Abstract
Centrosomes—as well as the related spindle pole bodies (SPBs) of yeast—have been extensively studied from the perspective of their microtubule-organizing roles. Moreover, the biogenesis and duplication of these organelles have been the subject of much attention, and the importance of centrosomes and the centriole–ciliary apparatus for human disease is well recognized. Much less developed is our understanding of another facet of centrosomes and SPBs, namely their possible role as signalling centres. Yet, many signalling components, including kinases and phosphatases, have been associated with centrosomes and spindle poles, giving rise to the hypothesis that these organelles might serve as hubs for the integration and coordination of signalling pathways. In this review, we discuss a number of selected studies that bear on this notion. We cover different processes (cell cycle control, development, DNA damage response) and organisms (yeast, invertebrates and vertebrates), but have made no attempt to be comprehensive. This field is still young and although the concept of centrosomes and SPBs as signalling centres is attractive, it remains primarily a concept—in need of further scrutiny. We hope that this review will stimulate thought and experimentation.
Keywords: cell cycle, mitosis, DNA damage, signalling hub, centrosome, spindle pole body
1. Introduction
Centrosomes function as the main microtubule-organizing centres (MTOCs) of animal cells [1]. Each centrosome consists of centrioles embedded in a pericentriolar matrix of proteins (PCM). The PCM comprises not only γ-tubulin ring complexes important for microtubule (MT) nucleation [2], but also a large number of proteins, many of which display coiled-coil domains [3,4]. Centrioles are cylindrical MT-based structures that display an evolutionarily conserved ninefold symmetry [5]. Of the two centrioles typically present in any one centrosome, the older one (the fully mature centriole) carries characteristic subdistal and distal appendages. Subdistal appendages are important for mature centrioles to function as MT-anchoring centres during interphase [6,7], whereas distal appendages enable mature centrioles to function as basal bodies for the formation of cilia and flagella [8]. Considerations of phylogeny suggest that evolutionary conservation of centrioles correlates with the need of an organism to form ciliary structures during its life cycle [9,10]. As yeast do not form cilia, they do not rely on centriole-based centrosomes for the organization of MTs. Instead, MTOC function in yeast is provided by spindle pole bodies (SPBs). SPBs are multilayered structures associated with the nuclear envelope and thus structurally distinct from centrosomes [11]. However, centrosomes and SPBs are functionally equivalent organelles in that both use a similar, γ-tubulin-based mechanism for MT nucleation. As a consequence, centrosomes and SPBs exert a strong influence on MT-based processes, notably intracellular transport and organelle distribution, cell shape and migration, as well as bipolar spindle formation and cell division.
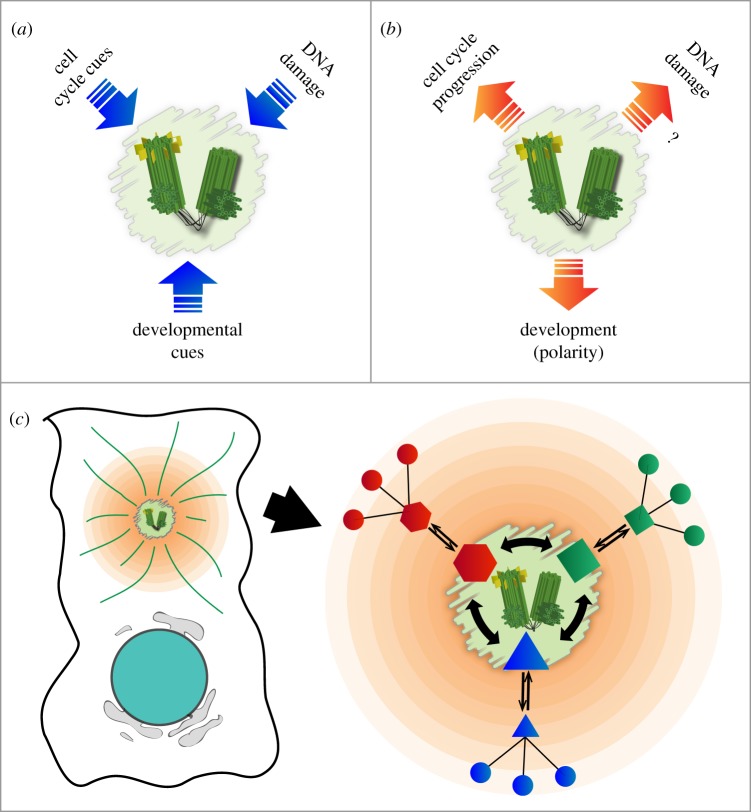
Deregulation of centrosome structure and number has long been implicated in chromosomal instability and carcinogenesis [12–16]. Moreover, mutations in genes coding for centriolar and centrosomal proteins have been causally linked to human disease, notably ciliopathies and brain diseases [17,18]. As a result of these findings, recent years have seen a surge of interest in the structure and function of centrosomes, centrioles and SPBs. With the exception of multiciliated epithelial cells, where hundreds of centrioles are present, centrosomes and SPBs exist as only one or two copies per cell, depending on cell cycle stage. As these organelles play important roles in spindle formation and chromosome segregation, their numbers as well as their MT-nucleating activities must be precisely controlled during the cell cycle. Additionally, centrosome separation prior to entry into mitosis [19–21], and the reversible formation of a primary cilium [22,23] also depend on cell cycle stage. Thus, it has long been recognized that centrosomes and SPBs are subjected to extensive regulation by cell cycle cues, developmental stimuli and DNA damage response pathways (figure 1a). Conversely, the centrosome has been proposed to serve as solid-state platform for the integration and coordination of various signalling pathways, and similar arguments have been made for yeast SPBs (figure 1b). In contrast to cytosolic signalling that merely depends on diffusion, the spatial concentration of signalling molecules at centrosomes and SPBs may enhance the kinetics of biochemical reactions and/or generate concentration gradients important for local responses (figure 1c). The anchoring of signalling proteins may also facilitate crosstalk between different signalling pathways and thereby contribute to the coordination of physiological responses (figure 1c). Moreover, asymmetries between individual centrosomes and SPBs provide opportunities for differential responses, potentially leading to differences in cell fate [22,24,25]. Finally, the mobility of centrosomes and their associated signalling molecules is expected to enhance the efficacy of signalling, as exemplified by events at the immune synapse [26].
Figure 1.
Centrosomes as signalling centres. It is well established that centrosomes (and SPBs) respond to multiple external signals (a). Increasing attention is being devoted to the alternative scenario, where centrosomes (and SPBs) function as signalling centres and solid-state platforms to influence cell physiology (b). The evolutionary advantages of using centrosomes (and SPBs) to locally concentrate signalling proteins could be manifold, as described in the main text. (c) Emphasizes the integration of multiple signalling pathways, the enhanced kinetics of biochemical reactions and the build-up of concentration gradients. (Online version in colour.)
An important corollary of the concept of centrosomes and SPBs as signalling centres is that structural and/or numerical aberrations in these organelles should affect cell physiology, cellular responses to stress and cell cycle progression [27–29]. Structural centrosome aberrations are typical of many cancers and believed to be important for cancer progression [16]. It may be rewarding to explore to what extent these structural alterations affect not only the MT-organizing capabilities of centrosomes, but also their signalling functions.
In this review, we assess a number of selected reports that bear on the notion of centrosomes and SPBs as signalling hubs. First, we summarize the evidence supporting such a role for yeast SPBs, with particular emphasis on the regulation of mitotic entry and exit. Then, we examine a few examples of centrosome-related signalling in the invertebrates Caenorhabditis elegans and Drosophila melanogaster. Finally, we extend our discussion to the centrosome of vertebrate cells. Reflecting the abundance of evidence relating to phosphorylation, this review focuses on the role of kinases and phosphatases at centrosomes and SPBs. Other post-translational modifications, including ubiquitylation, hydroxylation and acetylation, also occur at these organelles [30–34]; it seems safe to predict that the impact of these modifications will likely attract increasing interest in the future.
2. The importance of yeast spindle pole bodies for the regulation of mitosis
(a). Entry into mitosis
The cell cycle control machinery is centred on cyclin-dependent kinases (Cdks) and conserved from yeast to human. The traverse of M phase (mitosis and cytokinesis) depends on Cdk1 (also known as Cdc2 in fission yeast; CDC28 in budding yeast), which in mammals functions in association with A- and B-type cyclins. Activation of Cdk1 at the G2/M transition is controlled by antagonistic kinase and phosphatase activities. Core to this regulatory system is inhibitory phosphorylation imposed on Cdk1 by Wee1-related kinases, and relief of this inhibition by Cdc25 phosphatases [35]. Thus, entry into M phase requires both inhibition of Wee1-related kinases and activation of Cdc25 phosphatases (figure 2a). This is brought about by Polo-like kinase 1 (Plk1; Plo1 in fission yeast), which phosphorylates both Cdc25 and Wee1 [37–40], with the result that Cdc25 is activated whilst Wee1 is inhibited—in fact targeted for degradation [41] (figure 2a,c). Importantly, the recruitment of Plk1 to both Wee1 and Cdc25 depends on prior phosphorylation of these enzymes by an initial pool of activated Cdk1. This means that commitment to M phase depends on a positive feedback loop that displays all the features of a rapid and bistable switch.
