To the Editor
A growing trend in protein quantification is a targeted mass spectrometry (MS)-based technology called multiple reaction monitoring (MRM) or selected reaction monitoring (SRM). Here, we present the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (http://assays.cancer.gov/), a public repository of well-characterized, MS-based, targeted proteomic assays.
In contrast to ‘shotgun’ MS, MRM involves targeted measurements of specific proteins of interest that can be context-dependent on the basis of the biological question being asked. MRM-based assays are specific, multiplexable and precise, and they can be standardized, reproduced and distributed across laboratories and instruments1,2. A recent international study demonstrated the feasibility and usefulness of scaling to develop MRM-based assays covering a large number of human proteins3.
Although hundreds of MRM-based assays have been published, the information is dispersed throughout the literature, and protocols for characterization of assay performances have not been standardized, making it difficult to evaluate the quality of published assays and, by extension, the results of those assays4. While databases (for example, SRMAtlas5, PASSEL6, GPMDB/MRM7 and QuAD8) or libraries9 are available to identify peptide analytes and transitions for development of MRM assays, there is no public database of analytically validated assays and standard operating protocols (SOPs), which are critical for standardizing and harmonizing proteomic results across the community. As a result, despite widespread capability to perform MRM assays, the benefits of MRM have not yet been realized by the biological and clinical research communities.
To address this need, the CPTAC of the US National Cancer Institute (NCI) has launched an Assay Portal (http://assays.cancer.gov/) to serve as a public repository of well-characterized, MS-based, targeted proteomic assays (Fig. 1a). The purpose of the CPTAC Assay Portal is to facilitate widespread adoption of targeted MS assays by disseminating SOPs, reagents and assay characterization data. A primary aim of the portal is to bring together clinicians or biologists and analytical chemists to answer hypothesis-driven questions using targeted, MS-based assays. Assay content is accessed through queries, enabling investigators to find assays to proteins relevant to their areas of interest. Detailed characterization data are available for each assay, enabling researchers to evaluate performance before launching assays in their own laboratories.
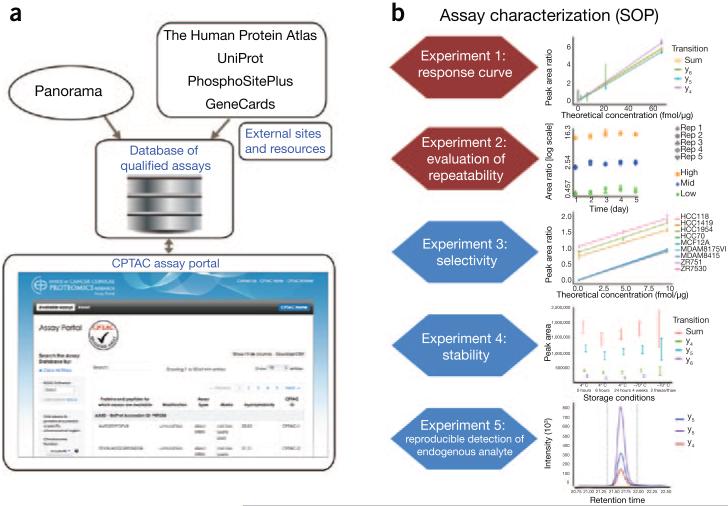
Figure 1.
Overview of the CPTAC assay portal. (a) The CPTAC Assay Portal enables query of a database of well-characterized, targeted MS-based assays. Bioinformatic annotations are pulled from outside sources to enable biologically relevant queries, as well as mapping of peptide analytes relative to sequence domains, isoforms, coding polymorphisms and post-translational modifications. Contributions from external sites are noted on the portal. The assay database is tied to Panorama, an open-source platform allowing for efficient collection and sharing of proteomics data in a vendor-neutral format (for example, currently supporting data from six vendors: AB SCIEX, Agilent, Bruker, Shimadzu, Thermo and Waters), facilitating upload and viewing of assay characterization data, and download of Skyline documents for implementing the assays. (b) CPTAC characterization is a set of experiments designed to help users evaluate assay performance. There are five recommended experiments. Experiments 1 and 2 are required for each assay; experiments 3–5 are optional. Experiment 1 is a response curve (dilution curve of peptides spiked into a representative background matrix) designed to evaluate linearity, the within-batch precision of the liquid chromatography–MRM-MS analysis of the analyte peptide in a complex mixture and the upper and lower limits of quantification, and to provide data on selectivity (through detection of interferences in the curve data). Experiment 2 (evaluation of repeatability, three replicates of peptides spiked at three concentrations—low, medium (mid) and high) determines the repeatability of measurements at multiple concentrations across 5 d. Experiment 3 tests selectivity by evaluating parallelism (a measure of the influence of matrix components on the quantification of peptides in a complex mixture) in multiple biological replicates of the matrix of interest. Experiment 4 evaluates the stability of peptides after sample preparation and helps the downstream user determine whether peptides can be left on the autosampler for a period of time or frozen before analysis. Experiment 5 aims to demonstrate that endogenous peptides can be quantified in a relevant matrix. y4, y5, y6, and Sum denote the transition ion plotted in each graph.
