Abstract
Centrosomes are the main microtubule-organizing centre of animal cells and are important for many critical cellular and developmental processes from cell polarization to cell division. At the core of the centrosome are centrioles, which recruit pericentriolar material to form the centrosome and act as basal bodies to nucleate formation of cilia and flagella. Defects in centriole structure, function and number are associated with a variety of human diseases, including cancer, brain diseases and ciliopathies. In this review, we discuss recent advances in our understanding of how new centrioles are assembled and how centriole number is controlled. We propose a general model for centriole duplication control in which cooperative binding of duplication factors defines a centriole ‘origin of duplication’ that initiates duplication, and passage through mitosis effects changes that license the centriole for a new round of duplication in the next cell cycle. We also focus on variations on the general theme in which many centrioles are created in a single cell cycle, including the specialized structures associated with these variations, the deuterosome in animal cells and the blepharoplast in lower plant cells.
Keywords: centrosome, centriole, blepharoplast, deuterosome
1. Introduction
Centrioles are cylindrical structures with a signature morphological motif of nine specialized microtubules symmetrically arranged about a central core (figure 1a). Mature centrioles are differentiated along their length for different functions. The distal end of the centriole is often specialized by the addition of appendages to organize microtubules and to nucleate a cilium [1,2], and interacts with the plasma membrane during ciliogenesis. The proximal end of the centriole is specialized to recruit a matrix of pericentriolar material proteins that support microtubule nucleation, and organization of microtubule arrays [3]. Centrioles are present in all eukaryotic species that form cilia and flagella, but are absent from higher plants and higher fungi which do not have cilia. Importantly, basal members of the plant and fungal groups do have centrioles and cilia, suggesting that these organelles are among the features that defined the earliest eukaryotic ancestor [4].
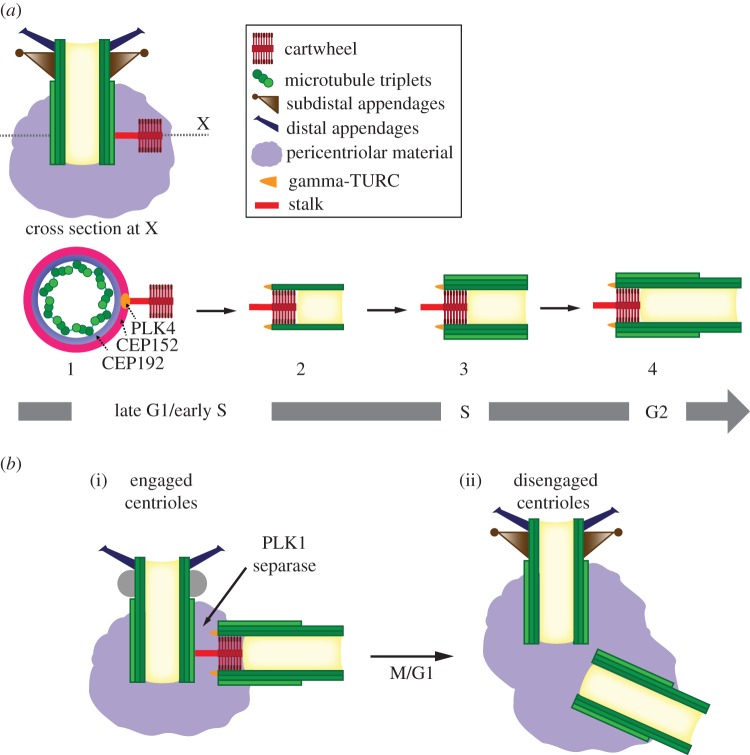
Figure 1.
Centriole assembly pathway in vertebrates. (a) Centrioles are cylindrical structures composed of nine microtubule triplets symmetrically arranged about a central core. The components important to the discussion here are indicated in the legend. Depicted is a longitudinal section of a mother centriole, which has two types of appendages, distal and subdistal, and lacks the internal cartwheel structure. The base of the mother centriole is embedded in the pericentriolar material. The formation of a procentriole has been initiated by assembly of the stalk and cartwheel from the side of the mother centriole. (a: 1–4) Stages of procentriole formation, depicted as viewed by cross section of centriole at X in longitudinal section. The mother centriole is not shown in (a: 2–4) for clarity, but is present and engaged to the procentriole throughout the process shown. (a-1) PLK4 accumulates at a single focus, in conjunction with CEP152 and CEP192, which are distributed in rings around the circumference of the centriole. PLK4 stimulates the assembly of a stalk and ninefold symmetric cartwheel that will provide structure to the procentriole and keep it engaged to the mother centriole. (a-2) Nine A-tubules are nucleated by the gamma-tubulin ring complex (gamma-TURC), in association with the cartwheel. These grow unidirectionally from the proximal to the distal end of the centriole. The A-tubules remain capped by the gamma-TURC throughout the assembly process, eventually being lost at the end of mitosis. (a-3) The B- and C-tubules form by a gamma-TURC-independent mechanism and grow until they reach the length of the A-tubule. (a-4) The distal end of the centriole is formed by elongation of the A- and B-tubules, creating a structurally distinct distal domain. (b) Centriole disengagement in the transition from M-G1. (i) A centrosome in metaphase of mitosis, with engaged mother centriole and procentriole. (ii) A centrosome in G1, after mitosis, with disengaged mother and daughter centrioles. The cartwheel has disassembled from the daughter centriole. Note that the subdistal appendages disassemble during mitosis, but the constituent proteins remain associated with the centriole. They are depicted as undifferentiated spheres in the mitotic centriole in place of the subdistal appendages. (Online version in colour.)
The structural complexity of the centrioles is reflected in the large number of centriole proteins identified by a combination of genomic and proteomic approaches in the past decade [5–19]. Some of the centriole proteins are conserved in all organisms with centrioles, suggesting that centrioles in divergent eukaryotes are likely to derive from a common ancestral structure [4,20,21]. By contrast, some centriole proteins are unique to a subset of organisms and tissues and these differences are probably owing to the diversity of contexts in which centrioles assemble and function.
In dividing cells, centrioles duplicate once per cell cycle, adjacent to a pre-existing centriole. What specifies the structure of a centriole, its location, orientation and copy number has been a longstanding question since the initial observations of the events of centriole duplication. The ‘parts list’ for the centriole is probably nearly complete, and the more recent challenge has been to address how these parts fit together to assemble centrioles. In this review, we first discuss the underlying mechanisms for the morphological duplication of centrioles, and how the complex ultrastructure of centrioles with ninefold symmetry and a well-defined length is established. We next describe progress in our understanding of how cells ensure centriole copy number in successive cell division and also how these control mechanisms are modified in different contexts.
