated ancestors. We then discuss the evolution of centrosome structure and function within the animal lineage.
Keywords: centrosome, evolution, cytoskeleton, microtubule, eukaryote
1. Introduction
Centrosomes are membrane-free organelles that serve as main microtubule-organizing centres in distinct eukaryotic lineages. Through their ability to organize microtubules, they are involved in cell polarity and cell division, and play key roles in the development of most animal species [1,2]. In animal cells, the centrosome is composed of two centrioles surrounded by the pericentriolar material (PCM). The PCM has a precise organization, which derives from the hierarchical recruitment of a small number of large coiled-coil proteins around the centrioles [3,4]. The centrioles ensure the stability and the duplication of the centrosome, whereas the PCM anchors microtubule-nucleating complexes and cell cycle regulators. Centrosome duplication occurs by the assembly of two new centrioles in the immediate vicinity of the pre-existing centrioles, and a pair of centrioles is inherited by each daughter cell following mitosis. The timing of centrosome duplication may vary from one species to another, but it is always precisely coupled to cell cycle progression [5–9].
The centrosome can also migrate to the cell periphery, where the older centriole, called the mother centriole, can dock at the plasma membrane and nucleate the assembly of a cilium. Most mammalian cell types assemble a primary cilium in G1 or G0, which is typically a non-motile cilium involved in sensory functions such as cell–cell signalling or flow sensing [10–13]. Non-motile cilia are also critical for the function of sensory neurons in a range of animal species, including photoreceptors as well as olfactory, mechanosensory or chemosensory neurons [14–16]. In other cell types, the mother centriole can template the assembly of a motile cilium or flagellum, for instance, in the zebrafish kidney or in most sperm cells [17–19]. In this configuration, the mother centriole is functionally equivalent to the basal bodies that nucleate cilia or flagella in a range of unicellular eukaryotes. Centrioles and basal bodies are highly similar at the ultrastructural level, and indeed the key factors for centriole assembly are conserved in the genomes of all ciliated organisms [20–24]. This supports that centrioles and basal bodies are related and most probably derive from a centriolar structure present in the last common ancestor of all eukaryotes. The ancestral function of centrioles was likely to nucleate cilia, as evidenced by the co-distribution of centrioles and cilia across the eukaryotic tree [25]. Cilia are involved in locomotion through either beating or gliding motility, and in addition have sensory functions in diverse eukaryotes, suggesting an ancient association between motion and sensory perception [26,27].
In contrast to basal bodies, centrosomes are by definition central organelles. The term centrosome (etymologically, central body) is a generic term to design any isolable single-copy organelle having in common three basic properties: (i) to generally maintain itself at the cell centre due to its microtubule-nucleating/anchoring activity, (ii) to duplicate once during the cell cycle and (iii) to be physically associated with the nucleus [26]. The fact that the centrosome and the basal body complex can interconvert in many types of animal cells suggests that the centrosome evolved by internalization of the basal body complex. At which point during evolution and in how many different lineages this happened is not clear, however.
Besides animals, organisms with life cycle stages where cilia are absent and internalized centrioles organize the microtubule cytoskeleton include brown algae and Amoebozoa such as Physarum polycephalum [28]. In addition, some lineages completely lost centrioles and cilia during evolution but assemble organelles that are functionally equivalent to animal centrosomes. These include higher fungi, Amoebozoa such as Dictyostelium discoideum, diatoms and probably Ichthyosporea. The centrosomes of higher fungi are called spindle pole bodies (SPB). In Saccharomyces cerevisiae, the SPB is a multilayered cylindrical organelle embedded within the nuclear envelope [29]. The outer layer connects cytoplasmic microtubules, whereas the inner layer organizes spindle microtubules during mitosis, which occurs without disruption of the nuclear envelope. During cell division, a new SPB is assembled on a structure attached to the old SPB called the half-bridge [30]. The centrosome in the fission yeast Schizosaccharomyces pombe has a less obvious internal structure than its budding yeast counterpart, and is embedded in the nuclear envelope only during mitosis. It duplicates through the formation of a half-bridge similar in its structure and molecular composition to S. cerevisiae half-bridge, however [31]. In Amoebozoa such as D. discoideum, the centrosome is called a nuclear-associated body (NAB). The NAB consists of a match-boxed shaped three-layered core surrounded by a corona of amorphous material. Microtubules are organized from the corona, which is thus regarded as equivalent to the PCM of animal centrosomes. Duplication of the NAB occurs in a very unusual way: early in mitosis, the central layer disappears, and the two outer layers peel apart and separate to organize the mitotic spindle. At the end of telophase, the layers fold in two and a central layer and a corona are reformed [32,33]. In diatoms, the microtubule centre (MC) is closely associated with the interphase nucleus and the Golgi apparatus. The MC itself does not duplicate, but it is closely associated to a cubic laminar structure called the polar complex, which forms prior to mitosis and splits into two ‘polar plates’ that form the poles of the mitotic spindle [34]. Finally, Ichthyosporea (also called Mesomycetozoa) assemble spindle pole bodies reminiscent of the SPB of higher fungi [35,36]. Ichthosporean SPBs are plaque-like structures apposed to the nuclear envelope in close proximity to the Golgi apparatus. By electron microscopy, interphase cells have a single SPB organizing cytoplasmic microtubules and cells preparing for mitosis have two SPBs, one at each end of the nucleus [35]. The SPB cycle has not been studied in these organisms however, and it is not known how duplication occurs.
Despite this great diversity of shape, acentriolar centrosomes in fungi and Dictyostelium share some components with animal centrosomes, which suggest a common evolutionary origin [37–41]. In this paper, we will explore the evolutionary history of centrosomes by comparing microtubule cytoskeleton ultrastructure and centrosome composition in different eukaryotic lineages in the light of recent phylogenetic studies. We will attempt to bring insights into whether these centrosomes derive from an ancestral centrosome or evolved independently from the flagellar apparatus of distinct unicellular ancestors. This is an important point as it can affect the way we understand the similarities and differences between widespread model organisms. Because ultrastructural and molecular data are otherwise limited, we will focus on the lineage comprising Dictyostelium, higher fungi and animals, called Amorphea. Finally, we will discuss the evolution of centrosome function in animals.
2. Phylogenetic relationships between species assembling centrosomes
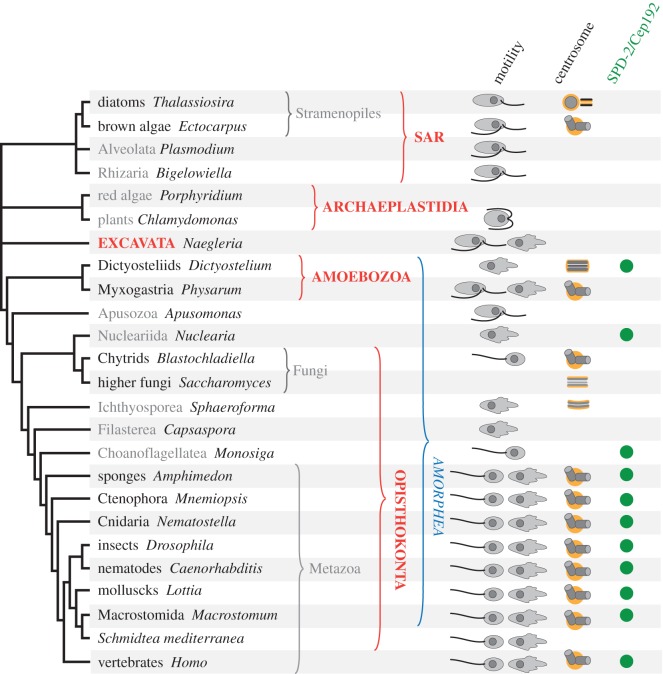
In the past decade, important progress has been made towards deciphering the relationships between the main eukaryotic lineages. This is due to the increased use of phylogenomic analyses, which are based on large sets of genes or complete genome sequences, as well as increased taxon sampling. Major questions remain unanswered, in particular the position of the root of the eukaryotic tree. Nevertheless, the existence of five supergroups is now widely accepted (figure 1) [42,43]. Among them, the most robustly predicted by phylogenetic analyses is the supergroup Opisthokonta, which comprises animals, fungi and the smaller groups Choanoflagellatea, Filasterea, Ichthyosporea and Nucleariida [44–47]. Choanoflagellates, which are unicellular or colonial free-living uniflagellates, are considered the closest living relatives of animals [45,47–51]. Filasterea comprise only two known species of filose amoebae completely lacking centrioles and cilia: the free-living marine protist Ministeria vibrans and Capsaspora owczarzaki, an endosymbiont of Biomphalaria glabrata, the intermediate snail host for intestinal schistosomiasis [46,51–53]. Ichthyosporeans comprises unicellular species living in parasitic or commensal relationships with animals, mostly fish. Species in this group are typically round cells with a thick cell wall. When known, their life cycle involves the formation of a plasmodium that can lead to the production of flagellated or amoeboid zoospores, although most taxa lack any type of motility [35,36]. Nucleariida, the sister group to fungi, contains filose amoebae that do not assemble centrioles or cilia and completely lack cytoplasmic microtubules [45,54–56].