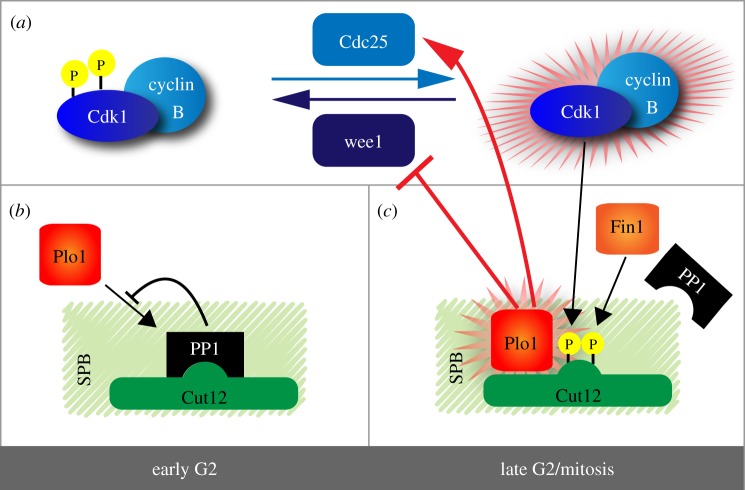
Figure 2.
Activation of Cdk1/cyclin complexes at the fission yeast SPB. (a) General scheme of Cdk1/cyclin B activation at the G2/M transition. (b,c) Role of fission yeast SPB-component Cut12 in the assembly of components involved in the activation of Cdk1 at the G2/M transition. For details, see text and [36]. (Online version in colour.)
Cdk1/cyclin B (also referred to as MPF in the early literature) has been observed at SPBs and centrosomes in many organisms [42–45]. These early observations already suggested that MTOCs might function as signalling platforms to promote mitotic entry, in addition to their roles in the organization of the MT network. The notion of MTOCs as signalling hubs has subsequently been strengthened, primarily through work on the yeast SPBs. This point is illustrated best by considering a series of studies on the SPB of the fission yeast Schizosaccharomyces pombe. Early work began with a screen that aimed at finding mutations that would allow cells to divide in the absence of Cdc25 phosphatase, which is essential for mitotic entry in wild-type cells. Surprisingly, this screen resulted in the identification of a gain-of-function mutation in the SPB component Cut12 (also known as Stf1). This raised the question of how an alteration in a genuine SPB component could bypass the requirement for the Cdc25 phosphatase at the G2/M transition [46,47]. In the absence of Cut12, SPBs failed to activate MT nucleation and they did not integrate into the nuclear envelope, resulting in mitotic arrest [48]. Interestingly, this lack of Cut12 could be compensated for by enhancing Cdc25 levels, suggesting that SPB activation defects might reflect local impairment of Cdk1 activation [49]. These results suggested that signalling events on the SPB were somehow important for triggering entry into mitosis, but it remained unclear how Cut12 and/or the SPB contribute to this process.
Recently, it has been uncovered that Plo1, the fission yeast homologue of vertebrate Plk1, holds many of the answers. As in other species, fission yeast Plo1 is involved in a positive feedback loop that ensures robust Cdk1/cyclin B activation at the G2/M transition. In a normal cell cycle, active Plo1 is observed at SPBs shortly before cells enter mitosis. However, in cells harbouring mutant versions of Cut12 that suppress the requirement for functional Cdc25, Plo1 associated with SPBs prematurely and, moreover, displayed increased kinase activity. This suggested that lack of Cdc25 could be overcome by temporally advancing Plo1 activation at SPBs, which might then cause inhibition of Wee1-related kinases and result in activation of Cdk1/cyclin B [50,51]. This in turn led Hagan and co-workers to ask how gain-of-function mutations in Cut12 could impact on the spatio-temporal regulation of Plo1. The authors found that Cut12 mutations mapped to a conserved motif implicated in the recruitment of protein phosphatase 1 (PP1). Mutation of this PP1 docking site in Cut12 decreased not only the affinity of PP1 for Cut12, but also advanced the localization of active Plo1 to SPBs [52]. This implies that PP1 association with Cut12 normally delays Plo1 localization to SPBs, presumably by interfering with a direct association between Plo1 and Cut12 (figure 2b) [51]. In turn, PP1 association with Cut12 was shown to be regulated by two conserved phosphorylation sites within the PP1 binding motif. Phosphorylation of these residues by Cdk1/cyclin B and Fin1, a kinase of the never in mitosis (NIMA) family, disrupted the association of PP1 with Cut12. As these phosphorylations occur late in G2 phase, this allows the recruitment of active Plo1 to SPBs and initial activation of SPB-associated Cdk1/cyclin B at the right time (figure 2c) [52]. This attractive model for the spatial control of Cdk1/cyclin B activation was confirmed by elegant re-targeting experiments demonstrating that activation of Cdk1/cyclin B or Plo1 at the SPB, but not at any other location, is sufficient to commit cells to mitotic entry [53].
(b). Exit from mitosis
The yeast SPB is also implicated in the control of mitotic exit. In particular, SPBs were recognized as important hubs for the assembly of signalling pathways known as the septum initiation network (SIN) in fission yeast and the mitotic exit network (MEN) in budding yeast, respectively. The SIN regulates key steps of cytokinesis, which in S. pombe comprises the formation and constriction of an actomyosin ring, septation and cell division, and asymmetry between the two SPBs has been implicated in the fine-tuning of these processes [54,55]. Central to the SIN are the GTPase Spg1, the three protein kinases Cdc7, Sid1, Sid2, as well as the inhibitory GAP complex Byr4–Cdc16. Importantly, the scaffolding protein complex Sid4–Cdc11 anchors the entire SIN cascade to SPBs [54]. This Sid4–Cdc11 scaffold is a stable integral part of SPBs and as such is thought to serve as a hub to assemble the various signalling components and regulators of the SIN [56]. In particular, Sid4 interacts with Plo1, a major activator of the SIN, whereas Cdc11 binds Spg1, thereby constitutively anchoring this GTPase to SPBs [56,57]. Spg1 drives the initiation of the SIN, and, when overexpressed, can trigger the onset of SIN signalling from any stage of the cell cycle [58]. In wild-type fission yeast, Spg1 is kept inactive during interphase by Byr4–Cdc16, and it is only activated upon entry into mitosis, when Byr4–Cdc16 dissociates from the SPBs [59,60]. Once activated, Spg1 is required for the activation of the three SIN kinases Cdc7, Sid1, Sid2. The terminal member of this cascade, Sid2, then translocates from the SPB to the site of division, where it drives cytokinesis [54]. As a rigorous test for the importance of the SPB in controlling SIN, fission yeast SPBs were subjected to laser ablation, a technically remarkable undertaking [61]. Ablation of both SPBs, but not a single SPB, was found to impair cytokinesis, indicating that the spatial assembly of SIN components on at least one of the two SPBs is indeed required for successful cytokinesis.
Similar to the fission yeast SIN, the components of the budding yeast MEN are also tightly associated with SPBs [62,63]. Moreover, the MEN of S. cerevisiae also operates through a GTPase-driven protein kinase cascade, the organization of which is similar to the one described above for the SIN. The MEN counterpart of the SIN scaffold Cdc11 is the protein Nud1, which acts as an SPB-associated platform onto which the various MEN components assemble. Specifically, Nud1 is responsible for the SPB localization of the kinase Cdc15, and Cdc15 in turn is required for the recruitment and activation of the effector kinase complex Mob1–Dbf2 [64]. Importantly, the SPB scaffolding proteins Nud1 and Cdc11 serve not only as assembly platforms for signalling components in budding and fission yeast, respectively, but they actively modulate downstream signalling events, depending on their phosphorylation status [65,66].
Collectively, the above-discussed studies provide ample evidence to support the notion that yeast SPBs play important roles in the spatial and temporal integration of signalling by cell cycle regulatory proteins.
