Abstract
The cardiac conduction system is a specialized tract of myocardial cells responsible for maintaining normal cardiac rhythm. Given its critical role in coordinating cardiac performance, a detailed analysis of the molecular mechanisms underlying conduction system formation should inform our understanding of arrhythmia pathophysiology and affect the development of novel therapeutic strategies. Historically, the ability to distinguish cells of the conduction system from neighboring working myocytes presented a major technical challenge for performing comprehensive mechanistic studies. Early lineage tracing experiments suggested that conduction cells derive from cardiomyocyte precursors, and these claims have been substantiated by using more contemporary approaches. However, regional specialization of conduction cells adds an additional layer of complexity to this system, and it appears that different components of the conduction system utilize unique modes of developmental formation. The identification of numerous transcription factors and their downstream target genes involved in regional differentiation of the conduction system has provided insight into how lineage commitment is achieved. Furthermore, by adopting cutting-edge genetic techniques in combination with sophisticated phenotyping capabilities, investigators have made substantial progress in delineating the regulatory networks that orchestrate conduction system formation and their role in cardiac rhythm and physiology. This review describes the connectivity of these gene regulatory networks in cardiac conduction system development and discusses how they provide a foundation for understanding normal and pathological human cardiac rhythms.
Keywords: conduction system, sinus node, atrioventricular node, Purkinje fiber, transcription factor
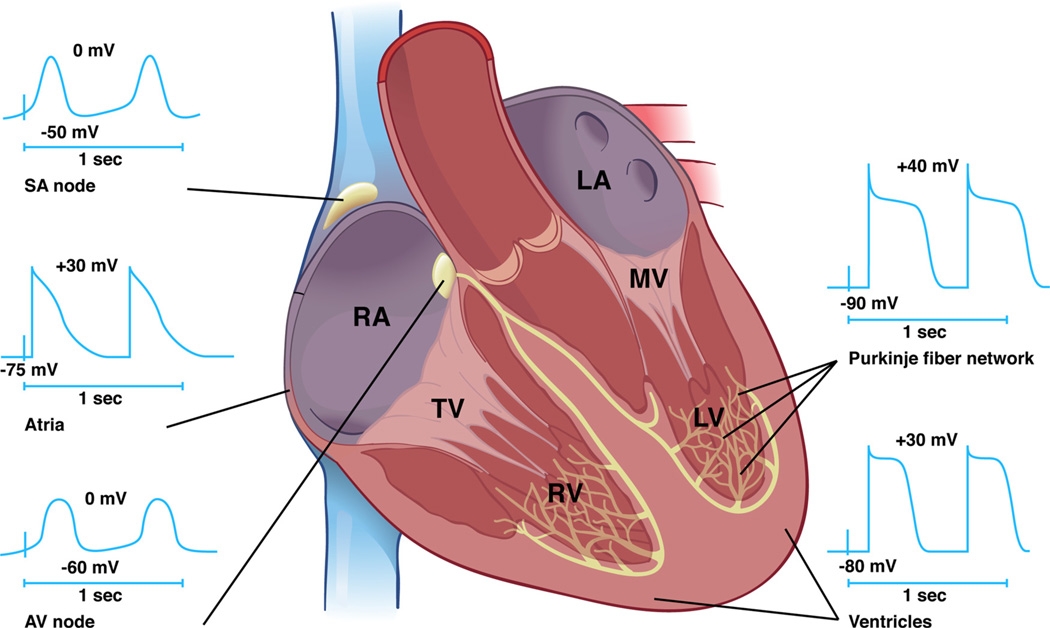
The cardiac conduction system is a specialized tract of cells responsible for initiation and propagation of normal cardiac rhythm (Figure 1). The cardiac impulse arises in the sinuatrial node (SAN) located at the junction of the superior vena cava and the crista terminalis of the right atrium. The SAN depolarizes neighboring atrial cardiomyocytes, and the impulse is propagated throughout both atria leading to subsequent atrial contraction. Before the onset of atrial systole, however, the impulse funnels toward the atrioventricular node (AVN), the only electric connection between the atria and ventricles in the mature heart. The AVN delays impulse propagation so that atrial contraction and late ventricular filling is completed before ventricular systole to optimize cardiac performance. After exiting the AVN, the impulse accelerates through the AV bundle (AVB) and conducts via asymmetrical right and left bundle branches (BB). Terminal arborization of the bundle branches gives rise to the Purkinje fiber (PF) network to directly depolarize working ventricular cardiomyocytes resulting in ventricular contraction and expulsion of blood through the right and left ventricular outflow tracts.