Figure 1.
Current view of the eukaryotic tree highlighting the phylogenetic groups mentioned in the text (adapted from [42]). The five supergroups are in bold capitals. Features of each group are indicated when present in at least some of its members: the type of motility (amoeboid or flagellar), the presence of a centrosome and the presence of an SPD-2/CEP192 orthologue. (Online version in colour.)
There is a general consensus that the supergroup Amoebozoa is the sister group to Opisthokonta [42,47,57–59]. Amoebozoa constitute a large ensemble of amoebae and flagellates that includes Entamoeba histolytica, a pathogen responsible for amoebic dysentery, and slime moulds such as D. discoideum and P. polycephalum. Opisthokonts and Amoebozoa together are called unikonts or Amorphea. The first term reflects the hypothesis that these organisms derive from a monociliated ancestor [60]. Because this hypothesis is now considered unlikely, the term Amorphea, which relates to the fact that most cells in this group have no fixed shape unless constrained by an extracellular layer, was recently proposed instead [42].
When one considers the distribution of known centrosomes across the tree of eukaryotes, these organelles occur most frequently within the amorphean lineage (figure 1). The animal centrosome, the SPBs of higher fungi and ichthyosporeans, and the NAB of dictyosteliids are found within Amorphea. Besides Amorphea, centrosomes are present in brown algae and diatoms, which both belong to a phylum called Stramenopiles, or Heterokonta [42] (figure 1). The centrosome of brown algae appears to share a similar structure with the animal centrosome, but very little is known about its molecular composition [28]. The composition of diatom polar complex and MC also remains unknown [34]. Stramenopiles belong to the SAR supergoup, which is called after the three main phyla that form it, i.e. Stramenopiles, Alveolates and Rhizaria [42]. The SAR subgroup contains a wide variety of forms, most of which have been little studied. The best-known members of this supergroup include alveolates such as ciliates and the causative agents of malaria and toxoplasmosis (Plasmodium falciparum and Toxoplasma gondii, respectively) or Phytophtora infestans, a stramenopile responsible for the late blight that caused the Irish potato famine. Because of the scarcity of information on brown algae and diatoms, and the uncertainties regarding the relative position of species within the SAR subgroup, it is difficult at this point to discuss the evolutionary origin of these centrosomes. We will thus focus on centrosome evolution within Amorphea in the rest of this paper.
3. Evolution of cytoskeleton architecture in Amorphea
Centrosomes are found in a variety of Amorphea, including the early-branching supergroup Amoebozoa, which suggests they could derive from a structure already present in a common ancestor of all Amorphea. By comparing cytoskeletal diversity within extant lineages, it is possible to infer ancestral traits and thus get a clearer picture of what the ancestral microtubule cytoskeleton was like. From such a model, it is then possible to make assumptions on how subsequent trait changes during the evolution shaped the cytoskeletons of modern species. The flagellar apparatus is usually well conserved between related species and comparing its ultrastructure has been widely used for solving phylogenetic relationships between species [61,62]. Major changes in cytoskeleton architecture can nevertheless result from adaptation to different ecological niches, in particular with respect to motility and feeding modes. For instance, centrioles and cilia are entirely missing in Filasteria and Nucleariida, which use actin-based filopodia to crawl along a substrate (figure 1) [45,51,52,55,56,63]. A possible scenario is that the last common ancestor of all Amorphea or even of all eukaryotes was capable of both flagellar and actin-based motility, and different lineages lost one or the other (or both) while adapting to their specific environment [26,27,64–66]. It is thus necessary when attempting to reconstitute the evolutionary history of the microtubule cytoskeleton to consider these aspects as well. In the next section, we will discuss the ultrastructure of the amorphean ancestor and describe the main ultrastructural features of the different amorphean lineages, from Amoebozoa to animals.
(a). The amorphean ancestor
In flagellated eukaryotes, basal bodies are often associated with microtubule roots and fibres, which are typically distinct for the different basal bodies composing the flagellar apparatus. Basal bodies and corresponding flagella go through different maturation stages before adopting their final position at least one cell cycle later, a process called flagellar transformation. Comparing root architecture and flagellar transformation between major eukaryotic groups allows identification of broad patterns of cytoskeleton homology [61,62]. These analyses support that the last common ancestor of Amorphea was a flagellate similar in its architecture to a modern group of protists called excavates, and that both lineages were overall unchanged relative to the last common ancestor of all eukaryotes (figure 1) [62,66]. Typical excavates are surface-dwelling phagotrophic biflagellates that form a ventral feeding groove. The basic architecture of their flagellar apparatus consists of a posterior basal body associated with two main microtubule roots (called R1 and R2) and an anterior basal body anchoring a distinct set of roots (R3 and sometimes R4) (figure 2). The posterior basal body nucleates a flagellum that beats within the ventral groove to facilitate prey capture and the associated R1 and R2 roots reinforce the edges of the groove. The anterior basal body nucleates a second flagellum required for gliding-locomotion and the R3 root shapes the dorsal side of the cell. The number of microtubules within each microtubule root is specific and conserved within species. During flagellar transformation, the anterior basal body becomes a posterior basal body, and R3 (and R4 when present) are replaced by R1 and R2. This architecture is found across the eukaryotic tree, suggesting that all eukaryotes derived from a biflagellate ancestor assembling a complex, excavate-like flagellar apparatus. In particular, it is observed in two amorphean lineages, Amoebozoa and Apusozoa, suggesting that the last common ancestor of all Amorphea was itself an excavate-like flagellate (figure 1) [62,66]. Partial or complete loss of the ancestral flagellar apparatus then occurred several times independently in eukaryotes [62].
Figure 2.
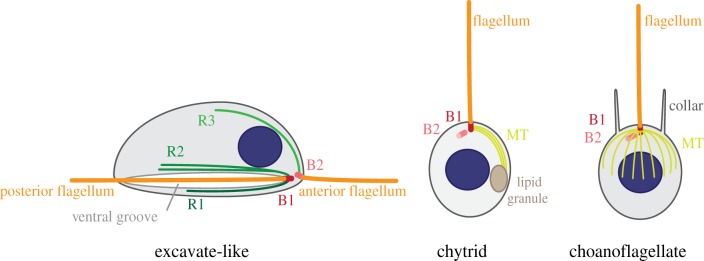
Schemes showing the main features of the microtubule cytoskeleton in some amorphean flagellated cells (adapted from [66–68]). The ancestral, excavate-like cytoskeleton seen in modern Myxogastria (for instance, P. polycephalum) and Apusuzoa is characterized by the presence of distinct sets of microtubule roots (R1, R2 and R3) associated with each basal body (B1 and B2), using the terminology introduced by Moestrup [61]. The ancestral microtubule roots are not conserved in chytrid zoospores and in choanoflagellates. In the most ancestral type of chytrids, a single bundle of microtubules emanates from one side of the old basal body (B1) and connects a large lipid granule. In choanoflagellates, microtubules radiate from a ring of electron-dense material encircling the old basal body (B1). (Online version in colour.)
