Abstract
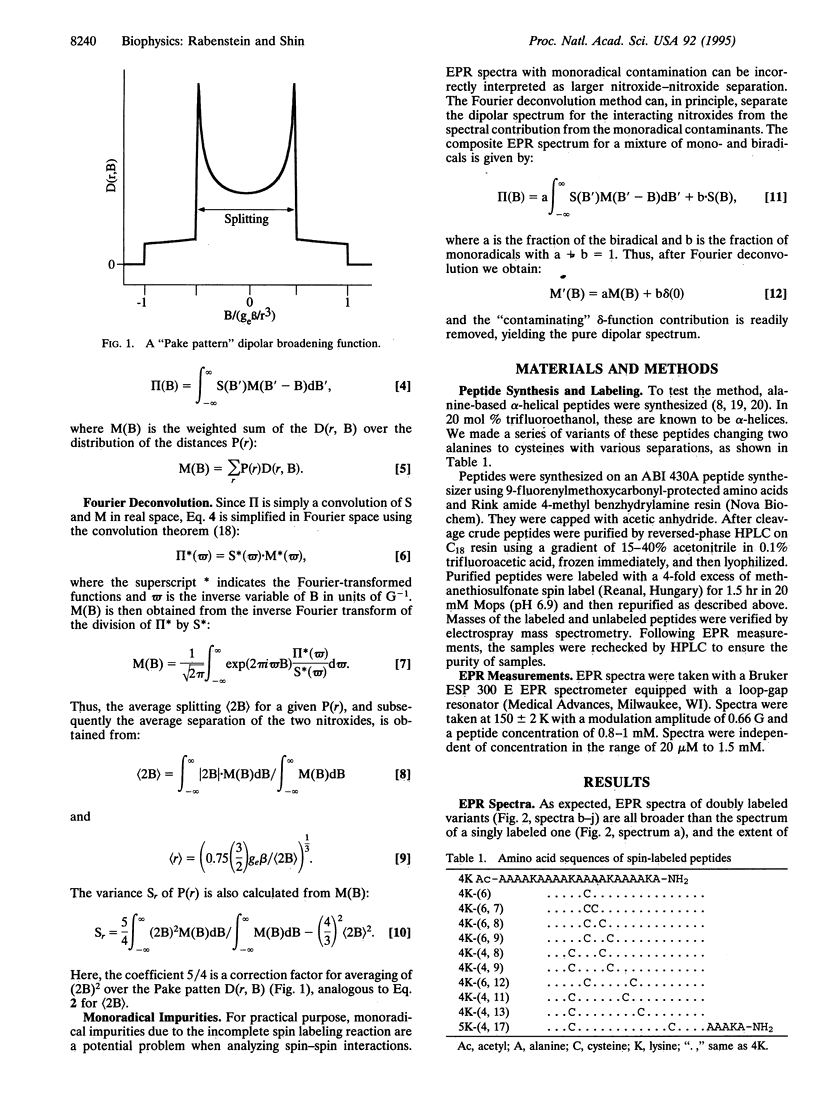
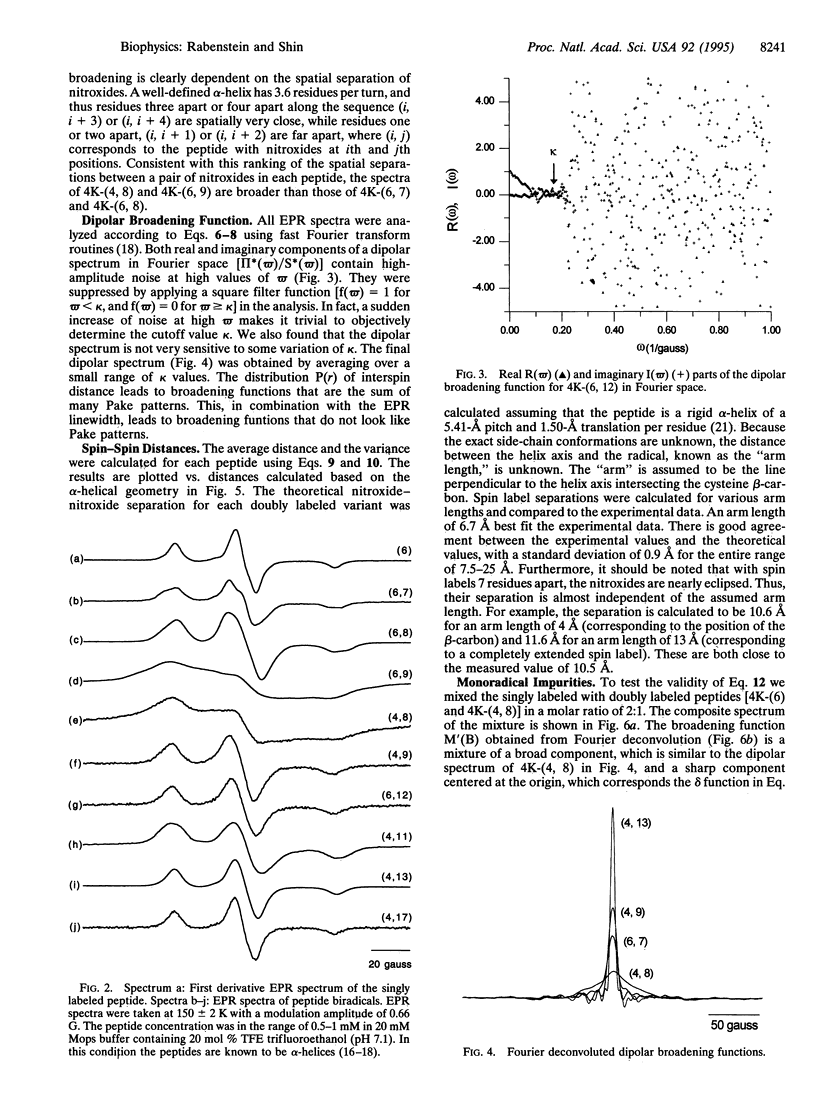
An EPR "spectroscopic ruler" was developed using a series of alpha-helical polypeptides, each modified with two nitroxide spin labels. The EPR line broadening due to electron-electron dipolar interactions in the frozen state was determined using the Fourier deconvolution method. These dipolar spectra were then used to estimate the distances between the two nitroxides separated by 8-25 A. Results agreed well with a simple alpha-helical model. The standard deviation from the model system was 0.9 A in the range of 8-25 A. This technique is applicable to complex systems such as membrane receptors and channels, which are difficult to access with high-resolution NMR or x-ray crystallography, and is expected to be particularly useful for systems for which optical methods are hampered by the presence of light-interfering membranes or chromophores.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbach C., Marti T., Khorana H. G., Hubbell W. L. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990 Jun 1;248(4959):1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- Anthony-Cahill S. J., Benfield P. A., Fairman R., Wasserman Z. R., Brenner S. L., Stafford W. F., 3rd, Altenbach C., Hubbell W. L., DeGrado W. F. Molecular characterization of helix-loop-helix peptides. Science. 1992 Feb 21;255(5047):979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Carraway K. L., 3rd, Cerione R. A. Fluorescent-labeled growth factor molecules serve as probes for receptor binding and endocytosis. Biochemistry. 1993 Nov 16;32(45):12039–12045. doi: 10.1021/bi00096a014. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Carlberg C., Diekmann S., Lilley D. M. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992 May 26;31(20):4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- Dale R. E., Eisinger J., Blumberg W. E. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys J. 1979 May;26(2):161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough R. H., Cantor C. R. The use of singlet-singlet energy transfer to study macromolecular assemblies. Methods Enzymol. 1978;48:347–379. doi: 10.1016/s0076-6879(78)48019-x. [DOI] [PubMed] [Google Scholar]