3. Centrosome-related signalling in invertebrates
(a). Centrosomes promote mitotic entry in early embryos of Caenorhabditis elegans
In the C. elegans one-cell embryo, the first mitotic division occurs according to a stereotypical temporal and spatial pattern. As in many other species, sperm contributes the first centrosome to the C. elegans zygote. As a consequence, centrosomes are initially positioned close to the posterior male pronucleus; they come into the vicinity of the female pronucleus only after pronuclear migration, when male and female pronuclei meet. In the wild-type embryo, the two pronuclei initiate the first mitotic division as soon as they meet, as reflected by synchronous nuclear envelope breakdown (NEBD; figure 3a). In striking contrast, when the two pronuclei are kept apart from each other through appropriate experimental manipulation, they undergo NEBD in an asynchronous manner, with the female pronucleus showing a substantial delay (figure 3b). This setting provides a unique opportunity for studying the role of centrosomes in regulating mitosis, with observation of NEBD as a visual read-out [67,68]. The results of these studies indicate that centrosomes accelerate the onset of mitotic entry, probably through local concentration of a diffusible factor that promotes NEBD in a distance-dependent manner [67,68]. In support of this view, perturbation of centrosome integrity abrogated the asynchrony in the timing of NEBD of separated pronuclei (figure 3c), whereas the overall time until initiation of NEBD was increased. An even more striking observation was made upon RNAi-mediated depletion of the protein ZYG-12, which is essential for the attachment of centrosomes to the nuclear envelope of the male pronucleus; under these circumstances, the centrosomes moved freely in the cytoplasm and stimulated NEBD whenever they came close to a pronucleus: the closer a centrosome approached either the male or female pronucleus, the sooner this pronucleus underwent NEBD (figure 3d) [67,68]. This stimulation of NEBD was independent of MT-nucleation activity, as neither γ-tubulin depletion nor nocodazole treatment inhibited the process [67,68]. Instead, the centrosomal factor inducing mitotic entry was proposed to be Aurora A, a protein kinase known to associate with centrosomes in both invertebrates and vertebrates. In fact, depletion of Aurora A caused similar phenotypic consequences as perturbation of centrosome integrity, i.e. a general delay in NEBD initiation and abrogation of the asynchrony in NEBD between the two pronuclei (figure 3e).
Figure 3.
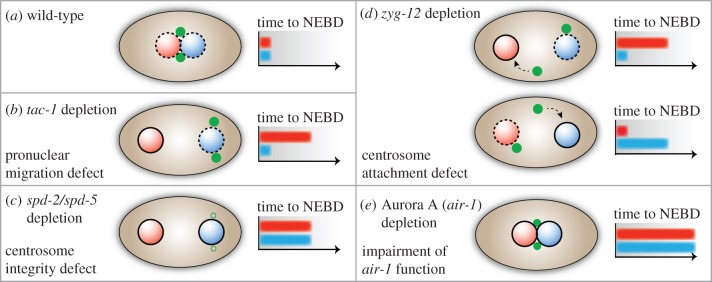
Centrosomes promote the onset of NEBD in the C. elegans one-cell embryo. The scheme depicts the role of centrosomes in facilitating NEBD in the C. elegans one-cell embryo (light grey/red, female pronucleus; dark grey/blue, male pronucleus; dashed black line, NEBD; filled black line, intact pronuclear membrane; black/green dots, centrosomes), as described in [67,68]. Light/red and dark grey/blue bars refer to the timing of NEBD for the female and male pronucleus, respectively. (a) The situations are described for wild-type embryos, (b) embryos with a defect in pronuclear migration, (c) centrosome integrity, (d) a centrosome attachment defect and (e) embryos depleted of Aurora A. (Online version in colour.)
How centrosome-associated Aurora A promotes NEBD is not fully understood. Aurora A has been implicated in the activation of Plk1 [69,70] and hence constitutes part of the positive feedback loop that drives activation of Cdk1/cyclin B. Thus, it is possible that Aurora A promotes NEBD through an indirect mechanism involving Cdk1/cyclin B [35]. Alternatively, or in addition, Aurora A might facilitate NEBD through direct phosphorylation of nuclear membrane components. In support of this latter possibility, a recent RNAi screen in C. elegans embryos defective for pronuclear migration points to an involvement of nucleoporins in Aurora A-mediated NEBD [71]. Depletion of several nucleoporins accelerated NEBD of separated female pronuclei, thereby decreasing the asynchrony in NEBD between the two pronuclei. Furthermore, at the onset of mitosis, some nucleoporins were locally removed from the nuclear envelope close to the site of centro-some association; this removal was dependent on intact centrosomes and active Aurora A, but not the presence of MTs [71]. Thus, it seems plausible that Aurora A can induce NEBD by directly or indirectly phosphorylating specific nucleoporins, leading to their dissociation from the nuclear envelope.
A next question to be addressed concerns the regulation of Aurora A abundance and activity at the C. elegans centrosome. According to one recent study, coordination between centrosome maturation timing and mitosis requires the UBXN-2 substrate adaptor for the AAA ATPase Cdc48/p97 [72]. RNAi-mediated depletion of UBXN-2 led to the accumulation of Aurora A at the centrosomes of one-cell embryos, indicating that UBXN-2/Cdc48/p97 negatively regulates centrosomal Aurora A levels. However, this ATP-dependent regulation of Aurora A association with centrosomes appears to be more relevant for the timing of centrosome separation and formation of astral MTs than for the timing of NEBD [72]. Instead, the timing of NEBD may depend primarily on the regulation of Aurora A activity at the centrosome. In this context, it is interesting to note that studies performed with Xenopus egg extracts identified the centrosome as a primary site for Aurora A activation [73].
(b). Centrosomes help to establish polarity in Caenorhabditis elegans embryos
In addition to their function in promoting NEBD, centrosomes have also been implicated in the establishment of polarity in the C. elegans embryo. This symmetry-breaking event is required for the asymmetric division of the one-cell embryo into two blastomeres of distinct sizes, each having a distinct fate. During polarization, the anterior–posterior cell axis is defined by the formation of two distinct cortical domains that are characterized by different polarity protein compositions and contractile properties [74–76]. Importantly, the initial cue for polarization is provided by the centrosome that enters the egg with the sperm. Although the critical contribution of a centrosome-associated function to the induction of polarity is well established, the exact nature of this polarity cue has been the subject of some debate. One key issue has been whether the centrosome contributes a diffusible factor, or, alternatively, acts through an MT-dependent process [77–80]. A priori, the two mechanisms are not mutually exclusive. Recently, it has been reported that polarity could be induced by centrosomes even when these were positioned far from the cell cortex, although the distance from the cortex clearly affected the time required for symmetry breaking [81]. As cytoplasmic MTs constrain centrosome movement near the cortex, these MTs would thus be expected to favour the polarity-inducing action of centrosome-associated factors at the cortex [81].
(c). Roles of centrosomes in Drosophila development
In an early test for the role of centrosomes during Drosophila development, the fate of flies lacking the centriole duplication factor DSAS-4 was examined [82]. Surprisingly, DSAS-4 mutants proceeded normally through most of development and advanced to morphologically normal adult flies, even though centrioles and centrosomes were undetectable in adult cells. Adult DSAS-4 mutant flies died shortly after birth, apparently because their sensory neurons lacked cilia [82]. This study clearly shows that flies are able to proceed through most of development by relying on a centrosome-independent pathway for bipolar spindle formation. However, when interpreting this finding, it is important to bear in mind that a maternally supplied pool of DSAS-4 allowed centrosome formation during the earliest stages of embryogenesis [82]. In the complete absence of centrosomes, Drosophila early embryogenesis is severely disrupted [83]. Most likely this reflects a key role for centrosomes in bipolar spindle formation and spindle positioning during asymmetric divisions, including possible effects on the balance between stem cells and differentiating cells [25,76,84–86]. Taken together, these studies confirm that centrosomes are not essential for all cell divisions, but are critical for asymmetric divisions during early development as well as for ciliogenesis.
Drosophila also provides a prime example for the involvement of centrosomes in the response to DNA damage. When encountering damaged or incompletely replicated DNA, Drosophila cells will activate a checkpoint response that delays cell cycle progression. This response relies on pathways that have been evolutionarily conserved in other eukaryotic species. In addition, however, Drosophila early embryos also manifest an intriguing DNA damage response that results in centrosome inactivation [87]. Early embryogenesis in Drosophila involves 13 extremely rapid nuclear divisions that proceed without cytokinesis, resulting in a syncytium. In these syncytial embryos, impaired DNA integrity was found to trigger intramitotic division failures that could be traced to the functional inactivation of spindle poles and centrosomes. This inactivation was characterized by loss of γ-tubulin from the centrosome, which led to anastral mitotic spindle assembly, mitotic delay and chromosome segregation failure. Most importantly, damaged nuclei associated with γ-tubulin-deficient centrosomes were detached from the cortex during cellularization and hence not incorporated into the developing embryo [87]. Subsequent studies revealed that this centrosome-related DNA damage response is mediated by Chk2, a key kinase implicated in ‘classical’ DNA damage response pathways [88]. Clearly, the pathway linking DNA damage to centrosome inactivation in Drosophila syncytial embryos represents a striking example for the importance of centrosomes in the safeguard of genome integrity.