Was the amorphean ancestor also capable of amoeboid motility? The widespread distribution of amoeboid motility among eukaryotes suggests that it originated early in their evolutionary history [64,66]. Switching between flagella-based swimming motility and actin-based gliding motility might be an ancient strategy for adapting to different environments. For instance, the excavate Naegleria gruberii can interconvert between a flagellate and an amoeba depending on whether it is placed in a liquid environment or associated with a solid substrate [64,69–72]. In Naegleria flagellates, the two centrioles anchor microtubule roots that contribute to cell morphology in association with a rigid cortex of actomyosin. In the amoeba, the entire microtubule cytoskeleton disassembles, including centrioles, and polymerizing actin is found in the cortex instead [72]. In Amorphea, interconversion between flagellates and amoebae occurs in Amoebozoa such as P. polycephalum and in sponge choanocytes (see §3b and 3d(iii)) [73,74]. In contrast to Naegleria amoebae, centrioles (Amoebozoa) and even cilia (choanocytes) are maintained during amoeboid phases, suggesting that the microtubule cytoskeleton might participate in the regulation of actin-based motility as in mammalian cells [75]. The amorphean centrosomes might thus have originated in unicellular ancestors capable of alternating between flagellated and amoeboid phases. The ability to organize dynamic arrays of cytoplasmic microtubules could have represented a benefit for optimizing amoeboid motility in ancestor cells. The widespread distribution of actin-based motility in Amorphea supports that it is indeed an ancestral trait. In agreement, many of the genes involved in the formation of actin-based protrusions are conserved across Amorphea, although they are not necessarily involved in motility [52,64]. For instance, many of the genes required for actin-based motility are conserved in choanoflagellates, which form filopodia for attachment to the substrate but not for motility [52]. It is also possible that formation of actin-based protrusions results from partially distinct mechanisms in Amoebozoa and in opisthokonts. Amoebozoa tend to form broad filopodia, called pseudopodia, whereas the ancestral opisthokont was probably forming slender filose protrusions [46]. In agreement, some key proteins for assembling filopodia in Metazoa are found in the genomes of Filasterea and choanoflagellates but not in Amoebozoa [52]. Thus, the ancestor of all Amorphea was likely an excavate-like flagellate, but whether it was also capable of amoeboid motility is unclear.
(b). At the bottom of the amorphean lineage: the Amoebozoa
Amoebozoa exhibit a variety of forms including amoebae, flagellates and sometimes more complex architectures such as fruiting bodies, multicellular slugs or multinucleated plasmodia. The excavate-like cytoskeleton architecture is still recognizable in modern flagellated species such as in the plasmodial slime mould P. polycephalum (Myxogastria) (figure 1) [62]. Physarum produces swarm cells that can interconvert between flagellates and amoebae [73]. The flagellates exhibit the R1 and R2 roots associated with the posterior basal body and the R3 root attached to the anterior basal body. In addition, the anterior basal body anchors a microtubule-organizing centre called the Mtoc1, which organizes an array of microtubules encircling the nucleus, called the flagellar cone [76]. Amoebae form a so-called proflagellar apparatus with the centrioles associated with reduced microtubule roots and the Mtoc1 [76,77]. Instead of the nuclear cone of flagellates, the Mtoc1 in amoebae nucleates a radial array of microtubules that appears similar to the interphase cytoskeleton of Dictyostelium or animal cells [73]. During mitosis, which happens in Physarum amoebae but not in flagellates, the microtubule roots and the Mtoc1 disappear and spindle microtubules are organized from centriole pairs present at each pole [77]. Thus, the proflagellar apparatus of Physarum amoebae shares similarities with the animal centrosome, the Mtoc1 playing the role of the PCM during interphase. The Mtoc1 could be an ancestral feature in Amoebozoa because similar structures are seen in distantly related species within this group [66,78]. The ancestor of all Amoebozoa possibly exhibited amoeboid stages resembling Physarum amoeboid swarm cells, in which the flagella were absent and the proflagellar apparatus, comprising a basal body complex and an Mtoc1-like structure, was acting as a centrosome. The NAB of dictyostelids would have derived secondarily from this ancestral proflagellar apparatus by retaining only a duplicating core and the microtubule-organizing activity of the Mtoc1.
(c). New relatives with ancient features: the Apusozoa
Apusozoa represent a small but widespread phylum of surface-dwelling biflagellates [79]. Like typical excavates, they move by ciliary gliding using their anterior flagellum, whereas the posterior flagellum forms within a ventral groove and serves for feeding. Apusozoa are similar to typical excavates and flagellated Amoebozoa with respect to microtubule cytoskeleton ultrastructure [62,80]. In addition, they form pseudopodia from their ventral surface, which are involved in prey capture but not motility [66]. Recent molecular phylogenies support that either the whole phylum, which comprises the genera Apusomonadida and Ancyromonadida, or Apusomonadida only, is the sister group to Opisthokonta, and hence that they branched off of the main amorphean lineage later than Amoebozoa [47,53,81] (figure 1). This suggests that the last common ancestor of Apusozoa and opisthokonts was itself an excavate-like biflagellate, and hence that Amoebozoa and opisthokonts evolved from two different flagellated ancestors. This would support that the centrosomes of Amoebozoa and animals evolved independently from the flagellar apparatus of distinct excavate-like ancestors and thus result from convergent evolution [62]. Further analysis of this clade will tell whether there is ground for an alternative scenario whereby Apusozoa would have evolved from a more complex ancestor by losing amoeboid motility.
(d). Dramatic changes in cytoskeleton architecture and cell motility in opisthokonts
In contrast to Amoebozoa and Apusozoa, no modern opisthokont exhibits the ancestral excavate-like cytoskeleton, suggesting that major modifications in cytoskeleton architecture occurred early in this lineage. Opisthokont flagellates propel themselves with a single posterior flagellum, a property that gave its name to the clade (opisthokont derives from the Greek words for posterior (opistho) and pole (kont), which refers to the flagellum) [82]. The flagellar apparatus of opisthokonts consists of a mature centriole nucleating the flagellum and an immature centriole in its vicinity. Opisthokonts lack the ancestral microtubule roots possibly in relation to the loss of a ventral feeding groove, which is never observed in this group. Typically, opisthokonts form arrays of cytoplasmic microtubules that radiate either from a pericentriolar matrix, or from conspicuous appendages decorating the old centriole [62,66].
(i). Flagellated fungi
At the base of the opisthokont lineage, flagellated zoospores are found in early-branching fungi such as chytrids, blastocladiales and zygomycetes. The flagellum and the centrioles were lost at least four times independently during the evolution of fungi, and the SPB took up microtubule organization in higher fungi [83]. In its simplest form, the flagellar apparatus of the most basal fungi, the chytrids, consists of two centrioles and a single bundle of cytoplasmic microtubules (figure 2). This bundle radiates from a spur of electron-dense material on one side of the old centriole and connects the flagellar apparatus to a membranous structure (called rumposome) that lies at the surface of a large lipid globule [84,85]. The microtubule cytoskeleton of chytrid zoospores thus appears little dynamic, much like in excavate-like cells but with a simpler architecture. Zoospores do not undergo mitosis, but instead encyst by losing the flagellum and forming a wall, which leads to the development of a zoosporangium. New zoospores form by repeated cycles of nuclear divisions followed by cellularization and flagellar assembly. In the developing sporangium of Rhizophydium spherotheca, a pair of centrioles is closely associated with each interphase nucleus in a position very comparable to the animal centrosome [86]. Cytoplasmic microtubules are nucleated from electron-dense material that coats the side of the centrioles. Prior to mitosis, two new centrioles form at right angles to the pre-existing centrioles and the two pairs of centrioles migrate apart. The nuclear envelope remains but spindle microtubules converge at the base of the oldest centriole within each pair through polar fenestrae [86]. Centrosomes in chytrid sporangia are thus highly reminiscent of animal centrosomes. Sporangia are not found outside fungi in the amorphean lineage however, which suggests that the common ancestor of fungi and animals was not forming these structures. Chytrid zoospores are highly amoeboid prior to encystment or following release from the sporangium [87], which could suggest that the fungal ancestor and possibly also the opisthokont ancestor were capable of amoeboid motility.
(ii). Animals' closest unicellular relatives: the choanoflagellates
The basic architecture of choanoflagellates is highly reminiscent of chytrid zoospores, with a single posterior flagellum that can serve for both motility and feeding [67,88–90]. In addition, a collar of actin-based microvilli surrounds the base of the flagellum and helps capture bacterial preys. Like in chytrids, the flagellar base consists of a mature centriole nucleating the flagellum and an immature centriole in its immediate vicinity. Cytoplasmic microtubules are organized from a ring of electron-dense material surrounding the mature centriole and run laterally under the plasma membrane, likely to support the collar of microvilli [67]. The flagellum does not disassemble during mitosis and in certain species the flagellar apparatus is duplicated prior to mitosis [89]. Choanoflagellates can form long actin-based protrusions resembling filopodia required for attachment to the substrate but only exhibit flagellar motility [52,90].
