Abstract
The centrosome is the main microtubule (MT)-organizing centre of animal cells. It consists of two centrioles and a multi-layered proteinaceous structure that surrounds the centrioles, the so-called pericentriolar material. Centrosomes promote de novo assembly of MTs and thus play important roles in Golgi organization, cell polarity, cell motility and the organization of the mitotic spindle. To execute these functions, centrosomes have to adopt particular cellular positions. Actin and MT networks and the association of the centrosomes to the nuclear envelope define the correct positioning of the centrosomes. Another important feature of centrosomes is the centrosomal linker that connects the two centrosomes. The centrosome linker assembles in late mitosis/G1 simultaneously with centriole disengagement and is dissolved before or at the beginning of mitosis. Linker dissolution is important for mitotic spindle formation, and its cell cycle timing has profound influences on the execution of mitosis and proficiency of chromosome segregation. In this review, we will focus on the mechanisms of centrosome positioning and separation, and describe their functions and mechanisms in the light of recent findings.
Keywords: centrosome separation, centrosome positioning, Eg5
1. Centrioles and centrosomes
The centrosome is the main microtubule-organizing centre (MTOC) of animal cells. It initiates the de novo assembly of microtubules (MTs) from tubulin subunits and anchors, and organizes MTs [1]. Depending on the cell cycle phase, centrosomes consist of one or two centrioles, which are surrounded by the pericentriolar material (PCM). Centrioles determine the replication status of centrosomes while the PCM carries most of the MT nucleation activity [2]. The centriole is typically approximately 0.5 μm in length and 0.2 μm in diameter with nine MT triplets [3,4]. In S-phase, the daughter centriole assembles adjacent to the mother centriole. The two centrioles are first connected in a perpendicular orientation by an unknown mechanism and are maintained this way until telophase/G1 when the two centrioles separate (disengagement).
Owing to the pivotal role of centrosomes in MT nucleation, it was assumed for many years that centrosomes are essential for the survival of cells, especially during chromosome segregation, by promoting formation of the mitotic spindle. Findings in Drosophila melanogaster demonstrate that the centrosome as a structural entity is not essential for cell viability [5]. In the absence of centrosomes, mitotic spindle formation is driven by MTs that assemble around chromatin by the activity of the small GTPase Ran. Ran-GTP locally activates MT-associated proteins including the γ-tubulin complex that initiate MT nucleation [6].
Although dispensable for spindle formation and chromosome segregation in flies, centrosomes are required for the formation of cilia. Flies without centrosomes die soon after birth due to lack of cilia [5]. Moreover, via the organization of astral MTs, centrosomes play an important role in correct spindle positioning, which determines the cell fate in asymmetric cell division. Elegant studies in D. melanogaster revealed that fly neuroblasts lacking centrosomes divide symmetrically in 15% of cases [5]. Also in D. melanogaster male germ stem cells, correct bipolar spindle orientation relies on the positioning of the mother centriole [7,8]. Moreover, the asymmetric inheritance of old and new mother centrioles in mouse cells is necessary for neuron development, implying that centrosomes are important for mouse development [9].
Recent data using vertebrate DT40 cells as a model [10] suggest that loss of centrioles causes delays in bipolar spindle formation, high rates of chromosome instability and aneuploidy. These data argue against the notion that centrosomes are dispensable for mitosis in vertebrate cells.
2. Centriole disengagement
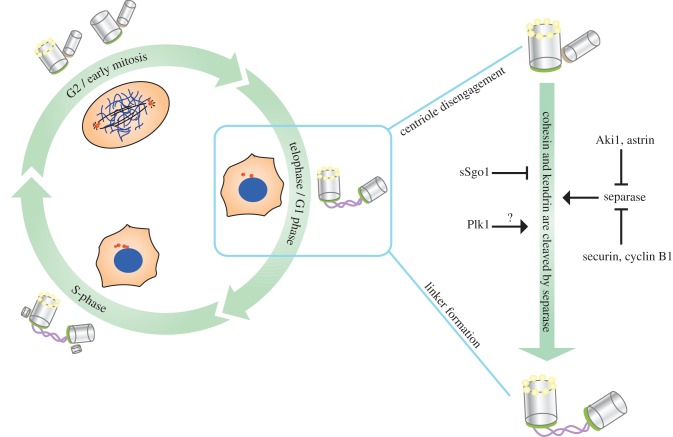
Canonical centriole duplication takes place in S-phase whereby the old mother centriole typically functions as an assembly platform for the new daughter centriole (figure 1). The new centriole assembles perpendicular to the mother centriole and the two centrioles stay connected by a poorly defined linker from S-phase until the end of mitosis [15]. The separation of centrioles, also termed as ‘centriole disengagement’, takes place in telophase/G1 [16]. Centriole disengagement is the licensing step for centriole duplication in the following S-phase and is therefore essential for centriole duplication [17,18].
Figure 1.
The centrosome cycle with special focus on centriole disengagement. The centrosome cycle starts in early S-phase when the daughter centriole assembles next to the mother centriole. Short daughter centrioles are formed in early S-phase and further elongate in G2. With cytokinesis each daughter cell inherits one centrosome with two joint centrioles. In late mitosis/G1, the two centrioles separate from each other. This process is called ‘centriole disengagement’ (depicted in the enlargement at the right) and is regulated by the protease separase. Cleavage of the cohesin subunit Rad21Scc1 and kendrin stimulates centriole separation [11,12]. It is unclear whether separase is regulated similarly at the centrosome and centromere. Securin and cyclin B1 are inhibitors of separase. At centrosomes a splice variant of Sgo1, sSgo1, additionally protects premature centriole disengagement. Moreover, Aki1 and astrin have been implicated as centrosome specific inhibitors of separase [13,14].
There is considerable complexity regarding the regulation of centriole disengagement (figure 1). One of the key proteins involved in this process is the protease separase (Espl1), which is well known for its role in sister chromatid segregation during the metaphase-to-anaphase transition. Sister chromatids are held together by the cohesin complex until the APC/C complex gets active. Upon the activation of separase, the cohesin subunit Scc1Rad21 is cleaved by separase, thus allowing the sister chromatids to be separated in anaphase [19,20]. A first hint for a function of separase at centrosomes came from the observation that a fraction of separase associates with centrosomes in mitosis [21]. The role of separase in centriole disengagement was first shown in Xenopus laevis egg extracts [17]. Human S-phase centrioles disengage upon incubation in X. laevis egg extracts with active separase. Following this study, construction of a human separase knockout cell line revealed that the lack of separase activity only partially prevented disengagement. However, chemical inhibition of polo-like kinase Plk1 in the separase knockout cell line completely prevented centriole disengagement. Thus, Plk1 and separase cooperate in centriole disengagement [22].