In contrast to vertebrate mitoses, the nuclear envelope remains largely intact during nuclear division in syncytial Drosophila embryos. Instead, it is fenestrated selectively at the poles to allow spindle formation. This implies that centrosomes should remain ‘attached’ to nuclear envelopes throughout mitosis. Recent work revealed that this attachment requires the cooperation between Drosophila Polo (the founding member of the Plk1 family) and protein phosphatase 2A (PP2A) in association with a B-type regulatory subunit known as Twins [89]. Moreover, Drosophila genetics also revealed an antagonism between Polo/PP2A and the protein kinase Greatwall [89,90]; contributing to the emerging recognition that, in many species, this kinase–phosphatase balance plays a key role in the regulation of the G2/M transition (reviewed in [91–94]). Although precise mechanistic details of the interactions between these proteins remain to be clarified, the abovementioned studies again highlight the importance of centrosomes and spindle poles for the spatial assembly of regulators of mitotic progression, reminiscent of the situation described in §2 for the fission yeast SPB.
Finally, Drosophila is also one of the first species in which centrosomes were implicated in the spatial control of cyclin B degradation at the metaphase to anaphase transition. Live-cell imaging of cyclin B-GFP fusion proteins in Drosophila embryos revealed that cyclin B-GFP already begins to vanish from spindle poles in late metaphase, before it disappears from spindle MTs [95]. Furthermore, in Drosophila centrosome fall off (cfo) mutant embryos, where centrosomes are detached from the mitotic spindles, cyclin B-GFP destruction was inhibited at the spindles, but not at the centrosomes [96]. Together, these studies suggest that cyclin B destruction is first triggered at centrosomes/spindle poles, and that propagation of the destruction wave to the spindle requires an intact connection between the centrosome and spindle MTs [96].
4. Centrosomes as signalling platforms in vertebrates?
(a). Cell cycle progression
Some of the earliest evidence for the ability of centrosomes to trigger cell cycle events stems from the demonstration that injection of purified centrosomes into eggs of both marine invertebrates and amphibians was sufficient to trigger parthenogenetic development [97,98]. These pioneering studies argue that centrosomes play a key role in the initiation of M phase in both vertebrates and invertebrates, similar to the conclusion drawn for yeast SPBs (see §2). Centrosomes have also been identified early on as an important hub for the localization of cAMP-dependent kinase [99,100]. Subsequently, this association was shown to be mediated by a specific targeting protein of the AKAP family [101–103], and the use of an elegant FRET biosensor demonstrates that locally confined regulation of cAMP signals at the centrosome impacts significantly on cell cycle progression [104]. Additionally, Cdk1/cyclin B, Cdc25B, Cdc25C, Plk1 and Aurora A have all been localized to centrosomes at the G2/M transition [43,105–110]. Thus, the entire signalling network implicated in the promotion of mitosis is present on vertebrate centrosomes. A phospho-specific antibody recognizing only the activated form of Cdk1/cyclin B was then used to demonstrate that initial activation of the complex occurs at the centrosome [111]. Recently, an elegant biosensor allowed careful analysis of the kinetics of Cdk1/cyclin B activation [112], and whereas the spatial resolution of this biosensor was limited, it would be interesting to explore the consequences of targeting a similar sensor to the centrosome. Interestingly, Cdc25B was also shown to act specifically on Cdk1/cyclin B at centrosomes [106,113]. Thus, centrosomes apparently facilitate the activation of Cdk1/cyclin B by bringing the necessary regulatory components into close spatial proximity, much like the situation discussed in §2 for yeast SPBs. However, while such a facilitating role appears plausible, the centrosome is not strictly essential for vertebrate somatic cells to enter mitosis: when centrosomes were removed by microsurgery or laser ablation, cells were still able to progress through mitosis with remarkably normal kinetics [114–116]. These results illustrate the efficiency of acentrosomal mechanisms for bipolar mitotic spindle formation, as noted already in the context of the phenotype associated with the Drosophila DSAS-4 mutant (see §3c). Tempering this conclusion, however, a recent study clearly demonstrates that genetic ablation of centrosomes from chicken DT40 B cells severely impaired cell cycle progression, indicating that centrosomes are critical for error-free segregation of large numbers of chromosomes [117].
Interestingly, the above mentioned studies on acentrosomal cells also revealed frequent defects in cytokinesis. Successful mitotic exit and cytokinesis rely on several key events. These include the timely onset of anaphase, triggered by cyclin degradation and consequent inactivation of Cdk1, as well as the formation of a central spindle, followed by assembly of an actomyosin ring beneath the cell cortex, ingression of the cleavage furrow and, finally, abscission [118]. The role of the mammalian centrosome in these late mitotic events is incompletely understood, but several intriguing observations have been reported. In particular, interest in a link between centrosomes and cytokinesis was kindled by the observation that, in some mammalian cells, the mother centriole moves towards the midbody (also known as the Flemming body) within the post-mitotic bridge [119]. Interestingly, this centriole movement appeared to correlate with abscission, the final step of cytokinesis, in that abscission occurred only after the centriole had moved back to the cell centre. These observations raised the possibility that centrioles provide a molecular cue to regulate abscission, reminiscent of the proposed role of the SPB in the MEN and SIN pathways in yeast [28]. Such a role would explain the observation of frequent cytokinesis and abscission failures in cells from which centrosomes had been experimentally removed [115,119]. However, although centriole movement towards the midbody clearly occurs in some cultured cells, it does not appear to be a universal feature associated with cell division.
An alternative, perhaps related, role for the centrosome in cytokinesis is suggested by the observation that several centrosomal proteins localize to the midbody towards the end of mitosis, and the list of these proteins seems to be growing [4]. Of particular interest is Cep55, a centrosome- and midbody-associated protein required for mitotic exit [120,121]. Cep55 depletion results in cytokinesis failure, and Cep55 localization to the midbody is controlled by Plk1. This led to a model according to which Plk1 negatively regulates the association of Cep55 with the midbody until Plk1 is degraded late in mitosis. This would then allow Cep55 to be recruited to the midbody to promote abscission. Another interesting centrosome-associated protein implicated in cytokinesis is centriolin [122,123]. This protein also localizes to the maternal centriole in interphase and to the midbody prior to cytokinesis, and it is also required for mitotic exit. Of note, centriolin shares a small region of homology with the fission yeast SIN scaffolding component Cdc11 [122]. How centriolin, Cep55 and other centrosome- and midbody-associated proteins cooperate to regulate mitotic exit in human cells remains to be elucidated, but they are expected to impact on the regulation of vesicle transport and MT severing [124,125].
Another interesting question is whether mitotic exit and cytokinesis in human cells could be regulated, at least in part, by orthologues of the yeast SIN/MEN components. Counterparts for at least some of these genes could readily be identified in both invertebrate and vertebrate species (notably the kinases Mst1 and 2 as well as the kinases Lats1 and 2, along with associated regulatory proteins termed Mob, and the Cdc14 phosphatase), but the Mst and Lats kinases are implicated primarily in the proliferation–inhibitory Hippo signalling pathway [126–128]. Nevertheless, it is interesting that both Mst and Lats kinases have been functionally linked to the centrosome cycle [129]. Moreover, the human Hippo pathway component Mob1 has recently been localized to both the centrosome and the midbody and accordingly was proposed to control abscission through modulation of MT stability [130].
Centrosomes have also been implicated in the control of the G1/S transition, but the nature of this control has initially been controversial. Early studies showed that cell cycle progression from G1 to S was impaired when centrosomes were removed from certain cell types by microsurgery or laser ablation [114,115,131]. This then led to the hypothesis that a G1/S checkpoint might monitor centrosome status. Although this idea was initially intriguing, subsequent work has largely discounted it. In particular, it has been demonstrated that different types of untransformed human cells, including RPE1, can proceed through G1/S in the absence of centrioles, regardless of how the centrioles were removed [116]. Furthermore, centrosome removal or impairment of centrosome integrity was shown to trigger a cellular stress response that involves the activation of p38 kinase and p53. The latter then induces the Cdk inhibitor p21Cip1, which readily explains the observed G1 arrests [28,29,116,132,133]. Thus, the idea of a specific centrosome-related G1 checkpoint has fallen out of favour, but centrosome integrity is likely to be important for avoidance of stress responses and efficient traverse of the G1 phase of the cell cycle.