The ultrastructure of choanoflagellates is also highly reminiscent of choanocytes, the feeding cells of sponges, which also assemble a collar of microvilli around the flagellum. Some colonial choanoflagellates even produce an extracellular matrix that resembles the gelatinous matrix present within sponges, the mesohyl. This led to the hypothesis, already formulated in the late 1800s by both James Clark and Haeckel, that animals evolved from a colonial choanoflagellate [66,91,92]. In support of it, molecular phylogenies have consistently found the choanoflagellates to be the sister group to Metazoa [45,47,49–51] and sponges are often considered the most basal animal taxon—although this is still highly debated [43,93,94]. The hypothesis that our unicellular ancestors were choanoflagellate-like seems hard to reconcile with the idea that the common ancestor was also capable of amoeboid motility. However, both Filasterea and animals exhibit actin-based motility, which suggests that their common ancestor was possibly undergoing amoeboid phases. In this scenario, choanoflagellates would have lost this trait secondarily, retaining filopodia only for attachment to the substrate [52,90]. The hypothesis that choanoflagellates are derived from ancestors with a more complex life cycle is not new [95,96]. Revisiting the synzoospore theory postulated by Zakhvatkin in 1949, Mikhailov et al. [96] proposed that Metazoa and choanoflagellates derived from an organism with a complex life cycle that included flagellated and amoeboid trophic cells. Choanoflagellates would correspond to forms with secondarily simplified life cycles, retaining only the flagellated trophic phase. In agreement with this idea, genes involved in integrin-mediated cell adhesion or in the regulation of tight junctions are found in the genomes of chytrids, Ichtyosporea and Filasterea but are absent from known choanoflagellate genomes [46,65]. Such genes, which are associated with multicellularity in metazoans, could have been required for the complex genetic regulations underlying stage transitions in response to changing environmental conditions in the ancestor of opisthokonts [96]. The microtubule cytoskeleton in this ancestor would have been more variable that in living choanoflagellates, possibly involving amoeboid stages with a basal body complex acting as a centrosome.
(iii). Early Metazoa
Sponge choanocytes are thought to represent the evolutionary link between choanoflagellates and animals. Like choanoflagellates, choanocytes bear a single flagellum surrounded by a collar of microvilli, with a mature centriole nucleating the flagellum and an accessory centriole (figure 2). Cytoplasmic microtubules are organized from an appendage called the basal foot that decorates the mature centriole, which is also seen in multiciliated cells in other animal lineages [97,98]. In addition, the mature centriole anchors a prominent striated ciliary rootlet that connects the base of the flagellum to the accessory centriole and to the nucleus [97]. In certain circumstances, choanocytes can reorganize their cytoskeleton and switch to amoeboid motility. Following dissociation, they gradually lose their spherical shape and reorganize their actin cytoskeleton to form a lamellipodium, while the flagellum becomes immotile [74]. Remarkably, dissociated cells can then re-aggregate to reform an animal and choanocytes possibly revert to their original shape and function. In addition, choanocytes can trans-differentiate into archeocytes, which are amoeboid stem cells that actively migrate within the mesohyl [99]. Choanocytes are thus capable of switching between flagellated and amoeboid forms by reorganizing their actin and microtubule cytoskeletons. Unfortunately, there is little available data on the architecture and function of the microtubule cytoskeleton during amoeboid motility in choanocytes. Further studies on choanocyte transformations could provide important insight into centrosome evolution and the transition to multicellularity.
(iv). Evolution of centrosome structure in animals
Animal cells exhibit a variety of microtubule cytoskeleton architectures, but overall the structure of the centrosome itself is conserved. Unduplicated centrosomes are composed of two centrioles assembled from microtubule triplets in a ninefold arrangement, surrounded by the PCM. Centrioles within the centrosomes of Drosophila somatic cells and in Caenorhabidtis elegans cells are shorter and composed of nine microtubule doublets and singlets, respectively, instead of triplets. The ninefold symmetry is even lost in some insects, which can nevertheless nucleate the assembly of cilia [100]. There are also differences in the appendages that decorate the centrioles. In vertebrate centrosomes, two sets of ninefold symmetrical appendages decorate the mother centriole. The distal appendages are involved in docking the mother centriole at the plasma membrane prior to ciliogenesis [101,102]. Ultrastructural data support that appendages similar to the distal appendages—often referred to as transition fibres—are present across eukaryotes, probably reflecting the ancient involvement of centrioles in ciliogenesis. In agreement with this, putative homologues of the distal appendage component Cep164 are encoded in the genomes of flagellated protists [102,103]. By contrast, subdistal appendages are not documented outside vertebrates. At the mother centriole, subdistal appendages stably anchor a subpopulation of microtubules which participate in centrosome positioning during interphase [104]. Subdistal appendages are structurally similar to the basal foot that decorates centrioles in sponge choanocytes and in multiciliated cells from a variety of animal species, and indeed their assembly relies on at least one common component [101,105,106]. Like subdistal appendages, the basal foot anchors microtubules required for controlling the position of the centrioles within the plane of the membrane, which in turn determines ciliary beating orientation [105,107,108]. Unlike subdistal appendages, basal feet are observed in all basal metazoan lineages, suggesting that it is an ancestral trait associated with planar ciliary beating [109–112]. Although subdistal appendages are clearly absent from Drosophila and C. elegans, it is nevertheless possible that less conspicuous microtubule-anchoring structures are associated with the mother centriole in other non-vertebrate species.
4. Is the molecular composition of the centrosome conserved?
In the past decade, a cohort of centrosome components was identified and the role of these proteins characterized in model organisms. In particular, pioneering studies in C. elegans and Drosophila uncovered a set of structural components and regulators of centriole assembly, many of which are conserved across ciliated eukaryotes [9,23,24,113,114]. The ability to isolate centrosomes also allowed the identification of additional centrosome components by proteomic approaches in animals, budding yeast and Dictyostelium [115–118].
Most of the core components of the human centrosome have orthologues in the genome of the planarian Schmidtea mediterranea, which completely lacks centrosomes but forms centrioles de novo in ciliated cell types. Inactivation of a majority of these conserved genes affects centriole assembly or the function of cilia in planarian ciliated cells, supporting that they are indeed centriolar components [103]. Only a handful of centrosome components are missing from the planarian genome, most notably homologues for SPD-2/Cep192 and centrosomin (CNN)/CDK5RAP2 (Drosophila/human proteins), which are both major components of the PCM. SPD-2/Cep192 proteins are recruited in the immediate vicinity of the centrioles and play a key role in organizing the PCM by recruiting CNN/CDK5RAP2 and microtubule-nucleating complexes [3,4,119]. Caenorhabidtis elegans and human SPD-2 homologues are also required for centriole duplication, but possibly indirectly through their ability to recruit PCM, as Drosophila SPD-2 is dispensable for centriole duplication [119–121]. Remarkably, an SPD-2 orthologue is encoded in the Dictyostelium genome and localizes to the corona that surrounds the NAB, which is functionally equivalent to the PCM [40]. In addition, SPD-2 homologues are found in the genomes of several chytrid species as well as in choanoflagellates, but not in other species, suggesting that the SPD-2 family of proteins originated in Amorphea [23]. Thus, although the phylogenetic position of Apusozoa suggests that Dictyostelium and animal centrosomes evolved independently from the flagellar apparatus of distinct excavate-like ancestors, they share a key centrosome protein. The presence of a SPD-2 orthologue in Dictyostelium even shows that the function of these proteins is not necessarily linked to the centriolar structure.
The apparition of SPD-2 in a common ancestor of all Amorphea might be a key event in centrosome evolution, as their functions appear so intimately linked. SPD-2 was then lost secondarily in certain Amorphea. For instance, the apusozoan Thecamonas trahens seems to lack an SPD-2 homologue, which could be due to the fact that Apusozoa exhibit an invariant, excavate-like cellular architecture that predates SPD-2. Higher fungi and the ichthyosporean Sphaeroforma arctica, which assemble SPBs, also lack an SPD-2 orthologue, however. In budding yeast, microtubule-nucleating complexes are anchored at the cytoplasmic and nuclear sides of the SPB by proteins called Spc72 and Spc110, respectively. Spc72 has no clear orthologue outside of saccharomycetes, but Spc110 shares homology with a PCM protein called pericentrin, which binds to the centrioles and recruits microtubule-nucleating complexes in animals [3,4,38,122].
By contrast, SPD-2 orthologues are found in the genomes of choanoflagellates, suggesting that these proteins can function in flagellates as well [23]. The architecture of the microtubule cytoskeleton in choanoflagellates is quite divergent compared with the ancestral excavate-like organization, and it is possible that SPD-2 is involved in organizing the radial array of cytoplasmic microtubules typical of choanoflagellates. Another possibility is that SPD-2 originally evolved to organize the poles of the mitotic spindle in Amorphea. In animals, SPD-2 family members are key to centrosome maturation, which leads to an increase in the microtubule-nucleation capacity of mitotic centrosomes required for centrosome-driven spindle assembly [121,123,124]. In chytrids and choanoflagellates, mitotic microtubules are nucleated in the immediate vicinity of the centrioles, which could involve SPD-2-dependent recruitment of microtubule-nucleating complexes. Further technical development of model systems such as the choanoflagellate Monosiga brevicollis and the chytrid Batrachochytrium dendrobatidis will hopefully help to address the role of SPD-2 proteins in our unicellular ancestors and clarify the evolutionary history of centrosomes in the amorphean lineage.