2. Morphological duplication of centrioles–centriole biogenesis
The structure of centrioles is remarkably conserved across the eukaryotic kingdom (figure 1a). All known centrioles are cylindrical in shape and are composed of a ninefold symmetric array of microtubules. However, among different organisms, these microtubule arrays can comprise singlet, doublet or triplet microtubules, and centriole size ranges from 100 to 250 nm in diameter and from 100 to 400 nm in length [4]. As usual in biology, there are some known exceptions to these rules, which presumably represent evolutionary adaptation of the conserved structure [22]. The morphological events of the centriole duplication cycle have been well defined through electron microscopy at the ultrastructural level [23–27] (figure 1). Centriole duplication begins at the G1–S transition, when a new daughter centriole, termed a procentriole, begins to grow orthogonally from the proximal end of each of the two existing centrioles, termed mother centrioles. Once formed, procentrioles elongate through S and G2 phases. At the end of mitosis, the mitotic spindle segregates the duplicated centriole pairs, so that each resulting daughter cell contains two centrioles. Here, we discuss what is known of how the morphological duplication of the defining features of centrioles is accomplished each cell cycle.
The first step in the centriole duplication cycle is the formation of a procentriole adjacent to the mother centriole (figure 1a). This occurs at only one site per centriole, to ensure centriole number control. We shall refer to this site as the origin of centriole duplication, by analogy to DNA replication. What defines the location of the origin of centriole duplication? Emerging evidence suggests that three centriole proteins, PLK4, SASS6 and STIL, have an instructive function in this process as they all localize at the site of procentriole assembly at the G1–S transition, when centriole assembly is initiated. SASS6 is a structural component of the cartwheel, the ninefold symmetric template structure internal to the proximal end of the centriole (see other chapter) [28–30], and STIL is associated with SASS6 [31–34].
Although SASS6 was previously reported to be the earliest marker at the site of procentriole assembly [35], two independent reports find that PLK4, a polo-like kinase required for centriole formation [36,37], localizes to a dot-like structure at this site prior to SASS6, suggesting that it recruits SASS6 [36,37] (figure 1a-1). In Caenorhabditis elegans, SAS-6 is recruited to the centriole by ZYG-1, a functional orthologue of PLK4, thus this might be an evolutionarily conserved mechanism of defining the site of centriole assembly [38]. Strikingly, PLK4 at the centriole changes from a ring-like localization around the centriole barrel (early G1) to the dot-like localization (G1–S transition), and this change coincides with initiation of downstream events of centriole duplication, further supporting the role of PLK4 in marking the origin of centriole duplication [36]. These studies identify PLK4 as the earliest marker that localizes to the site of procentriole assembly, and given that PLK4 is a protein kinase, the most straightforward model would be that PLK4 phosphorylation of downstream proteins in centriole assembly initiates the process. Although some proteins have been shown to be substrates of PLK4 in vitro, including some centriole duplication proteins [39–42], there is as yet no direct link between PLK4 kinase activity and centriole initiation.
If PLK4 defines the origin of centriole duplication, what targets PLK4 to the centrioles at the right time and place? Recent studies suggest that two PLK4-binding proteins are important in this process. PLK4 (or ZYG-1) recruitment to the centrioles depends on Asterless in Drosophila [43] and on SPD-2 in C. elegans [44,45]. Interestingly, in mammalian cells, the orthologues of these two proteins, CEP152 and CEP192, respectively, interact with PLK4 and cooperate in the recruitment of PLK4 to the centrioles [36,46] (figure 1a-1). This recruitment depends on electrostatic interactions between the positively charged polo-box domain of PLK4 and acidic regions of CEP192 and CEP152 [36,46]. Both CEP152 and CEP192 are distributed symmetrically around the circumference of the centriole barrel [37,47], thus, although these studies provide an appealing explanation for how PLK4 is recruited to centrioles, they do not address the fundamental question, which is how is a single site for initiation of a morphological event chosen from a radially symmetric surface? This is, in essence, a symmetry-breaking event, similar to that in choosing a single site for bud formation in budding yeast [48], or establishment of the axes of a C. elegans embryo [49,50]. In these cases, and presumably in centriole initiation, there is some form of cooperativity or positive feedback that results in asymmetric accumulation of the relevant proteins in a symmetric background. It is possible, then, that association of PLK4 with CEP192 and/or CEP152 has such cooperative properties, or that the kinase activity of PLK4 provides some positive feedback mechanism to that association. Importantly, such a model would depend critically on the concentration of PLK4 in the cell, and that the concentration be subsaturating with respect to its binding partners; as described below, the concentration of PLK4 is known to be critical to limiting duplication to one site.
Once the site of the origin of centriole duplication is defined on the mother centriole, the next step is formation of the cartwheel [22] (figure 1a-1). The ninefold symmetric structure of the cartwheel is derived from the intrinsic ninefold symmetry of SASS6 oligomers that form the cartwheel [28,30]. The mechanism of cartwheel formation is covered in another review in this theme issue. The cartwheel dictates the ninefold symmetry of centrioles and initiates sequential assembly of the nine triplet microtubules. Interestingly, in some cases (mammals included), the cartwheel is lost from mature centrioles [22,51], thus it is required to make, but not to maintain, the structure of the centriole. Cryoelectron tomography analysis of purified human centrosomes revealed that the centre, or hub, of the cartwheel is at the end of a stalk that is linked to the side of the proximal end of the mother centriole [51] (figure 1a-1). This stalk is likely to be the physical link that keeps the procentriole and mother centriole engaged until the end of mitosis. We will follow the convention of referring to the new centriole as a procentriole while it is engaged to the mother centriole, and as a daughter centriole, once it has become disengaged.
How are microtubules added to the ninefold symmetric cartwheel to form the microtubule triplets found in most species? The microtubules within the triplet are designated A-, B- and C-tubules, with A- being a complete microtubule and proximal to the interior of the centriole, and the B- and C-tubules each sharing a wall with the A- or B-tubules, respectively. Cryoelectron tomography of procentrioles [51] shows that A-tubules are capped at their minus end with a conical structure resembling the gamma-tubulin ring complex structure [52–54] (figure 1a-2). In agreement with this observation, gamma-tubulin was reported to localize to the core of the mammalian centriole, in addition to the pericentriolar material [55,56]. Functional studies in a wide range of organisms including Tetrahymena and Paramecium identified an important role for gamma-tubulin during centriole duplication, indicating that it has a highly conserved role in centriole assembly [57–59]. These results suggest that gamma-tubulin, in association with the cartwheel, nucleates the A-tubule, which then grows as the centriole elongates. Following the nucleation and elongation of the A-tubules, the B- and C- tubules form, but are not capped at their minus end, suggesting that their assembly is initiated by a different mechanism (figure 1a-3).