(b). The centrosome and the vertebrate DNA damage response
The role of the centrosome in DNA damage response has a chequered history. There are several intriguing hints for an important relationship [134], but definitive connections remain few and far between. One recurring theme is the finding that many proteins implicated in DNA damage response pathways, including Brca1, Brca2, Chk1, Chk2 and p53, seem to localize to centrosomes, usually in addition to cytoplasmic and/or nuclear localizations [28]. While some of these associations are undoubtedly real, it is important to bear in mind that artefactual stainings of centrosomes are not uncommon. These reflect either pre-existing anti-centrosome antibodies in rabbit sera or antibody cross-reactivity with centrosomal antigens [135]. Two contradictory studies on the purported centrosome association of the checkpoint kinase Chk1 may serve to illustrate this point: an initial, apparently convincing study reported that a specific, centrosome-associated population of Chk1 prevented premature activation of centrosomal Cdk1/cyclin B [136]. These data implicated Chk1 in the spatio-temporal regulation of cell division and, moreover, raised the possibility that the centrosome-associated pool of this enzyme might contribute to mediate the DNA-damage-induced inhibition of Cdk1/cyclin B. However, a subsequent study irrefutably showed that the reported centrosomal localization of Chk1 could be attributed to cross-reactivity of one particular antibody with a newly identified centrosomal protein, and that nuclear Chk1, not centrosomal Chk1, regulates Cdk1/cyclin B [137]. Likewise, the reported Cep63-dependent presence of Cdk1 at centrosomes [138] is almost certainly due to antibody cross-reactivity with Cep152 [139]. Thus, when interpreting immunocytochemical data reporting on unexpected centrosome-associations of particular proteins, it would seem wise to consider carefully the possibility of antibody-related artefacts [135,137,139,140].
Not only have proteins with well-established functions in DNA damage response pathways been reported to reside at centrosomes, but the opposite is also true: genuine centrosomal proteins have been implicated in DNA damage response. One early example is provided by a member of the centrin family of small, evolutionarily conserved calmodulin-related EF-hand proteins. Mammalian centrin-2 is considered the orthologue of the S. cerevisiae Cdc31 gene product, which is clearly required for SPB duplication [141]. Likewise, centrins are essential for the formation of basal bodies in ciliates [142,143]. Whether or not centrin-2 is required for centrosome duplication in human cells is controversial, but there is no question that this protein is a genuine component of vertebrate centrosomes [144–147]. Equally well established is the fact that centrin-2 interacts with xeroderma pigmentosum group C protein, a core component of nucleotide excision repair [148,149]. Much remains to be learned about the functions of centrin-2, but it is remarkable that genetic deletion of all centrin isoforms from chicken DT40 cells resulted in significant delays in nucleotide excision repair, but no obvious phenotypes associated with centrosome duplication or function [144].
In addition to centrins, other bona fide centrosome-associated components have also recently been implicated in DNA damage response pathways. These include pericentrin, a core component of the centrosomal PCM [150], and Cep164, a protein associated with distal appendages of centrioles [151]. Mutations in pericentrin were found to cause Seckel syndrome, a disorder characterized by reduced brain and body size, and cells from these patients showed an impairment of ATR (ATM and Rad3-related)-dependent checkpoint signalling [152]. Similarly, mutations in Cep164 were shown to cause nephronophthisis-related ciliopathies [153]. Upon DNA damage, Cep164 was reported to accumulate within nuclear foci that are implicated in the activation of ataxia telangiectasia mutated (ATM), and knockdown of Cep164 in zebrafish resulted in impaired DNA-damage response. These findings echo earlier reports on links between centrosome-associated proteins and DNA damage response pathways, as exemplified by Cdk5rap2/Cep215 [154] or the microcephaly-related protein MCPH1/Brit1 [155]. These emerging connections are most intriguing, but in order to understand their (patho)physiological significance, it will be indispensable to decipher the wiring of the underlying molecular mechanisms and pathways.
At this point, the DNA damage-induced and Chk2-mediated loss of γ-tubulin from Drosophila centrosomes probably remains the best understood link between the DNA damage response field and the centrosome field (see §3c). Accordingly, attempts have been made to extend these findings from Drosophila to vertebrates. Considering that vertebrate embryos do not go through syncytial stages followed by cellularization, strict conservation of the Drosophila mechanism in vertebrates was not to be expected and, indeed, loss of γ-tubulin was not generally observed in response to DNA damage. Nevertheless, DNA damage was found to affect vertebrate centrosomes in multiple ways. When Chinese hamster ovary cells were forced to enter mitosis in the presence of damaged or incompletely replicated DNA, centrosomes often underwent splitting or fragmentation, leading to multipolar divisions and severe mitotic abnormalities [156]. Similarly, centrosome splitting was observed in immortalized RPE1 cells in response to ionizing radiation and other DNA damaging treatments, leading to the proposal that centrosome splitting may represent a general response to potentiate centrosome amplification [157]. Indeed, centrosome amplification has been frequently observed in response to DNA damage, for example in ATM- or ATR-deficient human cells [158], or in G2 arrested, Rad51-deficient chicken DT40 cells [159]. The molecular mechanisms underlying centrosome amplification in response to DNA damage remain to be elucidated, but both Chk1 and centriolar satellites have been implicated [158,160]. Moreover, APC/C activity was shown to oscillate, leading to successive rounds of Cdk2 and separase activation in the arrested cells [161]. Although centrosome amplification does not necessarily require passage through mitosis [159], we emphasize that supernumerary centrosomes are expected to arise whenever cells with damaged DNA fail to arrest at the G2/M checkpoint and then advance to abortive divisions [16,140]. The physiological significance of centrosomal responses to DNA damage remains to be fully understood, but the possibility has been raised that centrosome fragmentation or amplification may constitute a safeguard mechanism to kill cells with DNA damage via induction of division failures [156,159].
5. Conclusion and perspectives
In this review, we have discussed a selection of reports that tend to support the view that centrosomes and SPBs may function as hubs or solid-state platforms for the integration of signalling pathways. We emphasize that we have made no attempt to be comprehensive. In particular, we have not addressed the evidence that attributes important roles to centrosomes in the replication of viruses and other intracellular pathogens, or in trafficking and turnover of cellular components. In addition, we are of the opinion that definitive evidence for an essential role of vertebrate centrosomes in any one signalling process is scarce. Considering that some cells go through the cell cycle without centrosomes just fine, this is probably not surprising. However, there is now strong evidence to indicate that SPBs contribute in major ways to the temporal and spatial organization of cell cycle regulatory components, and the same can be said for the centrosomes that act during early embryogenesis in invertebrates. Although it is tempting to extrapolate from these findings to vertebrates, it is to be expected that only truly advantageous functions have been conserved during evolution. At this time, several highly intriguing connections between centrosomes and DNA damage response pathways have been reported. Most of the dots still need to be filled in before we can possibly understand these connections, but this area certainly deserves further scrutiny. As the saying goes: ‘there is no smoke without fire’—or is there?
Acknowledgements
We thank all members of the Nigg laboratory for helpful discussions.
Funding statement
Work in the author's laboratory was supported by the University of Basel, the Swiss Cancer League (KFS-02657-08-2010) and the Swiss National Science Foundation (310030B_149641/1). C.A. was funded by a PhD fellowship from the Werner Siemens-Foundation (Zug, Switzerland).