5. Evolution of centrosome function in animals
In addition to a role in coordinating actin-based motility through its microtubule-organization capacity, the centrosome was probably involved in additional morphogenetic mechanisms in the common ancestor of animals. In most animal species, the first centrosome brought in by the sperm during fertilization triggers zygotic divisions and establishment of embryonic polarity [2]. The astral microtubules that emanate from the mitotic centrosomes and contact the cell cortex are essential for spindle positioning and thus for the control of cell division orientation. Embryonic cleavage in most animal species involves highly stereotypical patterns of cell divisions in controlled orientation. This is true for all the most basal lineages (i.e. sponges, ctenophores and cnidarians), suggesting that it is an ancestral mechanism [111,112,125–127]. Besides Metazoa, a role for centrosomes and astral microtubules in controlling spindle orientation has been well demonstrated in budding yeast. In this species, proper alignment of the spindle relies on an asymmetry in microtubule-nucleation capacity between the duplicated SPBs, which itself is correlated to centrosome age [128,129]. A similar mechanism was uncovered in animal stem cells, supporting that age asymmetry, which is inherent to the centrosome duplication cycle, is a key aspect of centrosome function [130–134]. This is not specific to the centrosome however, as age asymmetry between basal bodies, which underlies flagellar transformation in flagellated protists, is a key for maintaining cellular architecture through mitosis [135,136]. This property is likely a key reason for maintaining a duplicating organelle such as the centrosome in higher fungi that lost centrioles and cilia during evolution.
Besides its role in cell division orientation, the centrosome takes part in a range of morphogenetic processes by controlling cell polarity, nuclear positioning and primary cilia assembly in vertebrates. Nevertheless, development of relatively complex body patterns can occur in the complete absence of centrosomes, for instance, in the planarian S. mediterranea (figure 1). Planarian proliferating cells do not assemble a centriole-based centrosome or any other type of centrosome [103]. Planarians derive from flatworm ancestors that assembled centrosomes, as early-branching flatworms such as Macrostomum lignano do have centrosomes (figure 1) [137]. What triggered centrosome loss in the planarian ancestor is not known, but one hypothesis is that it was linked to decreased requirement for cell division orientation. For instance, early-branching flatworms exhibit a highly stereotypical pattern of embryonic cleavage, called spiral cleavage, in which each cell progeny forms at a specific position. By contrast, planarians exhibit a highly modified embryonic cleavage, called blastomere anarchy that does not seem to involve cell divisions in controlled orientation [138]. In adult planarians, mitosis only occurs in a population of pluripotent stem cells called neoblasts which are present throughout the body in the vicinity of the gastrovascular cavity [139]. Tissue homeostasis occurs by integration within existing tissues of neoblast progenies that have migrated away from the neoblast division site. In this context, mechanisms controlling cell division orientation could have become superfluous, resulting in the complete disappearance of the centrosome in planarian ancestors. Complex morphogenetic events also occur in Drosophila mutants unable to form centrosomes, although the centrosome is essential for early embryogenesis and proper control of cell division orientation in this species [140,141]. For instance, up to 30% of asymmetric divisions are abnormal in third instar larval brain cells of Drosophila Sas-4 mutants due to defects in spindle orientation [141].
Thus, the requirement for a centrosome is reduced in certain animal species, and this appears to be correlated with differences in the spectrum of developmental mechanisms at play. Centrioles are essential for ciliary assembly, but the presence of a centrosome is not the only way of controlling the number of cilia. For instance, planarians precisely assemble two centrioles in their male gametes although they derive from precursor cells devoid of centrosomes. By contrast, intrinsic age asymmetry, which provides one way of generating cell progenies with distinct fates, appears to be a unique property of the centrosome in animals.
6. Concluding remarks
The functions of animal centrosomes are intimately linked to the coordination of developmental processes. However, most centrosome components, including key PCM proteins, predate the evolution of Metazoa. The presence in the Dictyostelium genome of an orthologue of SPD-2/Cep192, a key PCM protein in animal cells, suggests that the centrosome is an ancestral trait in Amorphea. The functional conservation between the centrosomes of Dictyostelium, higher fungi and animals is also in favour of a common evolutionary origin of these organelles. The centrosome in all these species is involved in organizing the microtubule cytoskeleton during interphase and mitosis, regulating cell cycle progression and positioning the nucleus and the mitotic spindle. The different centrosomes duplicate once per cell cycle and are closely associated with the nuclear envelope. However, plotting what we know about the structure and molecular composition of the different centrosomes against the current scenarios of eukaryote evolution leads to a somewhat unexpected result: the more parsimonious hypothesis at this point is that these centrosomes result from convergent evolution. The main argument in favour of this second scenario is the phylogenetic position of Apusozoa. These excavate-like flagellates are more closely related to opisthokonts than to Amebozoa such as Dictyostelium. This suggests that the common ancestor of opisthokonts and Apusozoa was itself an excavate-like flagellate, and that opisthokonts and Amoebozoa evolved independently from the flagellar apparatus of distinct ancestors. Some key centrosome properties derive from properties of the ancestral basal body complex, such as duplication once per cell cycle, connection to the nucleus and role in organizing cytoplasmic microtubules. It is thus plausible that these characteristics were retained independently during evolution of the different centrosomes.
On the other hand, loss of ancestral traits leading to simplification of an ancestral morphology is not uncommon. The frequent idea that apparently simple organisms represent intermediates in the progressive construction of more complex ones was challenged as early as 1943 by André Lwoff [96] and more recently by molecular phylogenies [43,142,143]. For instance, the metazoan species exhibiting the simplest structure, Trichoplax adherens, probably does not represent the most ancient animal lineage but rather evolved from a more complex ancestor by losing ancestral traits [43,94]. Similarly, we now have a better picture of the ancestral eukaryotic cytoskeleton, and it turns out to be surprisingly complex. The last common ancestor of all eukaryotes was probably a biflagellate with a very elaborated microtubule cytoskeleton, an architecture that is still seen in many living species but was simplified in the evolution of many others [61,62]. The difficulty when trying to reconstitute the evolution of cellular architectures is that, due to a limited fossil record, we can only guess from the observation of living species what the sequence of events was. These species are all derived to a certain extent compared to their ancestors, and they probably represent only a fraction of the ancient variety of forms. Hence, it is possible that species thought to represent intermediate forms in evolution, such as Apusozoa and choanoflagellates might actually have lost ancestral traits, in particular amoeboid motility [96]. In this scenario, the different amorphean lineages would have evolved from ancestors capable of alternating between flagellates and amoebae. The different centrosomes in the amorphean lineage could thus all derive from a central, duplicating microtubule organizing centre organized around a centriole pair as in Physarum amoebae. Future work will hopefully help to establish more robust phylogenies and yield functional data in a greater variety of species. This should bring a better understanding of centrosome evolution and the conservation of its functions in the different amorphean lineages.
Acknowledgements
The author is grateful to the reviewers and the editors for helpful comments on this manuscript.