To appeal to biologists, bioinformaticians and analytical chemists, the CPTAC Assay Portal is organized into four levels. The first level of the portal is the landing page, which is designed to be relevant to biologists, enabling database query for validated assays to quantify proteins involved in specific cellular pathways or protein complexes, proteins whose genes map to specific chromosomal regions or proteins associated with specified Gene Ontology terms. The second level of the portal displays a protein-centric view onto which the positions of assay peptide analytes are mapped relative to features of interest, such as sequence domains, isoforms, coding polymorphisms and post-translational modifications. The third level provides detailed assay characterization data (accessible through Panorama (https://panoramaweb.org/)), allowing users to evaluate the expected analytical performance of each assay. Finally, the fourth level of the portal is designed to enable users to implement the assays in their laboratories. Users have the ability to download Skyline10 (a freely available Windows client application for building and analyzing targeted MS data) documents for assay implementation on most mass spectrometer platforms. For each assay, an SOP describing sample preparation and analysis is available for download, and a discussion board is provided for community-based information exchange on assay performance.
Data quality is a key emphasis and distinguishing feature of the portal. A framework for MRM assay ‘fit-for-purpose’ validation has been established by CPTAC, with input from the outside community solicited via a workshop jointly sponsored by the NCI and the National Heart, Lung, and Blood Institute4. Assays presented on the portal predominantly represent ‘Tier 2’ assays, as described in the workshop report. To ensure sufficient assay characterization and data quality, a guidance document describing the minimal characterization data required for assay inclusion in the CPTAC Assay Portal is available for download (https://assays.cancer.gov/guidance-document/). This guidance document outlines a list of characterizations that will help potential downstream users evaluate the utility of adopting and deploying the assays in their own work. Five experiments are outlined (Fig. 1b); experiments 1 and 2 are required for upload. Assays are entered through a web-based entry form and the uploading of data to Panorama. Characterization experiments are analyzed by scripts available through Panorama. Uploaded assays are subjected to manual review by site administrators to ensure data quality and the fulfillment of minimum requirements for assay characterization.
At launch, the portal contains >450 assays (from a recent scaled MRM development effort)3. CPTAC plans to add several hundred more assays over the next 2–3 years, and the portal will soon be open for contributions from the community. Of note, the portal is able to accept data from a variety of assay types, targeted MS methods and reference targeted data, such as parallel reaction monitoring (PRM)11,12, MRM with high resolution (MRM-HR) and standard metrics such as iRT peptides13, providing a versatile repository of well-characterized assays.
ACKNOWLEDGMENTS
This work was funded by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) of the US National Cancer Institute under grant nos. U24CA160034 (S.A.C., A.G.P.), U24CA160036 (D.W.C.), U24CA160019 (R.D.S., K.D.R.), U24CA159988 (D.C.L.) and U24CA160035 (M.J.C.E.), as well as US National Institutes of Health grant nos. R01GM103551 (M.J.M.) and U01CA164186 (M.J.M.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Addona TA, et al. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox HD, et al. Clin. Chem. 2014;60:541–548. doi: 10.1373/clinchem.2013.208538. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy JJ, et al. Nat. Methods. 2014;11:149–155. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr SA, et al. Mol. Cell. Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picotti P, et al. Nat. Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrah T, et al. Proteomics. 2012;12:1170–1175. doi: 10.1002/pmic.201100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig R, Cortens JP, Beavis RCJ. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 8.Remily-Wood ER, et al. Proteomics Clin. Appl. 2011;5:383–396. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Lazar IM. BMC Cancer. 2009;9:96. doi: 10.1186/1471-2407-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLean B, et al. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallien S, et al. Mol. Cell. Proteomics. 2012;11:1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escher C, et al. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]