- Fiori W. R., Miick S. M., Millhauser G. L. Increasing sequence length favors alpha-helix over 3(10)-helix in alanine-based peptides: evidence for a length-dependent structural transition. Biochemistry. 1993 Nov 16;32(45):11957–11962. doi: 10.1021/bi00096a003. [DOI] [PubMed] [Google Scholar]
- Langlois R., Lee C. C., Cantor C. R., Vince R., Pestka S. The distance between two functionally significant regions of the 50 S Escherichia coli ribosome: the erythromycin binding site and proteins L7/L12. J Mol Biol. 1976 Sep 15;106(2):297–313. doi: 10.1016/0022-2836(76)90087-5. [DOI] [PubMed] [Google Scholar]
- Lockhart D. J., Kim P. S. Electrostatic screening of charge and dipole interactions with the helix backbone. Science. 1993 Apr 9;260(5105):198–202. doi: 10.1126/science.8469972. [DOI] [PubMed] [Google Scholar]
- Marqusee S., Robbins V. H., Baldwin R. L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miick S. M., Martinez G. V., Fiori W. R., Todd A. P., Millhauser G. L. Short alanine-based peptides may form 3(10)-helices and not alpha-helices in aqueous solution. Nature. 1992 Oct 15;359(6396):653–655. doi: 10.1038/359653a0. [DOI] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Intrasubunit signal transduction by the aspartate chemoreceptor. Science. 1991 Dec 13;254(5038):1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Shin Y. K., Levinthal C., Levinthal F., Hubbell W. L. Colicin E1 binding to membranes: time-resolved studies of spin-labeled mutants. Science. 1993 Feb 12;259(5097):960–963. doi: 10.1126/science.8382373. [DOI] [PubMed] [Google Scholar]
- Stryer L., Haugland R. P. Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci U S A. 1967 Aug;58(2):719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Gerstein M., Oesterhelt D., Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993 Jan;12(1):1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley P., Gronenborn A. M., Christensen H., Wingfield P. T., Pain R. H., Clore G. M. Kinetics of folding of the all-beta sheet protein interleukin-1 beta. Science. 1993 May 21;260(5111):1110–1113. doi: 10.1126/science.8493553. [DOI] [PubMed] [Google Scholar]