In some organisms, centriole microtubules are singlets or doublets in most cells rather than triplets. For example, C. elegans centrioles have singlet microtubules. It is not clear what is required to make the specialized doublet and triplet microtubules. Recent cryo-electron microscopy (cryo-EM) studies of the centriole [60,61] clearly show that there are protein densities associated with the centriole microtubules, suggesting that proteins other than alpha- and beta-tubulin are required to make, or stabilize, them. It is interesting to note that although alpha-, beta- and gamma-tubulin are conserved in all eukaryotes, the other members of the tubulin family are not [62], even in organisms that have centrioles and cilia. This indicates that the other tubulin family members cannot be essential for duplicating the basic structure of the centriole, although they might be required for elaborations of the distal end of the centriole (see below).
Another aspect of the morphological duplication of the centrioles is the elongation of centrioles to a defined length (figure 1a-4), which is variable in different species and even in different cell types within a species. Given the morphology described above, in which the centriolar microtubules form on a central cartwheel structure [63,64], a simple hypothesis for length control is that the length of the cartwheel limits the elongation of the centriolar microtubules. This is consistent with the observation that some very long centrioles have a very long cartwheel [65]. In many organisms, the triplet microtubules extend beyond the cartwheel containing proximal zone of the centriole, and in some (mammals included), the A- and B-tubules extend even further, in a manner that requires POC5 [66]. It is this distal doublet region of the centriole to which the distal and subdistal appendages are attached. These steps of elongation occur in the context of the centriole duplication cycle, and it is likely that they, such as other events of duplication, are regulated processes, because the elongation of the centriole microtubules occurs much more slowly than microtubule polymerization in general. Lastly, a centriole of the appropriate length must be capped on the plus end by CP110 and associated proteins [63,64]. This cap must be removed to allow elongation of the axonemal microtubule doublets during cilium formation, but premature loss of the cap by depletion of a component allows centriole microtubules to elongate in the cytoplasm, beyond the centriole proper.
3. Centriole number control
The number of centrioles in most cells is controlled such that a G1 cell has a single pair of centrioles, one mother and one daughter. In the preceding text, we proposed that focal accumulation of a limiting initiator (PLK4) to the origin of centriole duplication might explain why only one procentriole forms next to each mother centriole. However, controlling the number of centrioles also requires that the level of that initiator be tightly regulated, and that there be a block to reduplication in a single cell cycle, which is not obviously explained by the above mechanism. In addition, any block to reduplication must be relieved at the appropriate time, so that the centrioles are ‘licensed’ to duplicate in the next cell cycle. This is exactly analogous to a replicating DNA molecule with a single origin of replication; that origin fires once and only once in a single S phase, and must be licensed for replication in the next cell cycle.
Consistent with the PLK4 focal accumulation model above, overexpression of PLK4 causes overduplication of centrioles in many cell types and species [67–71]. Remarkably, in the presence of existing mother centrioles, this overduplication occurs in the form of more than one centriole being initiated at each mother centriole, in a ‘centriole rosette’ [68,69]. This suggests that the additional PLK4 occupies more binding sites on the proximal end of the mother centriole, reaching the threshold for centriole initiation at multiple sites. In the absence of existing centrioles, such as in the eggs of Xenopus [72] and Drosophila [70] whose centrioles are naturally eliminated during development, overexpression of PLK4 induces de novo centriole formation, apparently overriding the need for a site on which to accumulate. Clearly, the amount of PLK4 is critical, and accordingly, the molecular mechanisms of PLK4 level control have been well studied. There is evidence for transcriptional regulation of PLK4 [73,74], but the main point of regulation seems to be the half-life of the PLK4 protein. Degradation of PLK4 is mediated by SCFβTrCP/ubiquitin-dependent proteolysis, which in turn depends on homodimerization and autophosphorylation of multiple sites on PLK4 [74–79]. Recent work in mammalian and Drosophila cells showed that phosphorylation of these sites depends on the kinase activity of PLK4, suggesting that PLK4 is a suicide kinase that initiates its own destruction [80,81]. If PLK4 is indeed a suicide kinase, how then does it achieve a high local concentration at the origin to initiate centriole assembly before it is degraded? PLK4 destruction entails autophosphorylation of multiple sites, including conserved phospho-residues within the downstream regulatory element and a phospho-cluster flanking this element [80,81]. The kinetics of these reactions is not known, but they occur in trans, thus would be sensitive to concentration. Perhaps PLK4 accumulation at the origin reaches a threshold to initiate centriole duplication, and only after that is the concentration of PLK4 sufficiently high to initiate destruction. Alternatively, the proteolysis machinery could be regulated such that it is locally inactive at the origin. In addition to regulation of the amount of PLK4, the activity of PLK4 is also regulated. The mitogen-activated protein kinase pathway in unfertilized Xenopus eggs was shown to block PLK4-dependent de novo centriole duplication [72], thus ensuring the paternal inheritance of centrioles. Moreover, the stress-activated protein kinase pathway and p53 regulate centriole duplication during the stress response in mammalian cells by cooperatively regulating PLK4 level and activity [82].
PLK4 is not the only centriole protein whose level must be regulated to ensure a single round of duplication. Several other core centriole proteins can stimulate the formation of extra centrioles when overexpressed, including STIL and SASS6 [16,34,35,83,84]. The extent of overduplication is less for these proteins than for PLK4 overexpression, but the importance of their regulation is evident from the fact that STIL and SASS6 both have KEN-box motifs, which are recognized by Cdh1 to target them to anaphase-promoting complex/cyclosome-dependent degradation during mitosis [31,35,83]. Interestingly, recent work demonstrated that CDK1 triggers relocalization of STIL from the centrosome to the cytosol for degradation [31]. SASS6 has another level of regulation, being targeted for destruction during G2 by the F-box protein FBXW5 [42], and this destruction is important for limiting duplication.
In addition to the control of the level of initiator proteins, there is also a centrosome-intrinsic block to reduplication which ensures that duplication is initiated only once per cell cycle. This block was revealed by cell fusion experiments in which cells from different stages of the cell cycle, containing duplicated or unduplicated centrioles were fused. In these experiments, a centriole that had previously duplicated (from a G2 cell, for example) was unable to duplicate in the same cytoplasm in which a previously unduplicated centriole (from a G1 cell) was able to duplicate [85], and thus that a centrosome ‘knew’ its duplication status and could not reduplicate until passage through mitosis licensed it for duplication in the next cycle. The nature of this centrosome-intrinsic block to reduplication was revealed by in vitro assays with engaged (G2) centrioles in Xenopus egg extracts, which showed that the tight orthogonal association of mother centriole and procentriole, termed engagement, blocks reduplication [86] (figure 1b). Disengagement of these centrioles from each other occurs at the end of mitosis, and is required to license duplication. In vertebrates, centriole disengagement requires the activity of PLK1, a mitotic kinase distantly related to PLK4, and separase, the protease responsible for sister chromatid separation at the metaphase-to-anaphase transition [87] (figure 1b). Having common regulatory proteins for both centriole duplication and chromosome segregation serves to couple these two cycles. This is important because, for example, disengagement of centriole pairs in a mitotic cell prior to chromosome segregation could result in multi-polar spindles, which interfere with normal mitotic chromosome segregation.