Even though cyclin B1 and securin are well known to be the key regulators of chromosome-bound separase [17,23]; regulation of separase at the centrosome seems to be more complex and may involve additional players, two of which were proposed to be astrin and Aki1. Both proteins were identified as inhibitors of centrosomal separase [13,14]. Consistently, depletion of either astrin or Aki1 caused premature separase activation, leading to the formation of multipolar mitoses with disengaged centrioles. Another player in controlling centriole disengagement is shugoshin (Sgo1), which prevents the premature separation of sister chromatids until separase gets active [24,25]. A splice variant of Sgo1, named as the small shugoshin (sSgo1), is predominantly associated with centrosomes. Expression of a dominant negative form of sSgo1 or depletion of the entire Sgo1 pool triggered centriole disengagement. At the same time, Plk1 activity is required for proper centrosome localization of sSgo1 [24]. Thus, sSgo1 has an ill-defined role in preventing premature centriole disengagement that is controlled by Plk1.
Involvement of separase in centriole disengagement pointed to a possible role of cohesin in this process. A number of reports indicate cohesin's localization to centrosomes [26–28]. Moreover, biochemical experiments with separase-resistant cohesin mutants suggest that the cohesin complex holds mother and daughter centrioles together until separase cleaves the cohesin subunit Scc1Rad21 [11,22]. However, there are many open questions concerning cohesin's role as the connector of centrioles. A comprehensive study demonstrating co-localization of cohesin subunits at the same centrosomal substructure through expression of functional cohesin-fluorescent protein fusions or through immunostaining of the endogenous proteins is lacking. Cohesin function in centriole engagement was derived from a heterologous system combining human centrosomes with X. laevis egg extracts. Demonstration of the role of cohesin in centriole engagement in a homologous system will be important. In addition, a recent report questioned this function of cohesin for centriole disengagement in Drosophila embryos [29]. Moreover, a recent study in C. elegans embryos showed that MT-dependent forces drive centriole disengagement and not primarily separase. Separase takes a role in disengagement only if the MT pulling forces are insufficient to separate centrioles [30].
Another puzzling observation in this context is the delay in the timing of centriole disengagement, which happens in telophase, in comparison to the timing of separase activation and sister chromatid separation at the metaphase-to-anaphase transition [16,23]. This means that separase either gets active at centrosomes later in the cell cycle or separase functions at the centrosome at metaphase-to-anaphase transition, but centriole disengagement is delayed due to additional mechanisms. Recently it was shown that separase cleaves the centrosomal protein pericentrin at the metaphase-to-anaphase transition and that cleaved pericentrin stays at the centrosome until the end of anaphase [12]. This may explain the lag between separase activation and centriole disengagement. In summary, although substrates of separase at the centrosomes have been unravelled, what cohesin embraces or how pericentrin regulates centriole disengagement is awaiting discovery.
3. Establishment of the centrosome linker
Following the disengagement of the two centrioles, a proteinaceous linker forms between the mother and the daughter centrioles to establish centrosome cohesion. This linker connects the two centrioles/centrosomes from G1 until the onset of mitosis when the linker disassembles and thus supports assembly of the mitotic spindle. How the establishment of the centrosomal linker is coupled to centriole disengagement is an appealing open question. Linker assembly is cell cycle regulated and requires a free proximal end of the centriole. With mitotic exit, linker proteins gain the ability to bind to the proximal end of the mother centriole [31,32]. However, at this stage the proximal end of the daughter centriole is still connected to the wall of the mother centriole and might be physically blocked. As soon as the two centrioles disengage, the proximal end of the daughter centriole becomes accessible and the linker proteins bind. Through a poorly understood self-assembly process additional linker proteins connect the pre-marked centriole ends giving rise to a flexible linker that joins the two centrosomes.
Proteins involved in the formation of the centrosomal linker were either identified as substrates of the NIMA-related kinase Nek2 that relieves the centrosome cohesion through phosphorylation or through siRNA screens for proteins that affect centrosome cohesion upon depletion (table 1). The best-known substrates of Nek2 are the so-called ‘centrosomal linker’ proteins C-Nap1 (Cep250) and rootletin [33,34]. C-Nap1 is found at the base of the mother centriole and two C-Nap1 pools are connected through rootletin fibres [41]. Nek2 efficiently phosphorylates C-Nap1 and rootletin in late G2 to allow their displacement from the centrosomes [34,41]. This ultimately leads to centrosome separation.
Table 1.
Centrosomal Linker Proteins.
| protein name | observation | references |
|---|---|---|
| C-Nap1 (Cep250) | C-Nap1 is found at the basal of the mother and disengaged daughter centrioles; it is the docking site for rootletin and interacts with Cep135 | [31,33] |
| rootletin | rootletin is a filamentous linker protein that connects two mother centrioles together | [32,34] |
| Cep68 | Cep68 co-localizes with rootletin; depletion causes disruption of centrosomal linker | [35] |
| Cep135 | Cep135 is important for the linker sustainability indirectly since it is required for C-Nap1 localization at the proximal end of centrioles; it binds to C-Nap1 via its C-terminal terminal half; the N-terminal half of Cep135 interacts with hSas-6 and CPAP | [36,37] |
| Cep215 (Cdk5Rap2) | Cep215 is important for centriole disengagement during mitosis and localizes to the base of the mother centriole together with the rootletin; depletion of Cep215 causes premature centrosome separation. Cep215 may not be a proper linker protein; it may influence linker integrity indirectly | [35,38] |
| LRRC45 | LRRC45 is a substrate of Nek2A as are C-Nap1 and rootletin, and is required for centrosome cohesion; like rootletin, it forms filaments between the centrosomes by binding to C-Nap1 | [39] |
| LGALS3BP | LGALS3BP localize to the filaments in between centrioles | [40] |
The human microceophaly protein Cep135 is a conserved centriolar protein that is associated at the proximal end of centrioles where it interacts with the centriole duplication factors hSAS-6 and CPAP [36]. Cep135 also interacts with the linker protein C-Nap1 (Cep250) [37]. Since Cep135 is permanently associated with centrioles, while the linker proteins are dissociated from centrioles during mitosis, Cep135 is likely the docking side to which linker proteins attach. Two separate domains of Cep135 support this docking model: the N-terminal part that targets the protein to centrioles and the C-terminal portion of Cep135 that is required for C-Nap1 interaction. Through these particular domains, the linker protein rootletin interacts with the N-terminal part of C-Nap1—the C-terminus of C-Nap1, in turn, interacts with Cep135 [33,34,37].