Figure 1. Functional anatomy of the cardiac conduction system.
The cardiac impulse is initiated in the SAN and propagates through the atria. The electric impulse is then delayed in the AVN before rapid conduction through the right and left bundle branches before terminating in the Purkinje fiber network, which directly interfaces with ventricular cardiomyocytes. Distinct time-voltage relationships are noted in the SAN, atria, AVN, PFs, and ventricles. RA indicates right atrium; LA, left atrium; TV, tricuspid valve; MV, mitral valve; RV, right ventricle; LV, left ventricle.
Even subtle perturbation of this finely orchestrated electric activation pattern can cause clinically important arrhythmias.1 Despite the fact that cardiac rhythm disturbances account for a significant proportion of morbidity and mortality attributable to cardiovascular disease in the general population, a comprehensive mechanistic understanding of arrhythmia pathophysiology remains incomplete. Such insight promises to inform the development of novel pharmacological agents targeted at ameliorating cardiac arrhythmias, especially given previous failures of antiarrhythmic agents in humans.2 Based on extensive characterization of electric excitability in neurons, initial studies aimed at elucidating arrhythmia mechanisms focused on the role of ion channels and gap junctions in establishing normal cardiac rhythm; indeed, a number of seminal human genetic linkage studies bolstered the merit of this approach.3–6 Although significant inroads have been made into the ionic mechanisms that orchestrate cardiac electric excitability and pacemaker function, the genetic blueprint responsible for differentiation, morphogenesis, and maintenance of the cardiac conduction system has only recently begun to emerge.
In this review, I outline our current understanding of the genetic networks that regulate cardiac conduction system development focusing on examples where mechanistic details have been analyzed in detail. The developmental basis of cardiac conduction system lineage commitment will be briefly reviewed before discussing transcriptional networks, and known genetic associations between conduction system regulatory mechanisms and human disease pathophysiology will be highlighted in the relevant sections.
Developmental Origins of the Cardiac Conduction System
Anatomic Observations
The inability to visually distinguish working cardiomyocytes from neighboring conduction system cells has been a major source of difficulty in advancing our knowledge of conduction system lineage specification. Pioneering anatomic studies first raised the possibility of a specialized network of cells dedicated to impulse generation and propagation within the heart.7 In search of the atrioventricular connection responsible for electric impulse propagation, Tawara identified the AVN and His-Purkinje system, which he elegantly illustrated in a series of classic monographs.8 Subsequently, Keith and Flack discovered the SAN and described its disposition within the right atrium.9 Based on these and other anatomic studies,10,11 3 criteria were set forth to distinguish conduction cells from working myocardium: (1) conduction cells must be histologically discrete from neighboring cells, (2) conduction cells should be traced from section to section, and (3) conduction cells should be insulated by fibrous tissue. Although crude by today’s standards, these criteria have stood the test of time and remain largely applicable today.
Histological Studies
Identifying cells destined to become part of the developing conduction system remained a challenge, however, owing to the lack of cell-type–specific markers and the dearth of fibrous tissue within the embryonic heart. Despite these shortcomings, Viragh and Challice provided a detailed histological analysis of the cardiac conduction system during mouse embryonic development based on their detailed knowledge of the anatomic disposition of the conduction system within the heart.12–15 They noted that a primitive AV conduction system (AVCS) forms as early as E8 by connecting the embryonic atrial myocardium with the trabecular system in the ventricles.13 Furthermore, a group of large PAS+ cells were noted in the dorsal AV canal (AVC) at E11 that were hypothesized to represent the AVN primordium; this structure formed concomitant with apoptosis occurring elsewhere in the AVC. By analyzing older embryos (E11–E12), they noted that the cells of the dorsal AVC proliferate and extend into the interventricular septum to form the primitive AVB and proximal BBs.12 At E12, connective tissue begins to surround and electrically isolate the conduction system, and the authors astutely postulated that failure to complete this process may contribute to the persistence of tracts that connect the atria and ventricles. They also examined the development of the SAN,14 noting that the primordium forms from cells located in the dorsal wall of the sinus horns in proximity to the right venous valve; these changes are observed at E8, even before the onset of visible contractions (E9) or the presence of a morphologically identifiable SAN (E11). Interestingly, these authors recognized the presence of nodal tissue within the left sinus horn (LSH) that regresses by E13 and becomes incorporated into the future coronary sinus. Later in mouse fetal gestation (E13–E16), the developing AVCS was noted to have progressively enriched glycogen content,15 suggesting from a cytological standpoint that lineage commitment had already occurred.