How do PLK1 and separase coordinate centriole disengagement with mitotic exit? A simple model is that there is a physical link between the mother centriole and procentriole and that PLK1 promotes separase-dependent cleavage of this link during mitotic exit. This is reminiscent of the PLK1-promoted separase-dependent cleavage of the chromatid linker cohesin to effect sister chromatid separation [88]. We note that a possible candidate for this link is the stalk, visible by cryo-EM [51] and shown in figure 1. Support for this model came from the demonstration that the protease activity of separase is required for disengagement [86,87], and from a study identifying cohesin itself as the disengagement-specific separase substrate [89]. This latter result was particularly appealing, because it furthered the mechanistic link between the centriole cycle and chromosome cycles.
What is the evidence for and against cohesin being the centriole engagement linker? The cleavage of cohesin by separase occurs at a defined site and it is possible to either mutate that site to make the protein non-degradable, or to replace that site with the recognition site for a different protease. Tsou et al. [87] expressed a non-degradable form of the cohesin subunit SSC1 and found that it prevented sister chromatid separation, as expected, but did not prevent centriole disengagement, suggesting that cohesin is not the engagement link. However, Schokël et al. [89] performed a similar experiment and obtained the opposite result, implicating cohesin. They then engineered cohesin subunits to contain cleavage sites for human rhinovirus or tobacco etch virus protease and showed that disengagement of centrioles could now be effected by those proteases in vitro and in vivo. These results would seem to argue compellingly that cohesin in some form is the engagement linker, but recent experiments in Drosophila showed that cohesin cleavage is insufficient for centriole disengagement [90]. Further, experiments in C. elegans show that cleavage is required for centriole disengagement only after meiosis II, and not for disengagement during mitotic cycles [91]. Lastly, Matsuo et al. [92] identified the centrosome protein pericentrin as a separase substrate, suggesting that it instead might be the important substrate of separase for centriole disengagement. The best that can be said at this point is that there is a physical link between the mother centriole and procentriole that keeps them engaged, but that the molecular identity of the engagement link, and the nature of its dissolution by PLK1 and separase, remain unclear.
How does disengagement license centrioles for duplication? A simple hypothesis is that centriole engagement suppresses further centriole assembly by sequestering at the origin one or more activities that are rate-limiting for centriole duplication, such as PLK4. Disengagement would either erase this origin mark on the mother centriole, or allow it to be re-used, allowing the formation of a new centriole. Consistent with this hypothesis, Loncarek et al. [93] showed that removal of a procentriole by laser ablation allows initiation of another procentriole on the mother centriole. Why does not the procentriole itself initiate a new centriole during S phase? Wang et al. [94] showed that PLK1-dependent modification of daughter centrioles during early mitosis, in addition to disengagement, is required to make them competent for centriole duplication in the next cell cycle, consistent with previous reports of a PLK1 requirement for reduplication in S phase [95].
We propose the following as a general model for centriole duplication control in animal cells. Note that, beginning from G1 phase of the cell cycle, a cell has a pair of disengaged centrioles: the mother centriole from the previous cell cycle and the daughter centriole, derived by disengagement of the procentriole from the mother at the end of mitosis. Both these centrioles are competent to duplicate, and, although they differ in other ways, they are the same with respect to duplication and will be referred to as ‘mother centriole’ below for the sake of simplicity.
(i) At the G1/S transition, the mother centriole acquires a single focus of PLK4 by a cooperative binding/positive feedback mechanism, creating an origin of duplication that initiates procentriole formation.
(ii) The mother centriole does not initiate a second procentriole, because PLK4 is limiting and cooperativity ensures that all free PLK4 goes to the existing single origin. The procentriole does not initiate a procentriole, because it is unmodified by PLK1, and thus is not competent to recruit the origin proteins.
(iii) At the G2/M transition, PLK1 modification of procentrioles makes them competent for recruiting pericentriolar material proteins involved in microtubule nucleation and organization, such as γ-tubulin [57,94]. Once this happens, centrioles become competent to organize microtubule-organizing centres and duplicate in the next cell cycle.
(iv) At mitosis, the two pairs of centrioles, each consisting of a mother centriole and an engaged procentriole, associate with the mitotic spindle. Passage through mitosis licenses the mother centriole by disengaging the procentriole, allowing a new focus to form, or the old to be re-used, and licenses the daughter by PLK1-dependent modification. Thus, in the ensuing G1, both centrioles received by a cell are competent for duplication.
We note that one glaring omission from this model is how the first step, formation of the centriole origin of duplication at the G1/S transition, is made to occur only at that time. This could involve the activity of the CDK2 kinase, which is activated at the G1/S transition. CDK2 was demonstrated to be required for centriole duplication in Xenopus eggs [96,97] and centriole overduplication in somatic cells [98,99]. However, CDK2 activity is dispensable for normal centriole duplication (and for cell cycle progression in general) in cycling cells [98,99], and for PLK4-induced de novo centriole formation in frog egg extracts [72]. This suggests that there is redundancy in Cdks that can initiate G1/S events—there is evidence that Cdk4 is involved in regulating centriole duplication [100]—and possibly that the requirement for Cdk activity can be bypassed by activating the pathway downstream of that requirement. Other, non-Cdk proteins might regulate the timing of centriole duplication by coupling it to the events of DNA replication. For example, the replication proteins MCM5 [101] and ORC1 [102,103] both inhibit centrosome reduplication, apparently by interacting with cyclin A/CDK2 and cyclin E/CDK2 at the centrosome.
4. Specific variations on the general duplication machinery
The abovementioned model accounts for centriole number control in most somatic cycling animal cells. There are two variations on this theme of number control that we consider here: multi-ciliated cells which form hundreds of centrioles during differentiation, and embryos in which centrioles are not derived from the sperm, which must generate centrioles de novo and achieve proper centriole number subsequently.