Another protein that is reported to be a linker protein is Cep68 [35], which is part of the rootletin network. Cep68 may be the protein that stabilizes rootletin fibres that self-assemble in head-to-tail orientation [35]. Recent evidence suggests the presence of two additional linker proteins named LLRC45 and LGALS3BP [38–40]. LLRRC45 was reported to interact with both C-Nap1 and rootletin. Thus, it could assist in the binding of rootletin fibres to the C-Nap1 platform at centrioles. Immuno-electron microscopy analyses showed the same fibre-like distribution of LGALS3BP as rootletin, raising the possibility that LGALS3BP interacts with linker components [40]. The Rho-associated protein kinase p160ROCK has been observed to localize to mother centrioles and to the linker, and inhibition of p160ROCK with the inhibitor Y-27632 increased the distance between mother and daughter centrioles in G1 cells. Moreover, p160ROCK is important for the centrosome attachment to the nuclear envelope (NE) during interphase, and inhibition of p160ROCK caused premature midbody targeting of the mother centriole during mitosis [42].
A lot is revealed about the composition of the centrosomal linker, but little is known about the connection between centriole disengagement and linker formation, the functional importance of the centrosomal linker and its precise regulation during the cell cycle.
4. Centrosome positioning
Cells in G1 phase have two disengaged centrioles that are connected by the centrosomal linker. Live cell imaging experiments with GFP-centrin marked centrioles showed that the mother centriole remains near the cell centre. By contrast, the daughter centriole is much more mobile and moves within the cytoplasm. Centriole movement is independent of the NE, but requires actin and the MT cytoskeleton [16].
What is the function of rapid daughter centriole movement in G1? In HeLa cells the repositioning of the mobile daughter centriole to the midbody correlates with the final phase of cell abscission, the cleavage of MTs adjacent to the midbody [16]. Interestingly, both centrioles have the ability to nucleate MTs, but it is only the mother centriole that stably anchors MTs through appendages at the distal end of the centriole. The daughter centriole develops these appendages only in the next mitosis. MTs nucleated by the mobile daughter centriole are released to the cell periphery, and this has important functions in migrating cells [43].
Centrosomes are important for organizing and positioning the Golgi apparatus of a cell. In mammalian cells, the Golgi is a single-copy organelle that localizes around the centrosome. After mitosis, Golgi fragments first assemble at the cell periphery. MTs organized by the centrosome then provide the tracks to transport Golgi fragments to the cell centre where the fragments fuse into one Golgi ribbon. Recently, it was shown that disturbing Golgi positioning by disconnecting it from the centrosome has a dramatic effect on directional cell migration [44]. Normally, the centrosome places the Golgi apparatus towards the leading edge of the cell. This positioning supports polarized secretion of cargo vesicles and is therefore essential for cell migration.
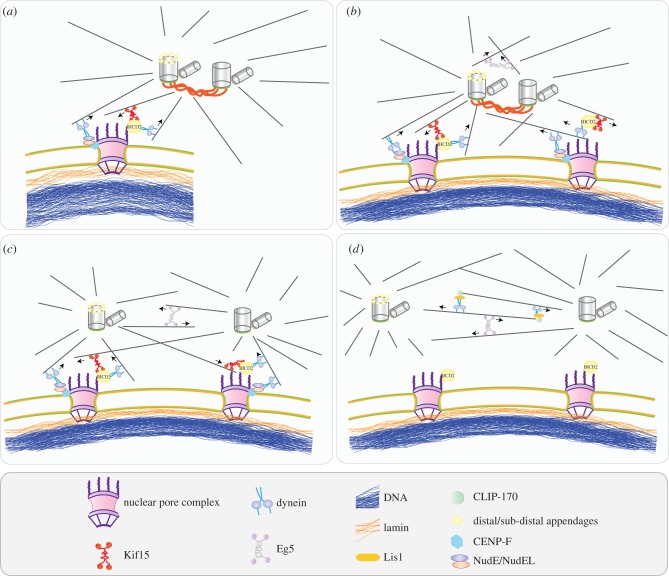
In G2 and prophase, centrosomes are anchored to the NE through at least two mechanisms (figure 2). First, RanBP2/Nup358 at the cytoplasmic side of the nuclear pore complex (NPC) recruits in G2 the adaptor protein bicaudal D (BICD2) which then tethers the dynein/dynactin complex to NPCs (figure 2a–c). Dynein/dynactin interacts with MTs organized by the centrosomes and through this mechanism positions the centrosomes at the NE [46]. The second pathway operates via centromere-associated CENP-F protein and the Nup133 subunit of the Nup107–160 nuclear pore subcomplex [45]. The N-terminal domain of Nup133 interacts with CENP-F which becomes positioned at the NE in G2/M. CENP-F in turn recruits NudE/NudEL (NudE and NudEL are related proteins that interact with cytoplasmic dynein) to the NE. Both pathways cooperate jointly in centrosome recruitment to the NE. Data from [45] indicate a role of the CEBP-F/Nup133 pathway in timely establishment of a properly positioned spindle.
Figure 2.
Centrosome positioning around the nucleus. (a) In G2 phase, the mother centrosome is attached to the NPC. Two mechanisms account for this attachment. First, Nup358, a component of the NPC, minus-end directed motor protein dynein and the adaptor protein BICD2 form a complex which attaches to astral MTs emanating from the mother centrosome. Second, the N-terminal domain of Nup133 interacts with CENP-F that becomes positioned at the NE in G2/M. Dynein binds via the NudE/NudEL/CENP-F complex. Recruitment of BICD2 or CENP-F to the NPC does not depend on each other, which implies that the two pathways are independent from each other [45]. (b) When both centrosomes attach to the nucleus, the pull-and-push forces created by dynein and the kinesin motor Kif1 separate the centrosomes, and the centrosomes start to slide on the nucleus by being attached to the next NPC. (c) When they are separated enough to form anti-parallel MTs, dynein and the plus-end directed motor protein Eg5 become involved in further separation of the centrosomes. Dynein and Eg5 create opposing forces in order to balance the separation. Eg5 pushes the centrosomes away from each other, although dynein tries to keep them together by forming a complex with plus-end protein CLIP-170 and adaptor protein Lis1. Dynein localizes to MTs arising from one centrosome and catches the plus-end of MTs from the other centrosome. By pulling the MTs, dynein creates an inward force opposite to Eg5. (d) Prophase centrosome separation is completed when the separated centrosomes detach from the nucleus.
The events we have described above concern the behaviour of centrosomes in cultured human cells. Interesting modes of centrosome positioning have also been observed in tissues. Cytotoxic T lymphocytes (CTLs) that destroy virally infected and tumorigenic cells present a fascinating example in which the centrosome moves to the immunological synapse and delivers lytic granules to the site where the CTL contacts the infected or transformed cell [47]. Interestingly, the centrosome contacts the plasma membrane during target-cell killing.