Lineage Tracing
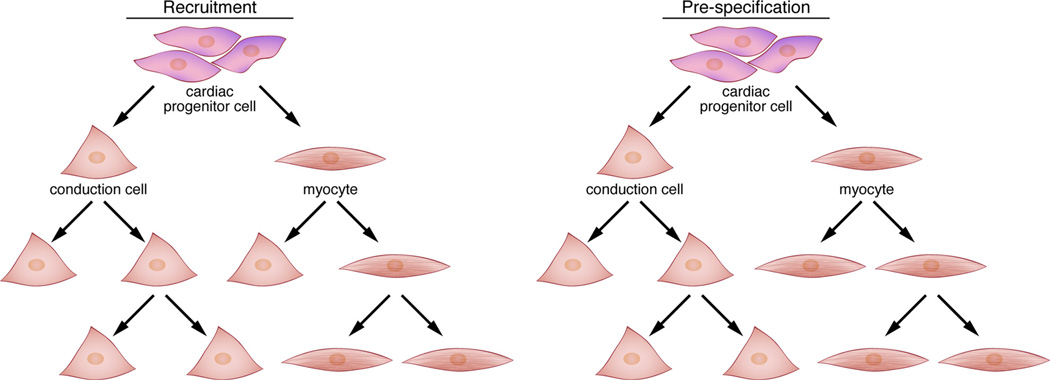
Taken together, anatomic and histological studies elegantly described the morphological and cytological changes that accompanied differentiation of the cardiac conduction system. However, the cell of origin for the conduction cell lineage remained elusive due to technical limitations. By adapting retroviral lineage tracing techniques in chick embryos, Gourdie, Mikawa, and colleagues were able to specifically address this fundamental question. From their studies, they concluded that peri-arterial PFs derive from nearby ventricular cardiomyoctyes under the inductive influence of endothelin-1 (ET-1) produced by localized arterial endothelium.16–21 Importantly, they demonstrated that cells of the avian conduction system derive from cardiomyocytes rather than nonmyocyte precursors. Furthermore, these studies supported the notion that avian PFs develop by continuous recruitment, rather than prespecification (Figure 2).
Figure 2. Models for cardiac conduction system development.
Two theoretical models are shown that provide a conceptual framework for understanding the mechanisms by which distinct components of the conduction system develop. In the recruitment model (left), a cardiac progenitor cell has the potential to give rise to a conduction cell or a working myocyte. As development proceeds, working myocytes retain bipotential characteristics and can continue to give rise to conduction cells, leading to the concept of ongoing recruitment. In the prespecification model (right), a cardiac progenitor cell can differentiate into a conduction cell or a myocyte, similar to the recruitment model. However, once a cell becomes committed to one of these fates, its progeny remain distinct from each other. During development of the SAN and AVN in particular, Tbx2 and Tbx3 are restricted to the primary myocardium (ie, inflow tract, AVC, outflow tract) and act to locally repress working myocyte differentiation while simultaneously reinforcing the nodal gene expression program. Note that the two models are not necessarily mutually exclusive.