References
- 1.Schatten H, Sun QY. 2010. The role of centrosomes in fertilization, cell division and establishment of asymmetry during embryo development. Sem. Cell Dev. Biol. 21, 174–184. ( 10.1016/j.semcdb.2010.01.012) [DOI] [PubMed] [Google Scholar]
- 2.Bornens M. 2012. The centrosome in cells and organisms. Science 335, 422–426. ( 10.1126/science.1209037) [DOI] [PubMed] [Google Scholar]
- 3.Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14, 1159–1168. ( 10.1038/ncb2597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148–1158. ( 10.1038/ncb2591) [DOI] [PubMed] [Google Scholar]
- 5.Azimzadeh J, Bornens M. 2007. Structure and duplication of the centrosome. J. Cell Sci. 120, 2139–2142. ( 10.1242/jcs.005231) [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA. 2007. Centrosome duplication: of rules and licenses. Trends Cell Biol. 17, 215–221. ( 10.1016/j.tcb.2007.03.003) [DOI] [PubMed] [Google Scholar]
- 7.Brownlee CW, Rogers GC. 2013. Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell Mol. Life Sci. 70, 1021–1034. ( 10.1007/s00018-012-1102-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sluder G, Khodjakov A. 2010. Centriole duplication: analogue control in a digital age. Cell Biol. Int. 34, 1239–1245. ( 10.1042/CBI20100612) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigg EA, Stearns T. 2011. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13, 1154–1160. ( 10.1038/ncb2345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goetz SC, Anderson KV. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344. ( 10.1038/nrg2774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeger-Nukpezah T, Golemis EA. 2012. The extracellular matrix and ciliary signaling. Curr. Opin. Cell Biol. 24, 652–661. ( 10.1016/j.ceb.2012.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Praetorius HA, Leipziger J. 2013. Primary cilium-dependent sensing of urinary flow and paracrine purinergic signaling. Sem. Cell Dev. Biol. 24, 3–10. ( 10.1016/j.semcdb.2012.10.003) [DOI] [PubMed] [Google Scholar]
- 13.Fry AM, Leaper MJ, Bayliss R. 2014. The primary cilium: guardian of organ development and homeostasis. Organogenesis 10, 62–68. ( 10.4161/org.28910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwen DP, Jenkins PM, Martens JR. 2008. Chapter 12 olfactory cilia: our direct neuronal connection to the external world. Curr. Top. Dev. Biol. 85, 333–370. ( 10.1016/S0070-2153(08)00812-0) [DOI] [PubMed] [Google Scholar]
- 15.Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. 2009. The primary cilium as a complex signaling center. Curr. Biol. 19, R526–R535. ( 10.1016/j.cub.2009.05.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil TA. 2012. Sensory cilia in arthropods. Arthropod Struct. Dev. 41, 515–534. ( 10.1016/j.asd.2012.07.001) [DOI] [PubMed] [Google Scholar]
- 17.Escalier D. 2006. Knockout mouse models of sperm flagellum anomalies. Hum. Reprod. Update 12, 449–461. ( 10.1093/humupd/dml013) [DOI] [PubMed] [Google Scholar]
- 18.Mencarelli C, Lupetti P, Dallai R. 2008. Chapter 4. New insights into the cell biology of insect axonemes. Intl Rev. Cell Mol. Biol. 268, 95–145. ( 10.1016/S1937-6448(08)00804-6) [DOI] [PubMed] [Google Scholar]
- 19.Malicki J, Avanesov A, Li J, Yuan S, Sun Z. 2011. Chapter 3–Analysis of cilia structure and function in zebrafish. Methods Cell Biol. 101, 39–74. ( 10.1016/B978-0-12-387036-0.00003-7) [DOI] [PubMed] [Google Scholar]
- 20.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527–539. ( 10.1016/S0092-8674(04)00412-X) [DOI] [PubMed] [Google Scholar]
- 21.Li JB, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552. ( 10.1016/S0092-8674(04)00450-7) [DOI] [PubMed] [Google Scholar]
- 22.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090–1098. ( 10.1016/j.cub.2005.05.024) [DOI] [PubMed] [Google Scholar]
- 23.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426. ( 10.1242/jcs.064931) [DOI] [PubMed] [Google Scholar]
- 24.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. 2010. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413. ( 10.1242/jcs.064873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175. ( 10.1083/jcb.201011152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azimzadeh J, Bornens M. 2005. The centrosome in evolution. In Centrosomes in development and disease (ed. EA Nigg), pp. 93–122. Weinheim, Germany: Wiley-VCH. [Google Scholar]
- 27.Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607, 130–140. ( 10.1007/978-0-387-74021-8_11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsaros C, Karyophyllis D, Galatis B. 2006. Cytoskeleton and morphogenesis in brown algae. Ann. Bot. 97, 679–693. ( 10.1093/aob/mcl023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilmartin JV. 2014. Lessons from yeast: the spindle pole body and the centrosome. Phil. Trans. R. Soc. B 369, 20130456 ( 10.1098/rstb.2013.0456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaspersen SL, Winey M. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1–28. ( 10.1146/annurev.cellbio.20.022003.114106) [DOI] [PubMed] [Google Scholar]
- 31.Paoletti A, Bordes N, Haddad R, Schwartz C, Chang F, Bornens M. 2003. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell 14, 2793–2808. ( 10.1091/mbc.E02-10-0661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daunderer C, Schliwa M, Graf R. 1999. Dictyostelium discoideum: a promising centrosome model system. Biol. Cell 91, 313–320. ( 10.1016/S0248-4900(99)80092-6) [DOI] [PubMed] [Google Scholar]
- 33.Gräf R. 2009. Microtubule organization in Dictyostelium. Encyclopedia of Life Sciences (ELS). Chichester, UK: John Wiley & Sons. [Google Scholar]
- 34.De Martino A, Amato A, Bowler C. 2009. Mitosis in diatoms: rediscovering an old model for cell division. Bioessays 31, 874–884. ( 10.1002/bies.200900007) [DOI] [PubMed] [Google Scholar]
- 35.Marshall WL, Berbee ML. 2013. Comparative morphology and genealogical delimitation of cryptic species of sympatric isolates of Sphaeroforma (Ichthyosporea, Opisthokonta). Protist 164, 287–311. ( 10.1016/j.protis.2012.12.002) [DOI] [PubMed] [Google Scholar]
- 36.Marshall WL, Celio G, McLaughlin DJ, Berbee ML. 2008. Multiple isolations of a culturable, motile ichthyosporean (Mesomycetozoa, Opisthokonta), Creolimax fragrantissima n. gen., n. sp., from marine invertebrate digestive tracts. Protist 159, 415–433. ( 10.1016/j.protis.2008.03.003) [DOI] [PubMed] [Google Scholar]
- 37.Baum P, Furlong C, Byers B. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl Acad. Sci. USA. 83, 5512–5516. ( 10.1073/pnas.83.15.5512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilmartin JV, Goh PY. 1996. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 15, 4592–4602. [PMC free article] [PubMed] [Google Scholar]
- 39.Kilmartin J. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211–1221. ( 10.1083/jcb.200307064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz I, et al. 2009. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum. Cell Motil. Cytoskeleton 66, 915–928. ( 10.1002/cm.20384) [DOI] [PubMed] [Google Scholar]
- 41.Mana-Capelli S, Graf R, Larochelle DA. 2009. Dictyostelium discoideum CenB is a bona fide centrin essential for nuclear architecture and centrosome stability. Eukaryot. Cell 8, 1106–1117. ( 10.1128/EC.00025-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adl SM, et al. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493. ( 10.1111/j.1550-7408.2012.00644.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brinkmann H, Philippe H. 2007. The diversity of eukaryotes and the root of the eukaryotic tree. Adv. Exp. Med. Biol. 607, 20–37. ( 10.1007/978-0-387-74021-8_2) [DOI] [PubMed] [Google Scholar]
- 44.Baldauf SL, Palmer JD. 1993. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl Acad. Sci. USA 90, 11 558–11 562. ( 10.1073/pnas.90.24.11558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steenkamp ET, Wright J, Baldauf SL. 2006. The protistan origins of animals and fungi. Mol. Biol. Evol. 23, 93–106. ( 10.1093/molbev/msj011) [DOI] [PubMed] [Google Scholar]
- 46.Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. 2008. Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE 3, e2098 ( 10.1371/journal.pone.0002098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paps J, Medina-Chacon LA, Marshall W, Suga H, Ruiz-Trillo I. 2013. Molecular phylogeny of unikonts: new insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist 164, 2–12. ( 10.1016/j.protis.2012.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL. 2008. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc. Natl Acad. Sci. USA 105, 16 641–166 46. ( 10.1073/pnas.0801667105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. 2008. A phylogenomic investigation into the origin of metazoa. Mol. Biol. Evol. 25, 664–672. ( 10.1093/molbev/msn006) [DOI] [PubMed] [Google Scholar]