Another instance where centrosome positioning and movement has an important function is during brain development in mice, where the neurons move inside-out towards the surface and where the nuclei show extended movements. Interestingly, centrosome–NE connections are important for nuclear movements. The integral membrane proteins SUN1 and SUN2 span the NE from the nuclear side and carry the SUN domain in the lumen of the NE. The SUN domain interacts with the KASH domain of Syne-1/nesprin-1 and Syne-2/nesprin-2 (several excellent reviews have been published on SUN-KASH interactions and topology—see [48–53]). These KASH domain proteins span the cytoplasmic side of the NE and the cytoplasmic domains of Syne-1/nesprin-1 and Syne-2/nesprin-2 mediate interactions with the centrosome via dynein/dynactin and kinesin complexes [54]. Syne-2 mutant mice display severe defects in learning and memory, emphasizing the importance of the centrosome–NE connection during neurogenesis.
The KASH-SUN domain tandem is also used in the social amoebae Dictyostelium discoideum to position the centrosome to the NE [55,56]. The Kin-I kinesin Kif9 carries a transmembrane domain at its C-terminus that places the C-terminal fragment of Kif9 in the lumen of the NE where it may bind to the SUN domain of Sun1. The motor domain of Kif9 interacts with centrosome-organized MTs, depolymerizes MTs and in this way places the centrosomes on the NE.
5. Centrosome disjunction: dissolution of the linker and control of Nek2 function at the centrosomes
During G2, before entry into mitosis, the centrosomal linker is dissolved by the so-called ‘centrosome disjunction’ process to allow centrosomes to separate and form the two poles of the bipolar spindle [57]. The best-studied kinase responsible for dissolving the centrosomal linker is the NIMA-related kinase Nek2 (figure 3). Nek2 shows an increased kinase activity starting in late S-phase and peaking in G2, which coincides with the timing of centrosome separation. Nek2A, which is a splice variant of Nek2, is degraded by the APC/C-proteasome system during prometaphase with a similar timing as cyclin A [63]. Overexpression of the active kinase leads to premature centrosome splitting independent of the cell cycle phase, whereas RNAi-mediated downregulation of Nek2 inhibits centrosome separation, albeit without significantly affecting cell cycle progression [57,64]. The function of Nek2A (a splice variant of Nek2) at the centrosomes is counteracted by protein phosphatase 1 (PP1) (figure 3). PP1 binds directly to a KVHF motif in the non-catalytic C-terminal region of Nek2A and dephosphorylates and thereby inactivates Nek2A in vitro. Conversely, Nek2A can phosphorylate and inhibit PP1 [65]. The tertiary complex consisting of C-Nap1, Nek2A and PP1γ is important for regulating the level of centrosome separation by C-Nap1 phosphorylation since C-Nap1 is a substrate not only of Nek2A but also PP1γ [65].
Figure 3.
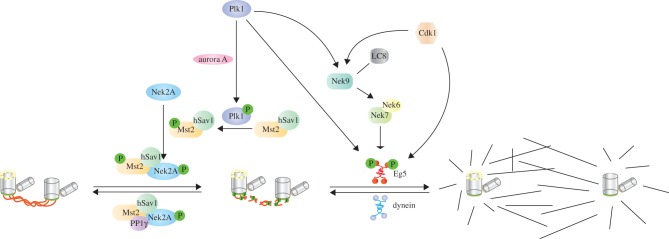
Molecules involved in centrosome separation. Aurora A is found on the top of the linker dissolution pathway. Plk1 is activated upon phosphorylation by aurora A. Plk1, in turn, phosphorylates Mst2 that is in complex with hSav1. Phosphorylation of Mst2 by Plk1 increases the activity of Nek2A and decreases the activity of PP1γ towards C-Nap1 and rootletin, by altering the complex formation between these proteins [58]. Phosphorylated C-Nap1 and rootletin diffuse away from the centrosome, and the centrosomes become ready for the separation. Plk1 additionally promotes centrosome separation directly by phosphorylating Eg5 and indirectly by regulating the Nek9-Nek6/7-Eg5 pathway. Nek9 is also regulated by LC8, which is a part of dynein complex and functions in linking dynein to different cargo proteins [59]. LC8 enhances the activation of auto-phosphorylated Nek9. Moreover, binding of LC8 impairs the interaction between Nek9 and Nek6 [60]. Cdk1 is also involved in centrosome separation by directly phosphorylating Eg5 [61,62].
The regulation of centrosome disjunction is not only at the level of substrates, namely C-Nap1 and rootletin, but also at the level of Nek2A and PP1γ. Nek2A and PP1γ regulate the activities of each other, but they are also subject to higher level regulation by components of the Hippo pathway (figure 3) and by epidermal growth factor (EGF) signalling ([58] and see §7). Being the main kinase responsible for centrosome disjunction, Nek2A is subject to a higher level of regulation by the components of the Hippo pathway. The Hippo pathway is a signalling network composed of many tumour suppressors and is well known for controlling cell proliferation and apoptosis in flies and humans [66]. Hippo pathway effector kinase sterile-20 like kinase Mst2 and the scaffold protein hSav1 regulate Nek2A [58,67]. Nek2A is in a complex with Mst2, the scaffold protein hSav1 and PP1γ. The phosphorylation of Nek2A by Mst2 targets it to the centrosome, which in turn causes centrosome disjunction by phosphorylating C-Nap1 and rootletin [67]. Mst2 kinase is shown to be a substrate of Plk1, which orchestrates the counteracting activities of PP1γ and Nek2A. Phosphorylation of Mst2 by Plk1 decreases the complex formation of Mst2-PP1γ-Nek2A [58]. Interestingly, many Hippo pathway components localize to the centrosomes, and they are reported to have crucial roles not only for centrosome disjunction, but also for centrosome duplication [68].
6. Centrosomes on the move: action of the motor proteins
Upon resolution of the linker, centrosomes move to two opposite poles to form the bipolar spindle. Surprisingly, inhibition of Nek2/Mst2 kinases does not perturb the formation of the bipolar spindles. This is explained by the redundant action of Mst2/Nek2 kinases and the kinesin-5 motor protein Eg5 on linker dissolution. Thus, centrosome separation can be considered as a two-step process, whereby the centrosomes become separated cooperatively by the Mst2-Nek2A and the Eg5 pathways [67,69] (figure 3). It is important to note that Eg5 is able to break the linker by force even in the absence of Mst2/Nek2 activity. In this case, linker proteins remain attached to the two centrioles in mitosis [67].