Multiple centrioles form near-simultaneously in some specialized cell types, including the multi-ciliated epithelial cells in animals and the ciliated sperm of land plants. Although the morphological events of centriole duplication in multi-ciliated cells have been well described by electron microscopy [104–108], the molecular pathway underlying these events has only recently been characterized (figure 2a). In multi-ciliated cells of vertebrates, such as mammalian tracheal epithelial cells [109], most of the genes for known centriole components and duplication factors are strongly upregulated. PLK4, for example, is upregulated more than 20-fold during differentiation of these cells. Transcriptional upregulation does not explain all of the properties of centriole amplification in these cells, however. Although some centrioles form around the pre-existing centrioles, the majority are assembled adjacent to deuterosomes, a structure unique to multi-ciliated cells that has no morphological resemblance to a centriole. Given that the centriole duplication pathway in these cells largely uses the same components as cycling cells [9], what is special about deuterosomes that they are able to initiate the assembly of multiple centrioles? Recent work shows that multi-ciliated cells specifically express DEUP1/CCDC67, a paralogue of the cycling cell centriole protein CEP63, that, such as CEP63, interacts with centriole duplication proteins [110–113]. DEUP1 localizes to deuterosomes and depletion of DEUP1 causes loss of deuterosomes, and greatly reduced centriole number. CCDC78 is another protein that localizes to deuterosomes [114] and is important for centriole assembly.
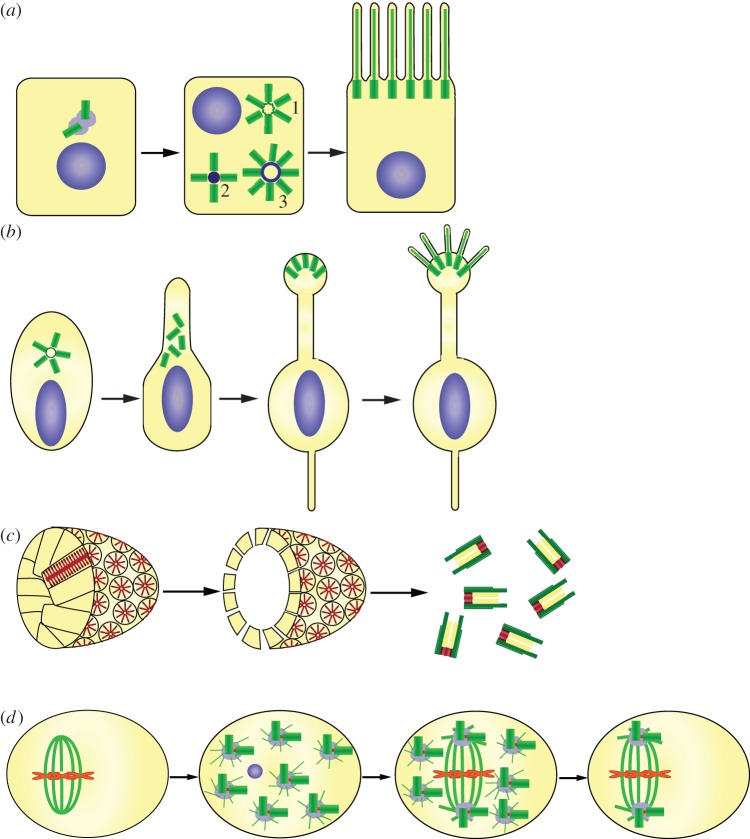
Figure 2.
Centriole number control in specialized cell types. (a) Multiple centrioles in animal multi-ciliated epithelial cells. Some centrioles form by initiation in a rosette around the mother centrioles (1), but most form on a deuterosome (2 and 3). The deuterosome ranges in size from centriole size (2) to much larger rings (3); it is not clear whether these represent a pathway of maturation of the structure. The centrioles are freed from the surface they grew from and migrate to the cell surface to nucleate motile cilia. (b) Multiple centrioles in olfactory sensory cilia. A neuronal cell forms approximately 10 centrioles, which nucleate sensory cilia from the end of the dendrite process. In this case, it is unknown whether centriole formation occurs strictly on mother centrioles or whether the deuterosome pathway is invoked. (c) Multiple centrioles in ciliated land plants. Sperm cells of some primitive plants are multi-ciliated and from centrioles from a blepharoplast (shown without surrounding cell). The blepharoplast consists of many radially arranged ninefold symmetric cylinders which resemble the cartwheel. The blepharoplast enlarges and ultimately disintegrates into many procentrioles which elongate and ultimately nucleate cilia on the surface of the sperm cell. (d) De novo formation and segregation of multiple centrioles during parthenogenesis. In insects of the hymenoptera order, parthenogenetic development is accompanied by the formation of many centrosomes de novo. Although depicted as centriole pairs, it is unclear whether centrosomes have single or paired centrioles at this stage. Only two of the centrosomes associate with the mitotic spindle, whereas the remainder degenerate, resulting in restoration of the appropriate number of centrioles per nucleus. (Online version in colour.)
We propose that multi-ciliated cells achieve the ability to form hundreds of centrioles simultaneously by a combination of a transcriptional programme that massively upregulates centriole components and the expression of specific proteins that modify the duplication pathway such that it becomes independent of cell cycle progression and of the requirement for a centriole to be the site of duplication. One aspect of centriole assembly in multi-ciliated cells that still is mysterious is whether the number of centrioles produced by deuterosomes is regulated, and, if so, how. This could be as simple as centriole number being dictated by the amount of limiting centriole assembly proteins, but might be more complex, linking the duplication process to apical membrane area and extracellular cues.
There are other situations in which multiple centrioles form, and the relationship between these and the deuterosome pathway is not clear. One example is the olfactory sensory neurons, which are the main sensory cells in the olfactory epithelium [115]. In these neurons, dendrites extend towards the nasal cavity, ending in a knob containing 10–30 centrioles from which cilia project to the epithelial surface. Ultrastructural studies suggested that these centrioles assemble near-simultaneously in the cell body and migrate to the dendritic knob [116,117], but whether the centrioles are nucleated from pre-existing centrioles or deuterosome-like structures is not known (figure 2b). Another example is the multi-ciliated sperm cells of primitive land plants [118–121]. Centriole formation during spermatogenesis in these plants is characterized by the appearance of a large structure, the blepharoplast, that consists of radially arranged ninefold symmetric structures (figure 2c). These resemble the cartwheel that initiates procentrioles [120], suggesting that these structures act as templates for procentriole formation. The blepharoplasts disintegrate at the end of mitosis, releasing procentrioles that elongate and form mature centrioles. The genome sequences of some of these plants have been determined, and some of the known centriole duplication proteins are conserved [20], suggesting that the blepharoplast represents an alternative manifestation of the same conserved centriole duplication machinery.