- 50.Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. 2002. The closest unicellular relatives of animals. Curr. Biol. 12, 1773–1778. ( 10.1016/S0960-9822(02)01187-9) [DOI] [PubMed] [Google Scholar]
- 51.Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325 ( 10.1038/ncomms3325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sebe-Pedros A, Burkhardt P, Sanchez-Pons N, Fairclough SR, Lang BF, King N, Ruiz-Trillo I. 2013. Insights into the origin of metazoan filopodia and microvilli. Mol. Biol. Evol. 30, 2013–2023. ( 10.1093/molbev/mst110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. 2012. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol. Biol. Evol. 29, 531–544. ( 10.1093/molbev/msr185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Steenkamp ET, Brinkmann H, Forget L, Philippe H, Lang BF. 2009. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol. Biol. 9, 272 ( 10.1186/1471-2148-9-272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patterson DJ. 1984. The genus Nuclearia (Sarcodina, Filosea, species composition and characteristics of the taxa. Archiv. Protistenkunde 128, 127–139. ( 10.1016/S0003-9365(84)80034-2) [DOI] [Google Scholar]
- 56.Yoshida M, Nakayama T, Inouye I. 2009. Nuclearia thermophila sp. nov. (Nucleariidae), a new nucleariid species isolated from Yunoko Lake in Nikko (Japan). Eur. J. Protistol. 45, 147–155. ( 10.1016/j.ejop.2008.09.004) [DOI] [PubMed] [Google Scholar]
- 57.Stechmann A, Cavalier-Smith T. 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297, 89–91. ( 10.1126/science.1071196) [DOI] [PubMed] [Google Scholar]
- 58.Richards TA, Cavalier-Smith T. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118. ( 10.1038/nature03949) [DOI] [PubMed] [Google Scholar]
- 59.He D, Fiz-Palacios O, Fu CJ, Fehling J, Tsai CC, Baldauf SL. 2014. An alternative root for the eukaryote tree of life. Curr. Biol. 24, 465–470. ( 10.1016/j.cub.2014.01.036) [DOI] [PubMed] [Google Scholar]
- 60.Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354. [DOI] [PubMed] [Google Scholar]
- 61.Moestrup Ø. 2000. The flagellate cytoskeleton: introduction of a general terminology for microtubular flagellar roots in protists. In Flagellates unity, diversity and evolution (eds Leadbeater BSC, Green JC.), pp. 69–94. London, UK: Taylor & Franscis. [Google Scholar]
- 62.Yubuki N, Leander BS. 2013. Evolution of microtubule organizing centers across the tree of eukaryotes. Plant J. 75, 230–244. ( 10.1111/tpj.12145) [DOI] [PubMed] [Google Scholar]
- 63.Schultheiss KP, Suga H, Ruiz-Trillo I, Miller WT. 2012. Lack of Csk-mediated negative regulation in a unicellular SRC kinase. Biochemistry 51, 8267–8277. ( 10.1021/bi300965h) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642. ( 10.1016/j.cell.2010.01.032) [DOI] [PubMed] [Google Scholar]
- 65.Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. 2010. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl Acad. Sci. USA 107, 10 142–10 147. ( 10.1073/pnas.1002257107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cavalier-Smith T. 2013. Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur. J. Protistol. 49, 115–178. ( 10.1016/j.ejop.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 67.Karpov SA, Leadbeater BS. 1998. Cytoskeleton structure and composition in choanoflagellates. J. Euk. Microbiol. 45, 361–367. ( 10.1111/j.1550-7408.1998.tb04550.x) [DOI] [Google Scholar]
- 68.Letcher PM, Powell MJ, Chambers JG, Longcore JE, Churchill PF, Harris PM. 2005. Ultrastructural and molecular delineation of the Chytridiaceae (Chytridiales). Can. J. Bot. 83, 1561–1573. ( 10.1139/b05-115) [DOI] [Google Scholar]
- 69.Fulton C. 1977. Intracellular regulation of cell shape and motility in Naegleria. First insights and a working hypothesis. J. Supramolecular Struct. 6, 13–43. ( 10.1002/jss.400060103) [DOI] [PubMed] [Google Scholar]
- 70.Fulton C. 1977. Cell differentiation in Naegleria gruberi. Annu. Rev. Microbiol. 31, 597–629. ( 10.1146/annurev.mi.31.100177.003121) [DOI] [PubMed] [Google Scholar]
- 71.Fulton C, Dingle AD. 1971. Basal bodies, but not centrioles, in Naegleria. J. Cell Biol. 51, 826–836. ( 10.1083/jcb.51.3.826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh CJ. 2007. The role of actin, actomyosin and microtubules in defining cell shape during the differentiation of Naegleria amebae into flagellates. Eur. J. Cell Biol. 86, 85–98. ( 10.1016/j.ejcb.2006.10.003) [DOI] [PubMed] [Google Scholar]
- 73.Uyeda TQ, Furuya M. 1985. Cytoskeletal changes visualized by fluorescence microscopy during amoeba-to-flagellate and flagellate-to-amoeba transformations in Physarum polycephalum. Protoplasma 126, 221–232. ( 10.1007/BF01281798) [DOI] [Google Scholar]
- 74.Gaino E, Magnino G. 1999. Dissociated cells of the calcareous sponge clathrina: a model for investigating cell adhesion and cell motility in vitro. Microsc. Res. Tech. 44, 279–292. () [DOI] [PubMed] [Google Scholar]
- 75.Luxton GW, Gundersen GG. 2011. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23, 579–588. ( 10.1016/j.ceb.2011.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright M, et al. 1988. Microtubule cytoskeleton and morphogenesis in the amoebae of the myxomycete Physarum polycephalum. Biol. Cell 63, 239–248. ( 10.1016/0248-4900(88)90061-5) [DOI] [PubMed] [Google Scholar]
- 77.Gely C, Wright M. 1986. The centriole cycle in the amoebae of the myxomycete Physarum polycephalum. Protoplasma 132, 23–31. ( 10.1007/BF01275786) [DOI] [Google Scholar]
- 78.Spiegel FW. 1981. Phylogenetic significance of the flagellar apparatus in protostelids (Eumycetozoa). Biosystems 14, 491–499. ( 10.1016/0303-2647(81)90053-8) [DOI] [PubMed] [Google Scholar]
- 79.Cavalier-Smith T, Chao EE. 2003. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J. Mol. Evol. 56, 540–563. ( 10.1007/s00239-002-2424-z) [DOI] [PubMed] [Google Scholar]
- 80.Karpov SA. 2007. The flagellar apparatus structure of Apusomonas proboscidea and apusomonad relationships. Protistology 5, 146–155. [Google Scholar]
- 81.Kim E, Simpson AG, Graham LE. 2006. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol. Biol. Evol. 23, 2455–2466. ( 10.1093/molbev/msl120) [DOI] [PubMed] [Google Scholar]
- 82.Cavalier-Smith T. 1988. Neomonada and the origin of animals and fungi. In Evolutionary relationships among protozoa (eds Coombs GH, Sleigh MA, Warren A.), pp. 375–407. London, UK: Kluwer. [Google Scholar]
- 83.James TY, et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822. ( 10.1038/nature05110) [DOI] [PubMed] [Google Scholar]
- 84.Barr DJ. 1981. The phylogenetic and taxonomic implications of flagellar rootlet morphology among zoosporic fungi. Biosystems 14, 359–370. ( 10.1016/0303-2647(81)90042-3) [DOI] [PubMed] [Google Scholar]
- 85.James TY, et al. 2006. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98, 860–871. ( 10.3852/mycologia.98.6.860) [DOI] [PubMed] [Google Scholar]
- 86.Powell MJ. 1980. Mitosis in the aquatic fungus Rhizophydium spherotheca (Chytridiales). Am. J. Bot. 67, 839–853. ( 10.2307/2442424) [DOI] [Google Scholar]
- 87.Barr DJ. 1992. Evolution and kingdoms of organisms from the perspective of a mycologist. Mycologia 84, 1–11. ( 10.2307/3760397) [DOI] [Google Scholar]
- 88.Hibberd DJ. 1975. Observations on the ultrastructure of the choanoflagellate Codosiga botrytis (Ehr.) Saville-Kent with special reference to the flagellar apparatus. J. Cell Sci. 17, 191–219. [DOI] [PubMed] [Google Scholar]
- 89.Karpov SA, Mylnikov AP. 1993. Preliminary observations on the ultrastructure of mitosis in choanoflagellates. Eur. J. Protistol. 29, 19–23. ( 10.1016/S0932-4739(11)80292-0) [DOI] [PubMed] [Google Scholar]
- 90.Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. 2011. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82. ( 10.1016/j.ydbio.2011.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nielsen C. 2008. Six major steps in animal evolution: are we derived sponge larvae? Evol. Dev. 10, 241–257. ( 10.1111/j.1525-142X.2008.00231.x) [DOI] [PubMed] [Google Scholar]
- 92.Richter DJ, King N. 2013. The genomic and cellular foundations of animal origins. Annu. Rev. Genet. 47, 509–537. ( 10.1146/annurev-genet-111212-133456) [DOI] [PubMed] [Google Scholar]
- 93.Philippe H, et al. 2009. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712. ( 10.1016/j.cub.2009.02.052) [DOI] [PubMed] [Google Scholar]