Eg5 is the principal force generator that drives centrosome separation. With the exception of C. elegans, inhibition or depletion of Eg5 motor induces monopolar spindles in all tested organisms [70]. Eg5 belongs to the kinesin-5 subfamily of motor proteins, which are homotetrameric, plus-end directed motors [71,72]. The motor domains of this subfamily face outwards [73], which allows sliding of the anti-parallel MTs in opposing directions [74]. In interphase cells, Eg5 is dispersed in the cytoplasm; however, in mitosis it localizes to the mitotic spindles and is enriched at the centrosomes [70]. Enrichment of Eg5 at the centrosomes has been shown to initiate the motor-driven centrosome separation process. A cascade of kinases controls this localizational switch (figure 3). At the top of this pathway are the cyclin-dependent kinase Cdk1 and the polo-like kinase Plk1 [75]. Cdk1 and Plk1 are necessary for the activation of NIMA-related kinase 9 (Nek9). Cdk1 presumably works as a priming kinase for Plk1, allowing Plk1 to bind and subsequently phosphorylate Nek9, thus activating it [75]. Nek9 is subject to another level of regulation by LC8. LC8 is a component of the dynein complex, which links dynein to cargo complexes. LC8 stimulates the auto-activation of Nek9 and inhibits the binding of Nek9 to Nek6 [60]. Further downstream of this activation, Nek6/Nek7 kinases eventually regulate centrosome separation; however, the exact mechanism of Nek6/Nek7 regulation is not fully understood. Eg5 phosphorylation by both Cdk1 and Nek6/Nek7 is crucial for controlling its MT-binding affinity and its localization to the spindle poles, respectively [61,75]. Although Cdk1 activity is required for proper MT binding of Eg5, Cdk1 is not essential for centrosome separation in DT40 chicken and HeLa human cell lines. However, without Cdk1 activity the separation of centrosomes is delayed for around 2 h [69]. The crosstalk between these pathways controlling Eg5 on centrosome separation remains to be elucidated.
Two lines of evidence suggest that Eg5 is not the sole motor protein responsible for the movement of the centrosomes. First, inhibition of Eg5 in metaphase does not cause collapse of the spindle into a monopolar structure [61,76]. Second, the movements of the two centrosomes are independent of each other and do not entirely depend on the anti-parallel sliding activity of Eg5, thus requiring an additional force generator [77]. Accordingly, dynein has been shown to be an important player of centrosome separation in human cells [78–80] (figure 2). Dynein in coordination with its partner protein Lis1 and the MT binding protein CLIP-170 antagonize Eg5 function at the centrosomes. CLIP-170 is a MT plus-end binding protein, and links the MTs to the subcellular structures, such as dynein and Lis1 [81]. Depletion of dynein, Lis1 or CLIP-170 restores bipolar spindle formation in the presence of substantial amounts of Eg5 inhibitor [79]. Initially, it was proposed that Eg5 and dynein simply work in an antagonistic fashion due to the fact that dynein localizes on anti-parallel MTs and causes spindle collapse when Eg5 is inhibited. However, recent data from titration experiments challenged this idea by suggesting that dynein creates an opposing force indirectly, not by force generation directly opposite to Eg5, in contradiction to the pull-and-push theory [82].
In addition, dynein localizes to the NE in G2 phase, and this pool of dynein is required for centrosome separation (figure 3). NE-bound dynein becomes essential for the separation of centrosomes when Eg5 is not active; however, it is functional even when Eg5 is not inhibited [83]. Dynein captures the MTs from the centrosomes and pushes the centrosomes along the NE to opposing sites. How MTs that had emanated from the centrosomes move the two centrosomes in opposite directions is still unclear.
Apart from dynein and Eg5, the more recently identified kinesin Kif15 (kinesin-12; figure 2) was shown to contribute to centrosome separation [84,85]. Depletion of Kif15 does not prevent the formation of bipolar spindles; however, it becomes essential when Eg5 is inhibited [85,86]. Kif15 localizes to the K-MTs after bipolar spindles are formed, and helps Eg5 to stabilize the bipolar spindles [85,87]. Moreover, in a cell line where Eg5 is not dominant for centrosome separation, Kif15 is recruited to the non-K-MTs and replaces the function of Eg5, thus initiating centrosome separation [87]. Kif15 has recently been shown not only to aid Eg5 in stabilizing the bipolar spindles but also to create an opposing force to prevent the formation of overly separated centrosomes [87]. Together, these data propose distinct functions for Kif15 in centrosome separation: it first helps Eg5 to form and maintain the bipolar spindle, and later it creates an opposing force to limit its length. In summary, the balance between the forces exerted at the centrosome is undoubtedly crucial for establishing the bipolar spindle.
When the centrosomes are separated, external factors help the cell to determine the direction of bipolar spindle [88]. The extracellular matrix (ECM) regulates the distribution of actin in interphase cells such that the actin binding proteins cortactin and ezrin are concentrated at membrane ruffles. These changes are also maintained in mitosis so that the ECM determines the direction of bipolar spindle formation by changing the actin cytoskeleton [88].
7. Timing is everything
As discussed in detail in §§5 and 6, centrosomes must be separated to support bipolar spindle assembly. What is the timing of centrosome separation relative to cell cycle progression and how is it controlled? Recent work has demonstrated that the timing of centrosome separation is not restricted to a specific stage of the cell cycle but varies with cell type. In non-cancer-derived RPE-1 cells, most of the cells enter mitosis with separated centrosomes [89]. Observation of HeLa cells revealed that half of the cells use the classical prophase pathway where centrosomes separated before NE breakdown (prophase pathway). The remaining half, however, enter mitosis without separated centrosomes and centrosome separation occurs after NE breakdown (prometaphase pathway) [90].
Another important determinant for the timing of centrosome separation is the growth conditions. Sherline & Mascardo [91] initially observed that the incubation of HeLa cells with the EGF results in rapid and premature separation of the centrosomes. Thirty years later, the biological outcome of this interesting observation was investigated in detail. Addition of EGF dissolves the centrosomal linker by increasing the local concentration of the upstream kinases Mst2 and Nek2 at the centrosomes, thus leading to premature centrosome separation [92]. Interestingly, this effect is cell type dependent since many cancer cells have a high degree of separated centrosomes due to elevated levels of EGF receptor (EGFR) expression. Regulation of centrosome disjunction therefore constitutes a previously unknown function of EGF signalling.
Despite accumulating evidence on the different routes of centrosome separation, whether or not the timing of centrosome separation is an important determinant for mitotic progression and chromosome segregation has not been directly addressed until recently. Time-lapse movies of centrosome and kinetochore movements in untransformed RPE-1 cells combined with computational modelling revealed that having separated centrosomes at the time of NE breakdown is advantageous for the cells [63]. When cells enter mitosis with separated centrosomes at NE breakdown, the metaphase spindle is assembled more rapidly than in cells where centrosomes separate after NE breakdown. Modelling of centrosome and chromosome movements suggested that the MT density should be increased within the spindle when the centrosomes are on opposite sides of the nucleus at NE breakdown [89], thus capturing kinetochores faster than in cells with joint centrosomes at the time of NE breakdown. Moreover, live cell imaging experiments combined with high-resolution confocal imaging of Ptk1 cells suggested that incomplete centrosome separation at the time of NE breakdown increases the likelihood of acquiring merotelic attachments [93]. Considering that merotelic attachments are the principal cause of chromosome missegregation in anaphase, these data highlight the importance of timing of centrosome separation in relation to NE breakdown.