References
- 1.Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34. ( 10.1016/S0955-0674(01)00290-3) [DOI] [PubMed] [Google Scholar]
- 2.Lüders J, Stearns T. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167. ( 10.1038/nrm2100) [DOI] [PubMed] [Google Scholar]
- 3.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574. ( 10.1038/nature02166) [DOI] [PubMed] [Google Scholar]
- 4.Jakobsen L, et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520–1535. ( 10.1038/emboj.2011.63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azimzadeh J, Marshall WF. 2010. Building the centriole. Curr. Biol. 20, R816–R825. ( 10.1016/j.cub.2010.08.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317–329. ( 10.2307/1619692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W-J, Soni RK, Uryu K, Tsou M-FB. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739. ( 10.1083/jcb.201101109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanos BE, Yang H-J, Soni R, Wang W-J, Macaluso FP, Asara JM, Tsou M-FB. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168. ( 10.1101/gad.207043.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175. ( 10.1083/jcb.201011152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bornens M. 2012. The centrosome in cells and organisms. Science 335, 422–426. ( 10.1126/science.1209037) [DOI] [PubMed] [Google Scholar]
- 11.Jaspersen SL, Winey M. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1–28. ( 10.1146/annurev.cellbio.20.022003.114106) [DOI] [PubMed] [Google Scholar]
- 12.Nigg EA. 2006. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer 119, 2717–2723. ( 10.1002/ijc.22245) [DOI] [PubMed] [Google Scholar]
- 13.Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. ( 10.1038/nature08136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zyss D, Gergely F. 2009. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 19, 334–346. ( 10.1016/j.tcb.2009.04.001) [DOI] [PubMed] [Google Scholar]
- 15.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032–1042. ( 10.1016/j.cell.2008.05.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigg EA. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825. ( 10.1038/nrc924) [DOI] [PubMed] [Google Scholar]
- 17.Nigg EA, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678. ( 10.1016/j.cell.2009.10.036) [DOI] [PubMed] [Google Scholar]
- 18.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. 2011. Centrosomes and cilia in human disease. Trends Genet. 27, 307–315. ( 10.1016/j.tig.2011.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574. ( 10.2307/1618725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. ( 10.1016/0092-8674(95)90142-6) [DOI] [PubMed] [Google Scholar]
- 21.Mardin BR, Schiebel E. 2012. Breaking the ties that bind: new advances in centrosome biology. J. Cell Biol. 197, 11–18. ( 10.1038/ncb1140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nigg EA, Stearns T. 2011. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13, 1154–1160. ( 10.1038/ncb2345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa H, Marshall WF. 2011. Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234. ( 10.1038/nrm3085) [DOI] [PubMed] [Google Scholar]
- 24.Anderson CT, Stearns T. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19, 1498–1502. ( 10.1016/j.cub.2009.07.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelletier L, Yamashita YM. 2012. Centrosome asymmetry and inheritance during animal development. Curr. Opin. Cell Biol. 24, 541–546. ( 10.1016/j.ceb.2012.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritter AT, Angus KL, Griffiths GM. 2013. The role of the cytoskeleton at the immunological synapse. Immunol. Rev. 256, 107–117. ( 10.1111/imr.12117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fry AM, Hames RS. 2005. The role of the centrosome in cell cycle progression. In Centrosomes in development and disease (ed. Nigg EA.), pp. 143–166. Weinheim, FRG: Wiley-VCH. [Google Scholar]
- 28.Doxsey S, McCollum D, Theurkauf W. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411–434. ( 10.1146/annurev.cellbio.21.122303.120418) [DOI] [PubMed] [Google Scholar]
- 29.Sluder G. 2005. Two-way traffic: centrosomes and the cell cycle. Nat. Rev. Mol. Cell Biol. 6, 743–748. ( 10.1038/nrm1712) [DOI] [PubMed] [Google Scholar]
- 30.Sankaran S, Starita LM, Groen AC, Ko MJ, Parvin JD. 2005. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol. Cell Biol. 25, 8656–8668. ( 10.1128/MCB.25.19.8656-8668.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser SC, Bensaddek D, Ortmann B, Maure J-F, Mudie S, Blow JJ, Lamond AI, Swedlow JR, Rocha S. 2013. PHD1 links cell-cycle progression to oxygen sensing through hydroxylation of the centrosomal protein Cep192. Dev. Cell 26, 381–392. ( 10.1016/j.devcel.2013.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, D'Angiolella V, Seeley ES, Kim S, Kobayashi T, Fu W, Campos EI, Pagano M, Dynlacht BD. 2013. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495, 255–259. ( 10.1038/nature11941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puklowski A, et al. 2011. The SCF–FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13, 1004–1009. ( 10.1038/ncb2282) [DOI] [PubMed] [Google Scholar]
- 34.Cuevas R, Korzeniewski N, Tolstov Y, Hohenfellner M, Duensing S. 2013. FGF-2 disrupts mitotic stability in prostate cancer through the intracellular trafficking protein CEP57. Cancer Res. 73, 1400–1410. ( 10.1158/0008-5472.CAN-12-1857) [DOI] [PubMed] [Google Scholar]
- 35.Lindqvist A, Rodríguez-Bravo V, Medema RH. 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185, 193–202. ( 10.1083/jcb.200812045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan IM, Grallert A. 2013. Spatial control of mitotic commitment in fission yeast. Biochem. Soc. Trans. 41, 1766–1771. ( 10.1042/BST20130190) [DOI] [PubMed] [Google Scholar]
- 37.Kumagai A, Dunphy WG. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273, 1377–1380. ( 10.1126/science.273.5280.1377) [DOI] [PubMed] [Google Scholar]
- 38.Abrieu AA, Brassac TT, Galas SS, Fisher DD, Labbé JCJ, Dorée MM. 1998. The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J. Cell. Sci. 111, 1751–1757. [DOI] [PubMed] [Google Scholar]
- 39.Qian YW, Erikson E, Taieb FE, Maller JL. 2001. The polo-like kinase Plx1 is required for activation of the phosphatase Cdc25C and cyclin B-Cdc2 in Xenopus oocytes. Mol. Biol. Cell 12, 1791–1799. ( 10.1091/mbc.12.6.1791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Vugt MATM, Brás A, Medema RH. 2004. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell 15, 799–811. ( 10.1016/j.molcel.2004.07.015) [DOI] [PubMed] [Google Scholar]
- 41.Watanabe N, Arai H, Iwasaki J-I, Shiina M, Ogata K, Hunter T, Osada H. 2005. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl Acad. Sci. USA 102, 11 663–11 668. ( 10.1073/pnas.0500410102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alfa CE, Ducommun B, Beach D, Hyams JS. 1990. Distinct nuclear and spindle pole body population of cyclin-cdc2 in fission yeast. Nature 347, 680–682. ( 10.1038/347680a0) [DOI] [PubMed] [Google Scholar]
- 43.Bailly E, Dorée M, Nurse P, Bornens M. 1989. P34cdc2 is located in both nucleus and cytoplasm - part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8, 3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riabowol K, Draetta G, Brizuela L, Vandre D, Beach D. 1989. The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell 57, 393–401. ( 10.1016/0092-8674(89)90914-8) [DOI] [PubMed] [Google Scholar]
- 45.Rattner JB, Lew J, Wang JH. 1989. p34cdc2 kinase is localized to distinct domains within the mitotic apparatus. Cell Motil. Cytoskeleton 17, 227–235. ( 10.1002/cm.970170309) [DOI] [PubMed] [Google Scholar]
- 46.Hudson JD, Feilotter H, Young PG. 1990. Stf1 - non-wee mutations epistatic to Cdc25 in the fission yeast Schizosaccharomyces pombe. Genetics 126, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bridge AJ, Morphew M, Bartlett R, Hagan IM. 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 12, 927–942. ( 10.1101/gad.12.7.927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tallada VA, Tanaka K, Yanagida M, Hagan IM. 2009. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J. Cell Biol. 185, 875–888. ( 10.1083/jcb.200812108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tallada VA, Bridge AJ, Emery PA, Hagan IM. 2007. Suppression of the Schizosaccharomyces pombe cut12.1 cell-cycle defect by mutations in cdc25 and genes involved in transcriptional and translational control. Genetics 176, 73–83. ( 10.1534/genetics.107.072090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. 1999. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell 10, 2771–2785. ( 10.1091/mbc.10.8.2771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacIver FH, Tanaka K, Robertson AM, Hagan IM. 2003. Physical and functional interactions between polo kinase and the spindle pole component Cut12 regulate mitotic commitment in S. pombe. Genes Dev. 17, 1507–1523. ( 10.1101/gad.256003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grallert A, et al. 2013. Removal of centrosomal PP1 by NIMA kinase unlocks the MPF feedback loop to promote mitotic commitment in S. pombe. Curr. Biol. 23, 213–222. ( 10.1016/j.cub.2012.12.039) [DOI] [PubMed] [Google Scholar]
- 53.Grallert A, Patel A, Tallada VA, Chan KY, Bagley S, Krapp A, Simanis V, Hagan IM. 2013. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat. Cell Biol. 15, 88–95. ( 10.1038/ncb2633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson AE, McCollum D, Gould KL. 2012. Polar opposites: fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 69, 686–699. ( 10.1002/cm.21044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krapp A, Simanis V. 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36, 411–415. ( 10.1042/BST0360411) [DOI] [PubMed] [Google Scholar]