- 94.Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 ( 10.1126/science.1242592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maldonado M. 2004. Choanoflagellates, choanocytes, and animal multicellularity. Inverteb. Biol. 123, 1–22. ( 10.1111/j.1744-7410.2004.tb00138.x) [DOI] [Google Scholar]
- 96.Mikhailov KV, et al. 2009. The origin of Metazoa: a transition from temporal to spatial cell differentiation. Bioessays 31, 758–768. ( 10.1002/bies.200800214) [DOI] [PubMed] [Google Scholar]
- 97.Gonobobleva E, Maldonado M. 2009. Choanocyte ultrastructure in Halisarca dujardini (Demospongiae, Halisarcida). J. Morphol. 270, 615–627. ( 10.1002/jmor.10709) [DOI] [PubMed] [Google Scholar]
- 98.Dawe HR, Farr H, Gull K. 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120, 7–15. [DOI] [PubMed] [Google Scholar]
- 99.Funayama N. 2010. The stem cell system in demosponges: insights into the origin of somatic stem cells. Dev. Growth Differ. 52, 1–14. ( 10.1111/j.1440-169X.2009.01162.x) [DOI] [PubMed] [Google Scholar]
- 100.Riparbelli MG, Dallai R, Callaini G. 2010. The insect centriole: a land of discovery. Tissue Cell 42, 69–80. ( 10.1016/j.tice.2010.01.002) [DOI] [PubMed] [Google Scholar]
- 101.Ishikawa H, Kubo A, Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7, 517–524. ( 10.1038/ncb1251) [DOI] [PubMed] [Google Scholar]
- 102.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330. ( 10.1083/jcb.200707181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF. 2012. Centrosome loss in the evolution of planarians. Science 335, 461–463. ( 10.1126/science.1214457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317–330. ( 10.1083/jcb.149.2.317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kunimoto K, et al. 2012. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200. ( 10.1016/j.cell.2011.10.052) [DOI] [PubMed] [Google Scholar]
- 106.Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. 2013. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 203, 417–425. ( 10.1083/jcb.201303071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ. 2011. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 195, 19–26. ( 10.1083/jcb.201106110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vladar EK, Bayly RD, Sangoram AM, Scott MP, Axelrod JD. 2012. Microtubules enable the planar cell polarity of airway cilia. Curr. Biol. 22, 2203–2212. ( 10.1016/j.cub.2012.09.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Holley MC. 1984. The ciliary basal apparatus is adapted to the structure and mechanics of the epithelium. Tissue Cell 16, 287–310. ( 10.1016/0040-8166(84)90050-8) [DOI] [PubMed] [Google Scholar]
- 110.Tamm S, Tamm SL. 1988. Development of macrociliary cells in Beroe. I. Actin bundles and centriole migration. J. Cell Sci. 89, 67–80. [DOI] [PubMed] [Google Scholar]
- 111.Boury-Esnault N, Efremova S, BÉZac C, Vacelet J. 1999. Reproduction of a hexactinellid sponge: first description of gastrulation by cellular delamination in the Porifera. Inverteb. Reprod. Dev. 35, 187–201. ( 10.1080/07924259.1999.9652385) [DOI] [Google Scholar]
- 112.Riesgo A, Taylor C, Leys SP. 2007. Reproduction in a carnivorous sponge: the significance of the absence of an aquiferous system to the sponge body plan. Evol. Dev. 9, 618–631. ( 10.1111/j.1525-142X.2007.00200.x) [DOI] [PubMed] [Google Scholar]
- 113.Azimzadeh J, Marshall WF. 2010. Building the centriole. Curr. Biol. 20, R816–R825. ( 10.1016/j.cub.2010.08.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gonczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425–435. ( 10.1038/nrm3373) [DOI] [PubMed] [Google Scholar]
- 115.Adams IR, Kilmartin JV. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145, 809–823. ( 10.1083/jcb.145.4.809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574. ( 10.1038/nature02166) [DOI] [PubMed] [Google Scholar]
- 117.Reinders Y, Schulz I, Graf R, Sickmann A. 2006. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J. Proteome Res. 5, 589–598. ( 10.1021/pr050350q) [DOI] [PubMed] [Google Scholar]
- 118.Muller H, et al. 2010. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 29, 3344–3357. ( 10.1038/emboj.2010.210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dix CI, Raff JW. 2007. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764. ( 10.1016/j.cub.2007.08.065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523. ( 10.1016/S1534-5807(04)00066-8) [DOI] [PubMed] [Google Scholar]
- 121.Zhu F, et al. 2008. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18, 136–141. ( 10.1016/j.cub.2007.12.055) [DOI] [PubMed] [Google Scholar]
- 122.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657. ( 10.1091/mbc.E03-11-0796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gomez-Ferreria MA, et al. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960–1966. ( 10.1016/j.cub.2007.10.019) [DOI] [PubMed] [Google Scholar]
- 124.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. 2010. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl Acad. Sci. USA 107, 21022 ( 10.1073/pnas.1014664107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martindale MQ, Henry JQ. 1997. Reassessing embryogenesis in the Ctenophora: the inductive role of e1 micromeres in organizing ctene row formation in the ‘mosaic’ embryo, Mnemiopsis leidyi. Development 124, 1999–2006. [DOI] [PubMed] [Google Scholar]
- 126.Fritzenwanker JH, Genikhovich G, Kraus Y, Technau U. 2007. Early development and axis specification in the sea anemone Nematostella vectensis. Dev. Biol. 310, 264–279. ( 10.1016/j.ydbio.2007.07.029) [DOI] [PubMed] [Google Scholar]
- 127.Fourrage C, Chevalier S, Houliston E. 2010. A highly conserved Poc1 protein characterized in embryos of the hydrozoan Clytia hemisphaerica: localization and functional studies. PLoS ONE 5, e13994 ( 10.1371/journal.pone.0013994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pereira G, Tanaka T, Nasmyth K, Schiebel E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20, 6359–6370. ( 10.1093/emboj/20.22.6359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Juanes MA, Twyman H, Tunnacliffe E, Guo Z, ten Hoopen R, Segal M. 2013. Spindle pole body history intrinsically links pole identity with asymmetric fate in budding yeast. Curr. Biol. 23, 1310–1319. ( 10.1016/j.cub.2013.05.057) [DOI] [PubMed] [Google Scholar]
- 130.Reina J, Gonzalez C. 2014. When fate follows age: unequal centrosomes in asymmetric cell division. Phil. Trans. R. Soc. B 369, 20130466 ( 10.1098/rstb.2013.0466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518–521. ( 10.1126/science.1134910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947–955. ( 10.1038/nature08435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Januschke J, Llamazares S, Reina J, Gonzalez C. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2, 243 ( 10.1038/ncomms1245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Habib SJ, Chen BC, Tsai FC, Anastassiadis K, Meyer T, Betzig E, Nusse R. 2013. A localized Wnt signal orients asymmetric stem cell division in vitro. Science 339, 1445–1448. ( 10.1126/science.1231077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. 2011. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J. Cell Biol. 193, 741–753. ( 10.1083/jcb.201009131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marshall WF. 2012. Centriole asymmetry determines algal cell geometry. Curr. Opin. Plant Biol. 15, 632–637. ( 10.1016/j.pbi.2012.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tyler S. 1981. Development of cilia in embryos of the turbellarian Macrostomum. Hydrobiologia 84, 231–239. ( 10.1007/BF00026184) [DOI] [Google Scholar]
- 138.Cardona A, Hartenstein V, Romero R. 2006. Early embryogenesis of planaria: a cryptic larva feeding on maternal resources. Dev. Genes Evol. 216, 667–681. ( 10.1007/s00427-006-0094-3) [DOI] [PubMed] [Google Scholar]
- 139.Eisenhoffer GT, Kang H, Sanchez Alvarado A. 2008. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327–339. ( 10.1016/j.stem.2008.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bettencourt-Dias M, et al. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207. ( 10.1016/j.cub.2005.11.042) [DOI] [PubMed] [Google Scholar]
- 141.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125, 1375–1386. ( 10.1016/j.cell.2006.05.025) [DOI] [PubMed] [Google Scholar]
- 142.Peyretaillade E, El Alaoui H, Diogon M, Polonais V, Parisot N, Biron DG, Peyret P, Delbac F. 2011. Extreme reduction and compaction of microsporidian genomes. Res. Microbiol. 162, 598–606. ( 10.1016/j.resmic.2011.03.004) [DOI] [PubMed] [Google Scholar]
- 143.Philippe H, et al. 2011. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature 470, 255–258. ( 10.1038/nature09676) [DOI] [PMC free article] [PubMed] [Google Scholar]