Why do the cells fail to assemble correct attachments? Computational models suggest the presence of a ‘blocking effect’ of the two centrosomes towards each other when they are in close proximity. As the centrosomes separate further, more and more chromosomes become available to the MTs emanating from both poles, and this increases the chances of correct syntelic attachments due to symmetric outgrowth of MTs from both spindle poles [94]. In line with these observations and models [89,90,92,94,95], EGF-induced early centrosome separation promotes a faster mitotic progression with fewer chromosome segregation errors [92]. Induction of early centrosome separation either by addition of EGF to the medium or by expression of the EGFR decreases the requirement of Eg5 for mitotic progression [92]. The relationship between EGFR signalling and Eg5 is noteworthy since both molecules are common targets of cancer therapy as discussed below.
Addition of EGF activates this signalling pathway by inducing the receptor dimerization and trans phosphorylation of the C-terminal tail of the EGFR, which upon phosphorylation becomes a scaffold for the recruitment of effector proteins. The downstream effector pathways of the EGFR signalling are overwhelmingly complex, and they contribute to different functions in the cell. Among EGFR effectors, Akt and GRK2 kinases are specifically involved in centrosome separation. While Akt is one of the best-studied effector kinases in EGFR signalling, the role of GRK2 in this pathway has only recently been worked out [92,95]. GRK2 belongs to the family of G-protein-coupled receptor (GPCR) kinases which are involved in a wide variety of cellular processes by activation of several signalling pathways. This family of kinases regulates the activity of GPCRs and contributes to their signalling activity by phosphorylating their intracellular domains and eventually causing impairment or desensitization of the receptor. Although most localize to the membranes, some members of this family of kinases were proposed to have distinct localizations and functions. Recent data have suggested that GRK2 is one such kinase that localizes to the centrosomes and mediates the signalling initiated by the addition of EGF. Upon EGF addition, GRK2 phosphorylates and activates Mst2, thereby initiating the separation of the two centrosomes.
Mitosis and the mitotic cytoskeleton, including the centrosome have long been thought of as attractive cancer targets. Indeed, inhibitors against many mitotic protein kinases that also localize to the centrosomes, such as Plk1, Cdk1, aurora kinase A and Chk2 are currently undergoing clinical trials [96]. Upcoming therapeutics also aim to target kinesins, the motor proteins responsible for accurate execution of cell division. Among these, Eg5 is the most widely studied kinesin and its inhibitors such as monastrol have entered clinical trials but shown little success because of unforeseen side effects [97]. Targeting mitotic kinases as well as kinesins is not a straightforward approach considering that they have several binding partners and functions. Drug development has typically targeted specific components of molecular networks altered in and essential for cancer cell viability. The weaknesses of this strategy are now becoming apparent given the ineffective or short-lived responses to many single agent therapeutics. Acquisition of specific resistance mutations by tumour cells probably underlies these shortcomings. A logical solution would thus be to target distorted signalling pathways using multiple agents, decreasing the likelihood of any single clone having the repertoire of individual resistance mutations necessary to withstand the combined therapy. For instance, promising results were recently presented from a phase I/II study which was investigating the proto-oncogene BRAF inhibitor dabrafenib in combination with mitogen-activated protein kinase MEK1/2 inhibitor trametinib. The results showed that the trametinib therapy was significantly more effective in patients previously treated with the BRAF inhibitor [98,99]. Similarly, targeting cancer types with high rates of early centrosome separation by combinatorial therapy against Eg5 and EGFR pathways might be plausible. As opposed to normal cells, the cells with elevated rates of centrosome separation seem to be less dependent on Eg5 for cell division and thus they become more resistant towards Eg5 inhibitors such as monastrol and S-trityl-l-cysteine [92]. However, targeting both the EGFR and Eg5 pathways with lower dosages than needed in single drug treatment seems to be most detrimental for these cells. In summary, combined targeting of Eg5 and EGFR in cells with elevated levels of centrosomes might prove not only to be effective but also avoid the potential side effects arising from targeting central mitotic kinases.
Acknowledgements
We apologize to those of our colleagues whose important contributions could not be acknowledged due to space constraints. We thank Dr A. Khmelinskii and C. Buccitelli for their comments on the manuscript.
Funding statement
The work of E.S. is supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) Schi 295/3. B.R.M. is funded by a Marie Curie Intra-European Fellowship for career development (IEF).
References
- 1.Bettencourt-Dias M, Glover DM. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463. ( 10.1038/nrm2180) [DOI] [PubMed] [Google Scholar]
- 2.Sluder G, Rieder CL. 1985. Centriole number and the reproductive capacity of spindle poles. J. Cell Biol. 100, 887–896. ( 10.1083/jcb.100.3.887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azimzadeh J, Bornens M. 2007. Structure and duplication of the centrosome. J. Cell Biol. 120, 2139–2142. ( 10.1242/jcs.005231) [DOI] [PubMed] [Google Scholar]
- 4.Bornens M, Azimzadeh J. 2007. Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 607, 119–129. ( 10.1007/978-0-387-74021-8_10) [DOI] [PubMed] [Google Scholar]
- 5.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125, 1375–1386. ( 10.1016/j.cell.2006.05.025) [DOI] [PubMed] [Google Scholar]
- 6.Kalab P, Heald R. 2008. The RanGTP gradient: a GPS for the mitotic spindle. J. Cell Biol. 121, 1577–1586. ( 10.1242/jcs.005959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita YM, Jones DL, Fuller MT. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550. ( 10.1126/science.1087795) [DOI] [PubMed] [Google Scholar]
- 8.Yamashita YM, et al. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518–521. ( 10.1126/science.1134910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Tsai J-W, Imai JH, Lian W-N, Vallee RB, Shi S-H. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947–955. ( 10.1038/nature08435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sir JH, Putz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 13, 747–756. ( 10.1083/jcb.201309038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schockel L, Möckel M, Mayer B, Boos D, Stemmann O. 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 13, 966–972. ( 10.1038/ncb2280) [DOI] [PubMed] [Google Scholar]
- 12.Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22, 915–921. ( 10.1016/j.cub.2012.03.048) [DOI] [PubMed] [Google Scholar]
- 13.Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178, 345–354. ( 10.1083/jcb.200701163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura A, Arai H, Fujita N. 2009. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J. Cell Biol. 187, 607–614. ( 10.1083/jcb.200906019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paintrand M, Moudjou M, Delacroix H, Bornens M. 1992. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108, 107–128. ( 10.1016/1047-8477(92)90011-X) [DOI] [PubMed] [Google Scholar]