- 56.Morrell JL, et al. 2004. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14, 579–584. ( 10.1016/j.cub.2004.03.036) [DOI] [PubMed] [Google Scholar]
- 57.Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20, 1259–1270. ( 10.1093/emboj/20.6.1259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11, 1519–1534. ( 10.1101/gad.11.12.1519) [DOI] [PubMed] [Google Scholar]
- 59.Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. 1998. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8, 947–954. ( 10.1016/S0960-9822(98)70394-X) [DOI] [PubMed] [Google Scholar]
- 60.Li CX, Furge KA, Cheng QC, Albright CF. 2000. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J. Biol. Chem. 275, 14 381–14 387. ( 10.1074/jbc.275.19.14381) [DOI] [PubMed] [Google Scholar]
- 61.Magidson V, Chang F, Khodjakov A. 2006. Regulation of cytokinesis by spindle-pole bodies. Nat. Cell Biol. 8, 891–893. ( 10.1038/ncb1449) [DOI] [PubMed] [Google Scholar]
- 62.Pereira G, Schiebel E. 2001. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr. Opin. Cell Biol. 13, 762–769. ( 10.1016/S0955-0674(00)00281-7) [DOI] [PubMed] [Google Scholar]
- 63.Bardin AJ, Amon A. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2, 815–826. ( 10.1038/35099020) [DOI] [PubMed] [Google Scholar]
- 64.Meitinger F, Palani S, Pereira G. 2012. The power of MEN in cytokinesis. Cell Cycle 11, 219–228. ( 10.4161/cc.11.2.18857) [DOI] [PubMed] [Google Scholar]
- 65.Rock JM, et al. 2013. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science 340, 871–875. ( 10.1126/science.1235822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feoktistova A, Morrell-Falvey J, Chen JS, Singh NS, Balasubramanian MK, Gould KL. 2012. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol. Biol. Cell 23, 1636–1645. ( 10.1091/mbc.E11-09-0792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hachet V, Canard C, Gönczy P. 2007. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev. Cell 12, 531–541. ( 10.1016/j.devcel.2007.02.015) [DOI] [PubMed] [Google Scholar]
- 68.Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. 2007. A microtubule-independent role for centrosomes and Aurora A in nuclear envelope breakdown. Dev. Cell 12, 515–529. ( 10.1016/j.devcel.2007.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macůrek L, et al. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123. ( 10.1038/nature07185) [DOI] [PubMed] [Google Scholar]
- 70.Seki A, Coppinger JA, Jang C-Y, Yates JR, Fang G. 2008. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658. ( 10.1126/science.1157425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hachet V, Busso C, Toya M, Sugimoto A, Askjaer P, Gonczy P. 2012. The nucleoporin Nup205/NPP-3 is lost near centrosomes at mitotic onset and can modulate the timing of this process in Caenorhabditis elegans embryos. Mol. Biol. Cell 23, 3111–3121. ( 10.1091/mbc.E12-03-0204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kress E, et al. 2013. The UBXN-2/p37/p47 adaptors of CDC-48/p97 regulate mitosis by limiting the centrosomal recruitment of Aurora A. J. Cell Biol. 201, 559–575. ( 10.1083/jcb.201209107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. 2010. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl Acad. Sci. USA 107, 21 022–21 027. ( 10.1073/pnas.1014664107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cowan CR, Hyman AA. 2004. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20, 427–453. ( 10.1146/annurev.cellbio.19.111301.113823) [DOI] [PubMed] [Google Scholar]
- 75.Noatynska A, Tavernier N, Gotta M, Pintard L. 2013. Coordinating cell polarity and cell cycle progression: what can we learn from flies and worms? Open Biol. 3, 130083 ( 10.1098/rsob.130083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gönczy P. 2008. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355–366. ( 10.1038/nrm2388) [DOI] [PubMed] [Google Scholar]
- 77.Cowan CR, Hyman AA. 2004. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature 431, 92–96. ( 10.1038/nature02825) [DOI] [PubMed] [Google Scholar]
- 78.Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. 2011. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat. Cell Biol. 13, 1361–1367. ( 10.1038/ncb2354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sonneville R, Gönczy P. 2004. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131, 3527–3543. ( 10.1242/dev.01244) [DOI] [PubMed] [Google Scholar]
- 80.Tsai M-C, Ahringer J. 2007. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J. Cell Biol. 179, 397–402. ( 10.1083/jcb.200708101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bienkowska D, Cowan CR. 2012. Centrosomes can initiate a polarity axis from any position within one-cell C. elegans embryos. Curr. Biol. 22, 583–589. ( 10.1016/j.cub.2012.01.064) [DOI] [PubMed] [Google Scholar]
- 82.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125, 1375–1386. ( 10.1016/j.cell.2006.05.025) [DOI] [PubMed] [Google Scholar]
- 83.Stevens NR, Raposo AASF, Basto R, St Johnston D, Raff JW. 2007. From stem cell to embryo without centrioles. Curr. Biol. 17, 1498–1503. ( 10.1016/j.cub.2007.07.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamashita YM, Fuller MT. 2008. Asymmetric centrosome behavior and the mechanisms of stem cell division. J. Cell Biol. 180, 261–266. ( 10.1083/jcb.200707083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Januschke J, Gonzalez C. 2008. Drosophila asymmetric division, polarity and cancer. Oncogene 27, 6994–7002. ( 10.1038/onc.2008.349) [DOI] [PubMed] [Google Scholar]
- 86.Lesage B, Gutierrez I, Martí E, Gonzalez C. 2010. Neural stem cells: the need for a proper orientation. Curr. Opin. Genet. Dev. 20, 438–442. ( 10.1016/j.gde.2010.04.013) [DOI] [PubMed] [Google Scholar]
- 87.Sibon OCO, Kelkar AA, Lemstra WW, Theurkauf WEW. 2000. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2, 90–95. ( 10.1038/35000041) [DOI] [PubMed] [Google Scholar]
- 88.Takada S, Kelkar A, Theurkauf WE. 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113, 87–99. ( 10.1016/S0092-8674(03)00202-2) [DOI] [PubMed] [Google Scholar]
- 89.Wang P, Pinson X, Archambault V. 2011. PP2A-twins is antagonized by greatwall and collaborates with polo for cell cycle progression and centrosome attachment to nuclei in Drosophila embryos. PLoS Genet. 7, e1002227 ( 10.1371/journal.pgen.1002227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rangone H, et al. 2011. Suppression of scant identifies Endos as a substrate of greatwall kinase and a negative regulator of protein phosphatase 2A in mitosis. PLoS Genet. 7, e1002225 ( 10.1371/journal.pgen.1002225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Glover DM. 2012. The overlooked greatwall: a new perspective on mitotic control. Open Biol. 2, 120023 ( 10.1098/rsob.120023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lorca T, Castro A. 2013. The Greatwall kinase: a new pathway in the control of the cell cycle. Oncogene 32, 537–543. ( 10.1038/onc.2012.79) [DOI] [PubMed] [Google Scholar]
- 93.Mochida S, Maslen SL, Skehel M, Hunt T. 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670–1673. ( 10.1126/science.1195689) [DOI] [PubMed] [Google Scholar]
- 94.Boke E, Hagan IM. 2011. Polo, greatwall, and protein phosphatase PP2A jostle for pole position. PLoS Genet. 7, e1002213 ( 10.1371/journal.pgen.1002213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang J, Raff JW. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18, 2184–2195. ( 10.1093/emboj/18.8.2184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wakefield JG, Huang JY, Raff JW. 2000. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr. Biol. 10, 1367–1370. ( 10.1016/S0960-9822(00)00776-4) [DOI] [PubMed] [Google Scholar]
- 97.Klotz C, Dabauvalle MC, Paintrand M, Weber T, Bornens M, Karsenti E. 1990. Parthenogenesis in Xenopus eggs requires centrosomal integrity. J. Cell Biol. 110, 405–415. ( 10.2307/1613919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Picard A, Karsenti E, Dabauvalle MC, Dorée M. 1987. Release of mature starfish oocytes from interphase arrest by microinjection of human centrosomes. Nature 327, 170–172. ( 10.1038/327170a0) [DOI] [PubMed] [Google Scholar]
- 99.Nigg EA, Schäfer G, Hilz H, Eppenberger HM. 1985. Cyclic-AMP-dependent protein-kinase type-II is associated with the Golgi-complex and with centrosomes. Cell 41, 1039–1051. ( 10.1016/S0092-8674(85)80084-2) [DOI] [PubMed] [Google Scholar]
- 100.De Camilli P, Moretti M, Donini SD, Walter U, Lohmann SM. 1986. Heterogeneous distribution of the cAMP receptor protein RII in the nervous system: evidence for its intracellular accumulation on microtubules, microtubule-organizing centers, and in the area of the Golgi complex. J. Cell Biol. 103, 189–203. ( 10.2307/1612053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Witczak O, Skålhegg BS, Keryer G, Bornens M, Taskén K, Jahnsen T, Orstavik S. 1999. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 18, 1858–1868. ( 10.1093/emboj/18.7.1858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M, Tassin A-M. 2003. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell 14, 4260–4271. ( 10.1091/mbc.E02-11-0773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. 2011. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193, 917–933. ( 10.1083/jcb.201011014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Terrin A, et al. 2012. PKA and PDE4D3 anchoring to AKAP9 provides distinct regulation of cAMP signals at the centrosome. J. Cell Biol. 198, 607–621. ( 10.1083/jcb.201201059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Debec A, Montmory C. 1992. Cyclin-B is associated with centrosomes in Drosophila mitotic cells. Biol. Cell 75, 121–126. ( 10.1016/0248-4900(92)90131-J) [DOI] [PubMed] [Google Scholar]
- 106.Dutertre S, et al. 2004. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell. Sci. 117, 2523–2531. ( 10.1242/jcs.01108) [DOI] [PubMed] [Google Scholar]