Centriole number control also differs from that of somatic cycling cells in embryos in which the centrioles are not derived from the sperm. In most animals, the sperm cell has one or two centrioles that are introduced into the egg upon fertilization, then duplicated, initiating the pathway described above for cycling cells. However, in some organisms, the embryo develops parthenogenetically, without sperm, thus centrioles must be formed de novo, and number control exerted after formation. For example, in insects of the hymenoptera order, hundreds of centrioles form de novo during the late stages of oogenesis [122,123] (figure 2d). These centrioles then recruit pericentriolar material and form microtubule asters. From these many centrosomes, only two become associated with the female pronucleus and form the mitotic spindle, whereas others degenerate, thus establishing the correct ratio of centrioles to nuclei. These observations are consistent with the re-emerging view that the mitotic spindle is important for both chromosome segregation and centrosome segregation [124]. In other example of de novo centriole formation, it is less clear how the appropriate number of centrioles is achieved; however, an alternative mechanism is raised by Naegleria, in which centrioles form de novo, in the developmental transition from an amoeba to a flagellate under stress conditions [125,126]. Here, it appears that only the desired number of centrioles (two) is formed, suggesting that number control can be exerted at the time of initiation of de novo centriole formation.
5. Concluding remarks
The past decade has seen great progress in defining the protein ‘parts list’ of the centrosome, and in identifying the proteins and activities that are required for centriole duplication. However, as in all of the great problems in biology, we still do not understand many of the fundamental properties of this remarkable organelle. We have chosen to focus this review on consideration of how the structural features of the centriole, which define it as a highly conserved component of eukaryotic cells, might be duplicated once per cell cycle. The importance of the cartwheel and its constituent proteins to our understanding of the events at the origin of centriole duplication, reviewed elsewhere in this volume, illustrates the critical importance of relating structure to function in the centriole. More studies combining in vitro reconstitution and structural analysis will be required to address the mechanism of centrosome assembly and determine what components are sufficient for the process, as opposed to those that are necessary, which is easier to determine using genetic manipulation. We believe that these questions, as well as those concerning the mechanism of centriole number control, will benefit from consideration of some of the special cases that we have highlighted here, in which centriole duplication in differentiated cells is uncoupled from cell cycle transitions.
References
- 1.Ishikawa H, Kubo A, Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7, 517–524. ( 10.1038/ncb1251) [DOI] [PubMed] [Google Scholar]
- 2.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330. ( 10.1083/jcb.200707181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luders J, Stearns T. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167. ( 10.1038/nrm2100) [DOI] [PubMed] [Google Scholar]
- 4.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175. ( 10.1083/jcb.201011152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574. ( 10.1038/nature02166) [DOI] [PubMed] [Google Scholar]
- 6.Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF. 2012. Centrosome loss in the evolution of planarians. Science 335, 461–463. ( 10.1126/science.1214457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829. ( 10.1016/j.devcel.2004.10.015) [DOI] [PubMed] [Google Scholar]
- 8.Fritz-Laylin LK, Assaf ZJ, Chen S, Cande WZ. 2010. Naegleria gruberi de novo basal body assembly occurs via stepwise incorporation of conserved proteins. Eukaryot. Cell 9, 860–865. ( 10.1128/EC.00381-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoh RA, Stowe TR, Turk E, Stearns T. 2012. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE 7, e52166 ( 10.1371/journal.pone.0052166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsen L, et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520–1535. ( 10.1038/emboj.2011.63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller LC, Romijn EP, Zamora I, Yates JR, III, Marshall WF. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090–1098. ( 10.1016/j.cub.2005.05.024) [DOI] [PubMed] [Google Scholar]
- 12.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, Winey M. 2007. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178, 905–912. ( 10.1083/jcb.200703109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Gönczy P. 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656–664. ( 10.1038/ncb1146) [DOI] [PubMed] [Google Scholar]
- 14.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523. ( 10.1016/S1534-5807(04)00066-8) [DOI] [PubMed] [Google Scholar]
- 15.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587. ( 10.1016/S0092-8674(03)00117-X) [DOI] [PubMed] [Google Scholar]
- 16.Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125. ( 10.1038/ncb1220) [DOI] [PubMed] [Google Scholar]
- 17.Leidel S, Gönczy P. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439. ( 10.1016/S1534-5807(03)00062-5) [DOI] [PubMed] [Google Scholar]
- 18.O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558. ( 10.1016/S0092-8674(01)00338-5) [DOI] [PubMed] [Google Scholar]
- 19.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Müller-Reichert T, Hyman AA. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873. ( 10.1016/j.cub.2004.04.012) [DOI] [PubMed] [Google Scholar]
- 20.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426. ( 10.1242/jcs.064931) [DOI] [PubMed] [Google Scholar]
- 21.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. 2010. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413. ( 10.1242/jcs.064873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425–435. ( 10.1038/nrm3373) [DOI] [PubMed] [Google Scholar]
- 23.Cavalier-Smith T. 1974. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J. Cell Sci. 16, 529–556. [DOI] [PubMed] [Google Scholar]
- 24.Dippell RV. 1968. The development of basal bodies in Paramecium. Proc. Natl Acad. Sci. USA 61, 461–468. ( 10.1073/pnas.61.2.461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuriyama R, Borisy GG. 1983. Cytasters induced within unfertilized sea-urchin eggs. J. Cell Sci. 61, 175–189. [DOI] [PubMed] [Google Scholar]
- 26.Robbins E, Jentzsch G, Micali A. 1968. The centriole cycle in synchronized HeLa cells. J. Cell Biol. 36, 329–339. ( 10.1083/jcb.36.2.329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorobjev IA, Chentsov YuS. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938–949. ( 10.1083/jcb.93.3.938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa D, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375. ( 10.1016/j.cell.2011.01.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17, 2169–2174. ( 10.1016/j.cub.2007.11.046) [DOI] [PubMed] [Google Scholar]
- 30.van Breugel M, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331, 1196–1199. ( 10.1126/science.1199325) [DOI] [PubMed] [Google Scholar]
- 31.Arquint C, Nigg EA. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24, 351–360. ( 10.1016/j.cub.2013.12.016.) [DOI] [PubMed] [Google Scholar]