- 16.Piel M, Meyer P, Khodjakov A, Rieder CL, Borners M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317–330. ( 10.1083/jcb.149.2.317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsou MF, Stearns T. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951. ( 10.1038/nature04985) [DOI] [PubMed] [Google Scholar]
- 18.Wang WJ, Soni RK, Uryu K, Bryan Tsou M-F. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739. ( 10.1083/jcb.201101109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067–1076. ( 10.1016/S0092-8674(00)81211-8) [DOI] [PubMed] [Google Scholar]
- 20.Uhlmann F, Lottspeich F, Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37–42. ( 10.1038/21831) [DOI] [PubMed] [Google Scholar]
- 21.Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. 2003. Processing, localization, and requirement of human separase for normal anaphase progression. Proc. Natl Acad. Sci. USA 100, 4574–4579. ( 10.1073/pnas.0730733100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsou MF, Wang W-J, George KA, Uryu K, Stearns T, Jallepalli PV. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344–354. ( 10.1016/j.devcel.2009.07.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. 2001. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726. ( 10.1016/S0092-8674(01)00603-1) [DOI] [PubMed] [Google Scholar]
- 24.Tsang WY, Dynlacht BD. 2008. sSgo1, a guardian of centriole cohesion. Dev. Cell 14, 320–322. ( 10.1016/j.devcel.2008.02.008) [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W. 2008. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell 14, 331–341. ( 10.1016/j.devcel.2007.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong RW, Blobel G. 2008. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl Acad. Sci. USA 105, 15 441–15 445. ( 10.1073/pnas.0807660105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K. 2001. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 276, 47 575–47 582. ( 10.1074/jbc.M103364200) [DOI] [PubMed] [Google Scholar]
- 28.Kong X, Ball AR, Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. 2009. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol. Biol. Cell 20, 1289–1301. ( 10.1091/mbc.E08-04-0419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira RA, Nasmyth K. 2013. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr. Biol. 23, R601–R603. ( 10.1016/j.cub.2013.04.003) [DOI] [PubMed] [Google Scholar]
- 30.Cabral G, Sanegre Sans S, Cowan CR, Dammermann A. 2013. Multiple mechanisms contribute to centriole separation in C. elegans. Curr. Biol. 23, 1380–1387. ( 10.1016/j.cub.2013.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayor T, Stierhof Y-D, Tanaka K, Fry AM, Nigg EA. 2000. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151, 837–846. ( 10.1083/jcb.151.4.837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. 2002. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 159, 431–440. ( 10.1083/jcb.200207153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Adamian M, Li T. 2006. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell 17, 1033–1040. ( 10.1091/mbc.E05-10-0943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171, 27–33. ( 10.1083/jcb.200504107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graser S, Stierhof YD, Nigg EA. 2007. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Biol. 120, 4321–4331. ( 10.1242/jcs.020248) [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Chang C-W, Hsu W-B, Tang C-JC, Lin Y-N, Chou E-J, Wu C-T, Tang TK. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32, 1141–1154. ( 10.1038/emboj.2013.56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim K, Lee S, Chang J, Rhee K. 2008. A novel function of CEP135 as a platform protein of C-NAP1 for its centriolar localization. Exp. Cell Res. 314, 3692–3700. ( 10.1016/j.yexcr.2008.09.016) [DOI] [PubMed] [Google Scholar]
- 38.Barrera JA, Kao L-R, Hammer RE, Seemann J, Fuchs JL, Megraw TL. 2010. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell 18, 913–926. ( 10.1016/j.devcel.2010.05.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He R, Huang N, Bao Y, Zhou H, Teng J, Chen J. 2013. LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Rep. 4, 1100–1107. ( 10.1016/j.celrep.2013.08.005) [DOI] [PubMed] [Google Scholar]
- 40.Fogeron ML, et al. 2013. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat. Commun. 4, 1531 ( 10.1038/ncomms2517) [DOI] [PubMed] [Google Scholar]
- 41.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574. ( 10.1083/jcb.141.7.1563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevrier V, Piel M, Collomb N, Saoudi Y, Frank R, Paintrand M, Narumiya S, Bornens M, Job D. 2002. The Rho-associated protein kinase p160ROCK is required for centrosome positioning. J. Cell Biol. 157, 807–817. ( 10.1083/jcb.200203034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abal M, Piel M, Bouckson-Castaing V, Mogensen M, Sibarita JB, Bornens M. 2002. Microtubule release from the centrosome in migrating cells. J. Cell Biol. 159, 731–737. ( 10.1083/jcb.200207076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. 2011. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193, 917–933. ( 10.1083/jcb.201011014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. 2011. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol. 192, 855–871. ( 10.1083/jcb.201007118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Splinter D, et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8, e1000350 ( 10.1371/journal.pbio.1000350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. 2006. Centrosome polarization delivers secretory granules to the immunological synapse. Nature 443, 462–465. ( 10.1038/nature05071) [DOI] [PubMed] [Google Scholar]
- 48.Starr DA. 2011. KASH and SUN proteins. Curr. Biol. 21, R414–R415. ( 10.1016/j.cub.2011.04.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, et al. 2012. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res 22, 1440–1452. ( 10.1038/cr.2012.126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke B. 2012. It takes KASH to hitch to the SUN. Cell 149, 961–963. ( 10.1016/j.cell.2012.05.004) [DOI] [PubMed] [Google Scholar]
- 51.Hiraoka Y, Dernburg AF. 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605. ( 10.1016/j.devcel.2009.10.014) [DOI] [PubMed] [Google Scholar]
- 52.Razafsky D, Hodzic D. 2009. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186, 461–472. ( 10.1083/jcb.200906068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. 2009. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol. Life Sci. 66, 1518–1533. ( 10.1007/s00018-008-8713-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187. ( 10.1016/j.neuron.2009.08.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leo M, Santino D, Tikhonenko I, Magidson V, Khodjakov A, Koonce MP. 2012. Rules of engagement: centrosome-nuclear connections in a closed mitotic system. Biol. Open 1, 1111–1117. ( 10.1242/bio.20122188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tikhonenko I, Magidson V, Gräf R, Khodjakov A, Koonce MP. 2013. A kinesin-mediated mechanism that couples centrosomes to nuclei. Cell Mol. Life Sci. 70, 1285–1296. ( 10.1007/s00018-012-1205-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faragher AJ, Fry AM. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 14, 2876–2889. ( 10.1091/mbc.E03-02-0108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mardin BR, Agircan FG, Lange C, Schiebel E. 2011. Plk1 controls the Nek2A-PP1gamma antagonism in centrosome disjunction. Curr. Biol. 21, 1145–1151. ( 10.1016/j.cub.2011.05.047) [DOI] [PubMed] [Google Scholar]