Building on these fundamental studies, the availability of transgenic mouse models allowing direct visualization of the developing cardiac conduction system provided the tools to more precisely describe the fate of these cells.22–25 Even though analysis of these transgenic lines provided unprecedented cellular resolution for the morphological events that take place during cardiac conduction system development, many of the findings were remarkably similar to earlier observations made with cruder approaches; nevertheless, investigators could now directly test which inductive cues influence conduction system morphogenesis in mice. Fishman et al demonstrated that Neuregulin-1 (NRG-1), rather than ET-1, could expand the developing Purkinje network in cultured mouse hearts.26 More importantly, perhaps, these lines have allowed investigators to directly assess the effect of specific genetic backgrounds on conduction system development using molecularly defined markers.27–29
The recent advent of sophisticated transgenic lines for direct lineage tracing in mice has opened up several additional avenues of investigation. For example, additional support for the notion that the conduction system originates from cardiomyocytes derives from experiments using Cre lines that mark cardiomyocyte progenitors (eg, myosin light chain-2v–Cre and Isl1-Cre), although it remains possible that these transgenes are activated only after conduction cells have differentiated.29–31 Interestingly, lineage tracing studies using neural crest-specific Cre lines have provided strong evidence that these cells do not contribute to the cardiac conduction system but rather to components of the surrounding annulus fibrosus and may affect conduction system maturation by non-cell-autonomous mechanisms.32–34 Subsequent fate-mapping studies utilizing Tbx2-Cre, Tbx3-Cre, Tbx18-Cre, and atrial natriuretic factor (ANF)-Cre lines have allowed several important conclusions regarding development of the cardiac conduction system.35–37 First, Tbx3-expressing cells in the atrium segregate early from ANF-expressing cells, suggesting that development of these cells follows a path of progenitor prespecification (Figure 2) rather than ongoing recruitment as ascribed to formation of PFs in the ventricular conduction system.36 Furthermore, Tbx18-expressing cells, a subset of which will comprise the SAN, are set aside from working atrial myocytes as early as E10; however, the observation that only a subset of Tbx18+ cells gives rise to the definitive SAN implies that ongoing recruitment also contributes to SAN formation.37 Second, Tbx2-expressing cells give rise to a large proportion of the basal left ventricle (in addition to AVC derivatives such as the AVN) but not the AVB,35 consistent with the fact that the AVC and basal left ventricle are in direct apposition during cardiac morphogenesis.38 Thus, the SAN and AVN differ not only in their functional properties but also in their developmental plasticity, with SAN progenitors seemingly confined to a specific fate from an early time point, whereas AVN progenitors may be more susceptible to the influence of local factors.35–37
Retrospective Lineage Analysis and Inducible Fate-Mapping
Although previous studies had hinted at the cell of origin16,17 and potential inductive factors18–21,26 involved in development of the ventricular conduction system (ie, AVB, BB, and PF network), recent experiments using cutting-edge techniques have provided additional valuable insights into this process. Kelly et al made use of an α-cardiac actin nlaacZ mouse line in combination with a Cx40 enhanced green fluorescent protein (eGFP) knock-in line to determine the relationship between cells fated to become ventricular conduction cells versus neighboring ventricular working myocytes.39 Retrospective clonal analysis revealed that both cell types derive from a common progenitor cell and that limited proliferation occurs even after ventricular conduction cells become restricted to their lineage (as determined by being Cx40+). They also made the provocative observation that lineage commitment is not symmetrical between the right and left ventricular conduction system,39 suggesting that the process by which these cells become fate-restricted fundamentally differs in each ventricular compartment. Interestingly, their results using a Cx40 Cre recombinase fused with estrogen receptor T2 (CreERT2) knock-in line suggested that Cx40-expressing cells labeled before E16.5 retain the ability to differentiate into working or conductive myocytes,39 reminiscent of the plasticity observed for Tbx2-expressing cells in the AVC.35 Moreover, since the Cx40 CreERT2 knock-in line confirmed many of the observations made by retrospective lineage analysis,39 the availability of several additional inducible Cre lines40,41 promises to further advance our understanding of lineage commitment in other components of the developing conduction system.
Transcriptional Hierarchies in Cardiac Conduction System Development
Many developmental cell fate decisions are made as a result of distinct transcriptional programs that elicit specific gene expression changes in committed cells.42 Interestingly, the organization of these transcriptional circuits often involves feed-forward loops that reinforce definitive developmental decisions,43 although this is clearly not always the case.35 Nevertheless, I have attempted to organize the genetic circuitry of each individual component of the developing cardiac conduction system into transcriptional hierarchies based on currently available data, paying particular attention to recently published studies.