- 32.Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. 2010. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188, 313–323. ( 10.1083/jcb.200910016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens NR, Roque H, Raff JW. 2010. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell 19, 913–919. ( 10.1016/j.devcel.2010.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang C-W, Wu K-S, Tang TK. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30, 4790–4804. ( 10.1038/emboj.2011.378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell 13, 203–213. ( 10.1016/j.devcel.2007.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TS, et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl Acad. Sci. USA 110, E4849–E4857. ( 10.1073/pnas.1319656110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open 1, 965–976. ( 10.1242/bio.20122337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell 25, 284–298. ( 10.1016/j.devcel.2013.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I. 2012. GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486–496. ( 10.1242/jcs.093930) [DOI] [PubMed] [Google Scholar]
- 40.Chang J, Cizmecioglu O, Hoffmann I, Rhee K. 2010. PLK2 phosphorylation is critical for CPAP function in procentriole formation during the centrosome cycle. EMBO J. 29, 2395–2406. ( 10.1038/emboj.2010.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729. ( 10.1083/jcb.201006049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puklowski A, et al. 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13, 1004–1009. ( 10.1038/ncb2282) [DOI] [PubMed] [Google Scholar]
- 43.Dzhindzhev NS, et al. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718. ( 10.1038/nature09445) [DOI] [PubMed] [Google Scholar]
- 44.Delattre M, Canard C, Gonczy P. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844–1849. ( 10.1016/j.cub.2006.07.059) [DOI] [PubMed] [Google Scholar]
- 45.Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. 2006. Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623. ( 10.1038/nature05318) [DOI] [PubMed] [Google Scholar]
- 46.Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126, 3223–3233. ( 10.1242/jcs.129502) [DOI] [PubMed] [Google Scholar]
- 47.Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148–1158. ( 10.1038/ncb2591) [DOI] [PubMed] [Google Scholar]
- 48.Johnson JM, Jin M, Lew DJ. 2011. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 21, 740–746. ( 10.1016/j.gde.2011.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonczy P, Rose LS. 2005. Asymmetric cell division and axis formation in the embryo. WormBook 1–20. ( 10.1895/wormbook.1.30.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyczak R, Gomes JE, Bowerman B. 2002. Heads or tails: cell polarity and axis formation in the early Caenorhabditis elegans embryo. Dev. Cell 3, 157–166. ( 10.1016/S1534-5807(02)00226-5) [DOI] [PubMed] [Google Scholar]
- 51.Guichard P, Chretien D, Marco S, Tassin AM. 2010. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29, 1565–1572. ( 10.1038/emboj.2010.45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erickson HP. 2000. Gamma-tubulin nucleation: template or protofilament? Nat. Cell Biol. 2, E93–E96. ( 10.1038/35014084) [DOI] [PubMed] [Google Scholar]
- 53.Keating TJ, Borisy GG. 2000. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat. Cell Biol. 2, 352–357. ( 10.1038/35014045) [DOI] [PubMed] [Google Scholar]
- 54.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. 2000. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365–370. ( 10.1038/35014058) [DOI] [PubMed] [Google Scholar]
- 55.Fuller SD, Gowen BE, Reinsch S, Sawyer A, Buendia B, Wepf R, Karsenti E. 1995. The core of the mammalian centriole contains gamma-tubulin. Curr. Biol. 5, 1384–1393. ( 10.1016/S0960-9822(95)00276-4) [DOI] [PubMed] [Google Scholar]
- 56.Moudjou M, Bordes N, Paintrand M, Bornens M. 1996. Gamma-tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109, 875–887. [DOI] [PubMed] [Google Scholar]
- 57.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. 2006. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505–515. ( 10.1083/jcb.200510028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz F, Beisson J, Rossier J, Dupuis-Williams P. 1999. Basal body duplication in Paramecium requires gamma-tubulin. Curr. Biol. 9, 43–46. ( 10.1016/S0960-9822(99)80045-1) [DOI] [PubMed] [Google Scholar]
- 59.Shang Y, Li B, Gorovsky MA. 2002. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 158, 1195–1206. ( 10.1083/jcb.200205101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guichard P, et al. 2013. Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr. Biol. 23, 1620–1628. ( 10.1016/j.cub.2013.06.061) [DOI] [PubMed] [Google Scholar]
- 61.Li S, Fernandez JJ, Marshall WF, Agard DA. 2012. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 31, 552–562. ( 10.1038/emboj.2011.460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dutcher SK. 2003. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr. Opin. Microbiol. 6, 634–640. ( 10.1016/j.mib.2003.10.016) [DOI] [PubMed] [Google Scholar]
- 63.Franz A, Roque H, Saurya S, Dobbelaere J, Raff JW. 2013. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 203, 785–799. ( 10.1083/jcb.201305109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011. ( 10.1016/j.cub.2009.05.016) [DOI] [PubMed] [Google Scholar]
- 65.Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gönczy P. 2012. Cartwheel architecture of Trichonympha basal body. Science 337, 553 ( 10.1126/science.1222789) [DOI] [PubMed] [Google Scholar]
- 66.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185, 101–114. ( 10.1083/jcb.200808082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bettencourt-Dias M, et al. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207. ( 10.1016/j.cub.2005.11.042) [DOI] [PubMed] [Google Scholar]
- 68.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146. ( 10.1038/ncb1320) [DOI] [PubMed] [Google Scholar]
- 69.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202. ( 10.1016/j.devcel.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 70.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science 316, 1046–1050. ( 10.1126/science.1142950) [DOI] [PubMed] [Google Scholar]
- 71.Peel N, Stevens NR, Basto R, Raff JW. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17, 834–843. ( 10.1016/j.cub.2007.04.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eckerdt F, Yamamoto TM, Lewellyn AL, Maller JL. 2011. Identification of a polo-like kinase 4-dependent pathway for de novo centriole formation. Curr. Biol. 21, 428–432. ( 10.1016/j.cub.2011.01.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brownlee CW, Klebba JE, Buster DW, Rogers GC. 2011. The protein phosphatase 2A regulatory subunit twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195, 231–243. ( 10.1083/jcb.201107086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184, 225–239. ( 10.1083/jcb.200808049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19, 43–49. ( 10.1016/j.cub.2008.11.037) [DOI] [PubMed] [Google Scholar]
- 76.Guderian G, Westendorf J, Uldschmid A, Nigg EA. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123, 2163–2169. ( 10.1242/jcs.068502) [DOI] [PubMed] [Google Scholar]
- 77.Holland AJ, et al. 2012. Polo-like kinase 4 controls centriole duplication but does not directly regulate cytokinesis. Mol. Biol. Cell 23, 1838–1845. ( 10.1091/mbc.E11-12-1043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. 2012. The autoregulated instability of polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26, 2684–2689. ( 10.1101/gad.207027.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188, 191–198. ( 10.1083/jcb.200911102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cunha-Ferreira I, et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23, 2245–2254. ( 10.1016/j.cub.2013.09.037) [DOI] [PubMed] [Google Scholar]