- 59.Rapali P, Szenes Á, Radnai L, Bakos A, Pál G, Nyitray L. 2011. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278, 2980–2996. ( 10.1111/j.1742-4658.2011.08254.x) [DOI] [PubMed] [Google Scholar]
- 60.Regue L, Sdelci S, Bertran MT, Caelles C, Reverter D, Roig J. 2011. DYNLL/LC8 protein controls signal transduction through the Nek9/Nek6 signaling module by regulating Nek6 binding to Nek9. J. Biol. Chem. 286, 18 118–18 129. ( 10.1074/jbc.M110.209080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Niggt EA. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. ( 10.1016/0092-8674(95)90142-6) [DOI] [PubMed] [Google Scholar]
- 62.Blangy A, Arnaud L, Nigg EA. 1997. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 272, 19 418–19 424. ( 10.1074/jbc.272.31.19418) [DOI] [PubMed] [Google Scholar]
- 63.Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20, 7117–7127. ( 10.1093/emboj/20.24.7117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fletcher L, Cerniglia GJ, Nigg EA, Yen TJ, Muschel RJ. 2004. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat. Res. 162, 128–135. ( 10.1667/RR3211) [DOI] [PubMed] [Google Scholar]
- 65.Helps NR, Luo X, Barker HM, Cohen PTW. 2000. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J. 349, 509–518. ( 10.1042/0264-6021:3490509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu FX, et al. 2013. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27, 1223–1232. ( 10.1101/gad.219402.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. 2010. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176. ( 10.1038/ncb2120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hergovich A, Cornils H, Hemmings BA. 2008. Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim. Biophys. Acta 1784, 3–15. ( 10.1016/j.bbapap.2007.07.017) [DOI] [PubMed] [Google Scholar]
- 69.Smith E, et al. 2011. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 30, 2233–2245. ( 10.1038/emboj.2011.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferenz NP, Gable A, Wadsworth P. 2010. Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 21, 255–259. ( 10.1016/j.semcdb.2010.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543. ( 10.1038/359540a0) [DOI] [PubMed] [Google Scholar]
- 72.Cole DG, Saxton WM, Sheehan KB, Scholey JM. 1994. A ‘slow’ homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 269, 22 913–22 916. [PMC free article] [PubMed] [Google Scholar]
- 73.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. 1996. A bipolar kinesin. Nature 379, 270–272. ( 10.1038/379270a0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118. ( 10.1038/nature03503) [DOI] [PubMed] [Google Scholar]
- 75.Bertran MT, Sdelci S, Regué L, Avruch J, Caelles C, Roig J. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30, 2634–2647. ( 10.1038/emboj.2011.179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waters JC, Cole RW, Rieder CL. 1993. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J. Cell Biol. 122, 361–372. ( 10.1083/jcb.122.2.361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanenbaum ME, Medema RH. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell 19, 797–806. ( 10.1016/j.devcel.2010.11.011) [DOI] [PubMed] [Google Scholar]
- 78.Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135, 399–414. ( 10.1083/jcb.135.2.399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanenbaum ME, Macůrek L, Galjart N, Medema RH. 2008. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27, 3235–3245. ( 10.1038/emboj.2008.242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gonczy P, Pichler S, Kirkham M, Hyman AA. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147, 135–150. ( 10.1083/jcb.147.1.135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pereira AL, et al. 2006. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell 17, 4526–4542. ( 10.1091/mbc.E06-07-0579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Florian S, Mayer TU. 2012. The functional antagonism between Eg5 and dynein in spindle bipolarization is not compatible with a simple push-pull model. Cell Rep. 1, 408–416. ( 10.1016/j.celrep.2012.03.006) [DOI] [PubMed] [Google Scholar]
- 83.Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. 2012. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31, 4179–4190. ( 10.1038/emboj.2012.272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vanneste D, Ferreira V, Vernos I. 2011. Chromokinesins: localization-dependent functions and regulation during cell division. Biochem. Soc. Trans. 39, 1154–1160. ( 10.1042/BST0391154) [DOI] [PubMed] [Google Scholar]
- 85.Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema R. 2009. Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr. Biol. 19, 1703–1711. ( 10.1016/j.cub.2009.08.027) [DOI] [PubMed] [Google Scholar]
- 86.Boleti H, Karsenti E, Vernos I. 1996. Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84, 49–59. ( 10.1016/S0092-8674(00)80992-7) [DOI] [PubMed] [Google Scholar]
- 87.Sturgill EG, Ohi R. 2013. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol. 23, 1280–1290. ( 10.1016/j.cub.2013.05.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thery M, Racine V, Pépin A, Piel M, Chen Y, Sibarita J-B, Bornens M. 2005. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7, 947–953. ( 10.1038/ncb1307) [DOI] [PubMed] [Google Scholar]
- 89.Magidson V, O'Connell CB, Lončarek J, Paul R, Mogilner A, Khodjakov A. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567. ( 10.1016/j.cell.2011.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184, 365–372. ( 10.1083/jcb.200809055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sherline P, Mascardo R. 1982. Epidermal growth factor-induced centrosomal separation: mechanism and relationship to mitogenesis. J. Cell Biol. 95, 316–322. ( 10.1083/jcb.95.1.316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mardin BR, Isokane M, Cosenza MR, Krämer A, Ellenberg J, Fry AM, Schiebel E. 2013. EGF-induced centrosome separation promotes mitotic progression and cell survival. Dev. Cell 25, 229–240. ( 10.1016/j.devcel.2013.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell 23, 401–411. ( 10.1091/mbc.E11-02-0095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaseda K, McAinsh AD, Cross RA. 2012. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open 1, 12–18. ( 10.1242/bio.2011012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.So CH, Michal A, Komolov KE, Luo J, Benovic JL. 2013. G protein-coupled receptor kinase 2 (GRK2) is localized to centrosomes and mediates epidermal growth factor-promoted centrosomal separation. Mol. Biol. Cell 24, 2795–2806. ( 10.1091/mbc.E13-01-0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mazzorana M, Montoya G, Mortuza GB. 2011. The centrosome: a target for cancer therapy. Curr. Cancer Drug Targets 11, 600–612. ( 10.2174/156800911795655949) [DOI] [PubMed] [Google Scholar]
- 97.Bergnes G, Brejc K, Belmont L. 2005. Mitotic kinesins: prospects for antimitotic drug discovery. Curr. Top. Med. Chem. 5, 127–145. ( 10.2174/1568026053507697) [DOI] [PubMed] [Google Scholar]
- 98.Kim KB, et al. 2013. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J. Clin. Oncol. 31, 482–489. ( 10.1200/JCO.2012.43.5966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flaherty KT, et al. 2012. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, 1694–1703. ( 10.1056/NEJMoa1210093) [DOI] [PMC free article] [PubMed] [Google Scholar]