- 107.Gopalan GG, Chan CSC, Donovan PJP. 1997. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J. Cell Biol. 138, 643–656. ( 10.2307/1618199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kimura M, Kotani S, Hattori T, Sumi N, Yoshioka T, Todokoro K, Okano Y. 1997. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to aurora of Drosophila and yeast Ipl1. J. Biol. Chem. 272, 13 766–13 771. ( 10.1074/jbc.272.21.13766) [DOI] [PubMed] [Google Scholar]
- 109.Roghi C, et al. 1998. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell. Sci. 111, 557–572. [DOI] [PubMed] [Google Scholar]
- 110.Golsteyn RM, Mundt KE, Fry AM, Nigg EA. 1995. Cell-cycle regulation of the activity and subcellular-localization of Plk1, a human protein-kinase implicated in mitotic spindle function. J. Cell Biol. 129, 1617–1628. ( 10.2307/1616934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jackman M, Lindon C, Nigg EA, Pines J. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5, 143–148. ( 10.1038/ncb918) [DOI] [PubMed] [Google Scholar]
- 112.Gavet O, Pines J. 2010. Progressive activation of cyclinB1–Cdk1 coordinates entry to mitosis. Dev. Cell 18, 533–543. ( 10.1016/j.devcel.2010.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lindqvist A, Källström H, Lundgren A, Barsoum E, Rosenthal CK. 2005. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J. Cell Biol. 171, 35–45. ( 10.1083/jcb.200503066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 291, 1547–1550. ( 10.1126/science.1056866) [DOI] [PubMed] [Google Scholar]
- 115.Khodjakov A, Rieder CL. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242. ( 10.2307/1620438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176, 173–182. ( 10.1083/jcb.200607073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sir J-H, Pütz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203, 747–756. ( 10.1083/jcb.201309038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barr FA, Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell 131, 847–860. ( 10.1016/j.cell.2007.11.011) [DOI] [PubMed] [Google Scholar]
- 119.Piel M, et al. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553. ( 10.1126/science.1057330) [DOI] [PubMed] [Google Scholar]
- 120.Fabbro M, et al. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9, 477–488. ( 10.1016/j.devcel.2005.09.003) [DOI] [PubMed] [Google Scholar]
- 121.Bastos RN, Barr FA. 2010. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 191, 751–760. ( 10.1083/jcb.201008108.dv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161, 535–545. ( 10.1083/jcb.200301105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gromley A, Yeaman C, Rosa J, Redick S, Chen C-T, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. 2005. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87. ( 10.1016/j.cell.2005.07.027) [DOI] [PubMed] [Google Scholar]
- 124.Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620. ( 10.1126/science.1201847) [DOI] [PubMed] [Google Scholar]
- 125.Chen C-T, Hehnly H, Doxsey SJ. 2012. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat. Rev. Mol. Cell Biol. 13, 483–488. ( 10.1038/nrm3395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pan D. 2010. The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505. ( 10.1016/j.devcel.2010.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu F-X, Guan K-L. 2013. The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371. ( 10.1101/gad.210773.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schroeder MC, Halder G. 2012. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin. Cell Dev. Biol. 23, 803–811. ( 10.1016/j.semcdb.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 129.Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. 2010. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176. ( 10.1038/ncb2120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Florindo C, Perdigão J, Fesquet D, Schiebel E, Pines J, ÁA Tavares. 2012. Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. J. Cell. Sci. 125, 3085–3090. ( 10.1242/jcs.097147) [DOI] [PubMed] [Google Scholar]
- 131.Maniotis A, Schliwa M. 1991. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell 67, 495–504. ( 10.1016/0092-8674(91)90524-3) [DOI] [PubMed] [Google Scholar]
- 132.Srsen V, Gnadt N, Dammermann A, Merdes A. 2006. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 174, 625–630. ( 10.1083/jcb.200606051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. 2006. Loss of centrosome integrity induces p38-p53-p21-dependent G1–S arrest. Nat. Cell Biol. 9, 160–170. ( 10.1038/ncb1529) [DOI] [PubMed] [Google Scholar]
- 134.Fletcher L, Muschel RJ. 2006. The centrosome and the DNA damage induced checkpoint. Cancer Lett. 243, 1–8. ( 10.1016/j.canlet.2006.01.006) [DOI] [PubMed] [Google Scholar]
- 135.Wilkinson CJ, Andersen JS, Mann M, Nigg EA. 2005. A proteomic approach to the inventory of the human centrosome. In Centrosomes in development and disease (ed. Nigg EA.), pp. 123–142. Weinheim, FRG: Wiley-VCH. [Google Scholar]
- 136.Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. 2004. Centrosome-associated Chk1 prevents premature activation of cyclin-B–Cdk1 kinase. Nat. Cell Biol. 6, 884–891. ( 10.1038/ncb1165) [DOI] [PubMed] [Google Scholar]
- 137.Matsuyama M, Goto H, Kasahara K, Kawakami Y, Nakanishi M, Kiyono T, Goshima N, Inagaki M. 2011. Nuclear Chk1 prevents premature mitotic entry. J. Cell. Sci. 124, 2113–2119. ( 10.1242/jcs.086488) [DOI] [PubMed] [Google Scholar]
- 138.Löffler H, et al. 2011. Cep63 recruits Cdk1 to the centrosome: implications for regulation of mitotic entry, centrosome amplification, and genome maintenance. Cancer Res. 71, 2129–2139. ( 10.1158/0008-5472.CAN-10-2684) [DOI] [PubMed] [Google Scholar]
- 139.Lukinavičius G, Lavogina D, Goenczy P, Johnsson K. 2013. Commercial Cdk1 antibodies recognize the centrosomal protein Cep152. Biotechniques 55, 111–114. ( 10.2144/000114074) [DOI] [PubMed] [Google Scholar]
- 140.Hut HMJ, Rembacz KP, van Waarde MAWH, Lemstra W, van Cappellen WA, Kampinga HH, Sibon OCM. 2005. Dysfunctional BRCA1 is only indirectly linked to multiple centrosomes. Oncogene 24, 7619–7623. ( 10.1038/sj.onc.1208859) [DOI] [PubMed] [Google Scholar]
- 141.Middendorp S, Paoletti A, Schiebel E, Bornens M. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl Acad. Sci. USA 94, 9141–9146. ( 10.1073/pnas.94.17.9141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stemm-Wolf AJ, Morgan G, Giddings TH, White EA, Marchione R, McDonald HB, Winey M. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell 16, 3606–3619. ( 10.1091/mbc.E04-10-0919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. 2005. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr. Biol. 15, 2097–2106. ( 10.1016/j.cub.2005.11.038) [DOI] [PubMed] [Google Scholar]
- 144.Dantas TJT, Wang YY, Lalor PP, Dockery PP, Morrison CGC. 2011. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J. Cell Biol. 193, 307–318. ( 10.1083/jcb.201012093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof Y-D, Nigg EA. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202. ( 10.1016/j.devcel.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 146.Salisbury JL, Suino KM, Busby R, Springett M. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287–1292. ( 10.1016/S0960-9822(02)01019-9) [DOI] [PubMed] [Google Scholar]
- 147.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell. Sci. 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- 148.Popescu A, Miron S, Blouquit Y, Duchambon P, Christova P, Craescu CT. 2003. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J. Biol. Chem. 278, 40 252–40 261. ( 10.1074/jbc.M302546200) [DOI] [PubMed] [Google Scholar]
- 149.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276, 18 665–18 672. ( 10.1074/jbc.M100855200) [DOI] [PubMed] [Google Scholar]
- 150.Delaval B, Doxsey SJ. 2010. Pericentrin in cellular function and disease. J. Cell Biol. 188, 181–190. ( 10.1083/jcb.200908114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Graser S, Stierhof Y-D, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330. ( 10.1083/jcb.200707181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Griffith E, et al. 2007. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 40, 232–236. ( 10.1038/ng.2007.80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chaki M, et al. 2012. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150, 533–548. ( 10.1016/j.cell.2012.06.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barr AR, Kilmartin JV, Gergely F. 2010. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 189, 23–39. ( 10.1083/jcb.200912163.dv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. 2007. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene 27, 139–144. ( 10.1038/sj.onc.1210595) [DOI] [PubMed] [Google Scholar]
- 156.Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. 2003. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol. Biol. Cell 14, 1993–2004. ( 10.1091/mbc.E02-08-0510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Inanc B, Dodson H, Morrison CG. 2010. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol. Biol. Cell 21, 3866–3877. ( 10.1091/mbc.E10-02-0124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. 2007. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 8, 603–609. ( 10.1038/sj.embor.7400962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23, 3864–3873. ( 10.1038/sj.emboj.7600393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Löffler H, Fechter A, Liu FY, Poppelreuther S, Kramer A. 2013. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene 32, 2963–2972. ( 10.1038/onc.2012.310) [DOI] [PubMed] [Google Scholar]
- 161.Prosser SL, Samant MD, Baxter JE, Morrison CG, Fry AM. 2012. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J. Cell. Sci. 125, 5353–5368. ( 10.1242/jcs.106096) [DOI] [PMC free article] [PubMed] [Google Scholar]