- 81.Klebba JE, Buster DW, Nguyen AL, Swatkoski S, Gucek M, Rusan NM, Rogers GC. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23, 2255–2261. ( 10.1016/j.cub.2013.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura T, Saito H, Takekawa M. 2013. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat. Commun. 4, 1775 ( 10.1038/ncomms2752) [DOI] [PubMed] [Google Scholar]
- 83.Arquint C, Sonnen KF, Stierhof YD, Nigg EA. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125, 1342–1352. ( 10.1242/jcs.099887) [DOI] [PubMed] [Google Scholar]
- 84.Vulprecht J, et al. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125, 1353–1362. ( 10.1242/jcs.104109) [DOI] [PubMed] [Google Scholar]
- 85.Wong C, Stearns T. 2003. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 5, 539–544. ( 10.1038/ncb993) [DOI] [PubMed] [Google Scholar]
- 86.Tsou MF, Stearns T. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951. ( 10.1038/nature04985) [DOI] [PubMed] [Google Scholar]
- 87.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344–354. ( 10.1016/j.devcel.2009.07.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M. 2002. The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol. Cell 9, 515–525. ( 10.1016/S1097-2765(02)00473-2) [DOI] [PubMed] [Google Scholar]
- 89.Schockel L, Mockel M, Mayer B, Boos D, Stemmann O. 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 13, 966–972. ( 10.1038/ncb2280) [DOI] [PubMed] [Google Scholar]
- 90.Oliveira RA, Nasmyth K. 2013. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr. Biol. 23, R601–R603. ( 10.1016/j.cub.2013.04.003) [DOI] [PubMed] [Google Scholar]
- 91.Cabral G, Sans SS, Cowan CR, Dammermann A. 2013. Multiple mechanisms contribute to centriole separation in C. elegans. Curr. Biol. 23, 1380–1387. ( 10.1016/j.cub.2013.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee K, Rhee K. 2012. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle 11, 2476–2485. ( 10.4161/cc.20878) [DOI] [PubMed] [Google Scholar]
- 93.Loncarek J, Hergert P, Magidson V, Khodjakov A. 2008. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10, 322–328. ( 10.1038/ncb1694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang WJ, Soni RK, Uryu K, Tsou MF. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739. ( 10.1083/jcb.201101109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loncarek J, Hergert P, Khodjakov A. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20, 1277–1282. ( 10.1016/j.cub.2010.05.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283, 851–854. ( 10.1126/science.283.5403.851) [DOI] [PubMed] [Google Scholar]
- 97.Lacey KR, Jackson PK, Stearns T. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl Acad. Sci. USA 96, 2817–2822. ( 10.1073/pnas.96.6.2817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. 2006. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene 25, 2943–2949. ( 10.1038/sj.onc.1209310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1, 88–93. ( 10.1038/10054) [DOI] [PubMed] [Google Scholar]
- 100.Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, Saavedra HI. 2010. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol. Cell Biol. 30, 694–710. ( 10.1128/MCB.00253-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferguson RL, Pascreau G, Maller JL. 2010. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J. Cell Sci. 123, 2743–2749. ( 10.1242/jcs.073098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793. ( 10.1126/science.1166745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hossain M, Stillman B. 2012. Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 26, 1797–1810. ( 10.1101/gad.197178.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anderson RG, Brenner RM. 1971. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J. Cell Biol. 50, 10–34. ( 10.1083/jcb.50.1.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dirksen ER. 1971. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 51, 286–302. ( 10.1083/jcb.51.1.286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalnins VI, Porter KR. 1969. Centriole replication during ciliogenesis in the chick tracheal epithelium. Z. Zellforsch Mikrosk. Anat. 100, 1–30. ( 10.1007/BF00343818) [DOI] [PubMed] [Google Scholar]
- 107.Sorokin SP. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3, 207–230. [DOI] [PubMed] [Google Scholar]
- 108.Steinman RM. 1968. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am. J. Anat. 122, 19–55. ( 10.1002/aja.1001220103) [DOI] [PubMed] [Google Scholar]
- 109.Vladar EK, Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 78, 31–42. ( 10.1083/jcb.200703064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V. 2013. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS ONE 8, e69986 ( 10.1371/journal.pone.0069986) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Comartin D, et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23, 1360–1366. ( 10.1016/j.cub.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 112.Sir JH, et al. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43, 1147–1153. ( 10.1038/ng.971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao H, et al. 2013. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat. Cell Biol. 15, 1434–1444. ( 10.1038/ncb2880) [DOI] [PubMed] [Google Scholar]
- 114.Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ. 2013. Deuterosome-mediated centriole biogenesis. Dev. Cell 27, 103–112. ( 10.1016/j.devcel.2013.08.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jenkins PM, McEwen DP, Martens JR. 2009. Olfactory cilia: linking sensory cilia function and human disease. Chem. Senses 34, 451–464. ( 10.1093/chemse/bjp020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cuschieri A, Bannister LH. 1975. The development of the olfactory mucosa in the mouse: electron microscopy. J. Anat. 119, 471–498. [PMC free article] [PubMed] [Google Scholar]
- 117.Cuschieri A, Bannister LH. 1975. The development of the olfactory mucosa in the mouse: light microscopy. J. Anat. 119, 277–286. [PMC free article] [PubMed] [Google Scholar]
- 118.Hepler PK. 1976. The blepharoplast of Marsilea: its de novo formation and spindle association. J. Cell Sci. 21, 361–390. [DOI] [PubMed] [Google Scholar]
- 119.Hodges ME, Wickstead B, Gull K, Langdale JA. 2012. The evolution of land plant cilia. New Phytol. 195, 526–540. ( 10.1111/j.1469-8137.2012.04197.x) [DOI] [PubMed] [Google Scholar]
- 120.Mizukami I, Gall J. 1966. Centriole replication. II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 29, 97–111. ( 10.1083/jcb.29.1.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaughn KC, Harper JD. 1998. Microtubule-organizing centers and nucleating sites in land plants. Int. Rev. Cytol. 181, 75–149. ( 10.1016/S0074-7696(08)60417-9) [DOI] [PubMed] [Google Scholar]
- 122.Ferree PM, McDonald K, Fasulo B, Sullivan W. 2006. The origin of centrosomes in parthenogenetic hymenopteran insects. Curr. Biol. 16, 801–807. ( 10.1016/j.cub.2006.03.066) [DOI] [PubMed] [Google Scholar]
- 123.Riparbelli MG, Stouthamer R, Dallai R, Callaini G. 1998. Microtubule organization during the early development of the parthenogenetic egg of the hymenopteran Muscidifurax uniraptor. Dev. Biol. 195, 89–99. ( 10.1006/dbio.1997.8841) [DOI] [PubMed] [Google Scholar]
- 124.Debec A, Sullivan W, Bettencourt-Dias M. 2010. Centrioles: active players or passengers during mitosis? Cell Mol. Life Sci. 67, 2173–2194. ( 10.1007/s00018-010-0323-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dingle AD, Fulton C. 1966. Development of the flagellar apparatus of Naegleria. J. Cell Biol. 31, 43–54. ( 10.1083/jcb.31.1.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fulton C, Dingle AD. 1971. Basal bodies, but not centrioles, in Naegleria. J. Cell Biol. 51, 826–836. ( 10.1083/jcb.51.3.826) [DOI] [PMC free article] [PubMed] [Google Scholar]