Sinuatrial Node
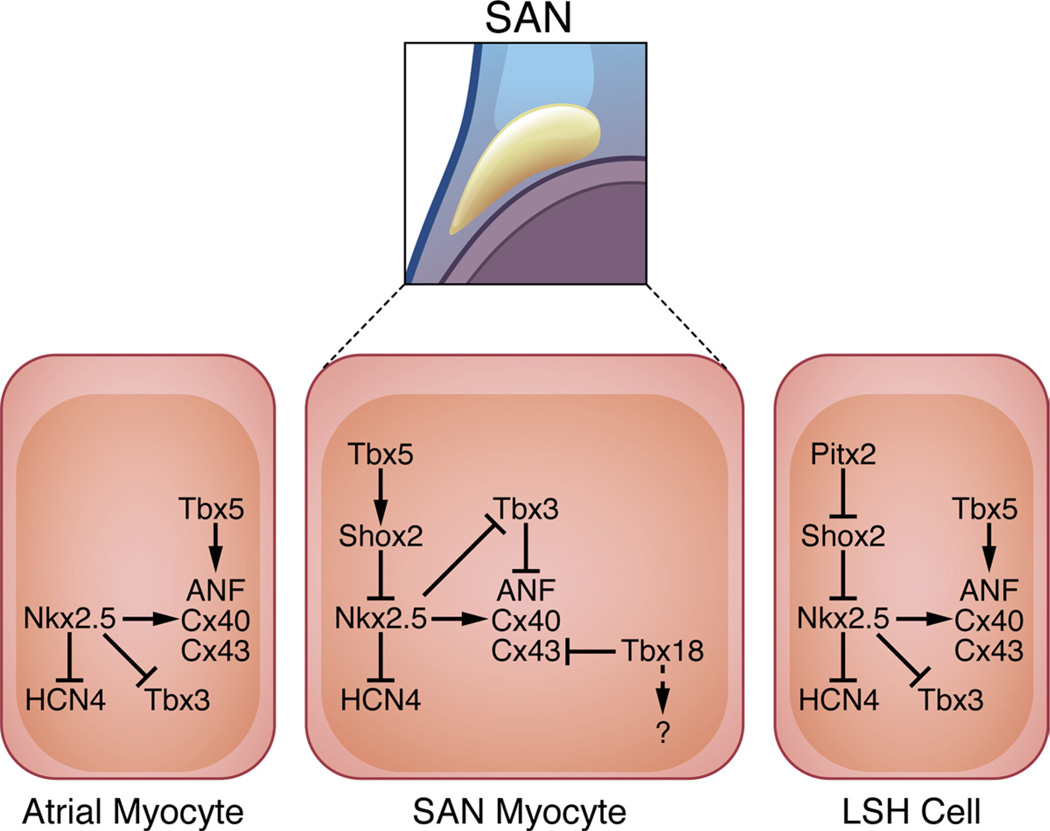
The SAN is absolutely essential for establishing dominant pacemaker activity in the developing heart, and its genetic circuitry has recently come under intense investigation (Figure 3). Based on the expression pattern of specific molecular markers, the SAN begins to develop from the embryonic sinus venosus myocardium at E9.5,44 concomitant with the first observed heart beat. At the top of the SAN transcriptional hierarchy sits Tbx5, which may function as a competence factor for cardiac morphogenetic events occurring toward the venous pole of the developing heart.45,46 In turn, Tbx5 has been shown to regulate SAN expression of Shox2,47 which is necessary for proper SAN formation and function.48,49 Shox2 null mice are embryonic-lethal, probably secondary to a slow heart rate as a result of severe hypoplasia of the SAN.48,49 Molecular analysis revealed that Nkx2.5, Cx40, Cx43, and ANF are ectopically expressed in the developing SAN of Shox2 knockout animals. Furthermore, Shox2 directly binds and represses an Nkx2.5 enhancer element, suggesting that Nkx2.5 is a direct downstream target of Shox2-dependent transcriptional repression.48 This is consistent with the fact that atrial Nkx2.5-expressing myocardium neither contributes to the SAN nor expresses SAN-specific markers Tbx3 and HCN4.50 Thus, Shox2 reinforces the SAN transcriptional program by directly repressing Nkx2.5 and Nkx2.5-dependent target genes (Cx40/Cx43/ANF), while preventing repression of Tbx3 and HCN4. In working atrial myocardium that lacks the repressive effects of Shox2, Nkx2.5 activates expression of chamber-specific genes (Cx40/Cx43/ANF) and represses SAN-specific genes (Tbx3/HCN4) to prevent ectopic pacemaker formation.51
Figure 3. Gene networks in the SAN.
Tbx18 is specifically expressed in cells of the SAN “head” region and is required for its morphogenesis in part by repressing Cx43, although the downstream effectors of this pathway remain largely uncharacterized. While Tbx5 activates chamber-specific genes in atrial cells along with Nkx2.5, Tbx5 likely acts as a competence factor for the SAN by activating Shox2 expression, which in turn directly represses Nkx2.5 transcription within this cell type. Nkx2.5 is typically expressed in a mutually exclusive manner in surrounding atrial myocytes, where it represses HCN4 and Tbx3 expression and activates transcription of the chamber-specific genes ANF, Cx40, and Cx43. Tbx3 is also specifically expressed in the SAN and works by preventing expression of chamber-specific genes. Left sinus horn (LSH) pacemaker tissue is prevented from forming by Pitx2, which inhibits Shox2 expression.
Aside from the Shox2 regulatory network, the transcriptional repressor Tbx3 has received considerable attention for its role in formation of the SAN. Indeed, Tbx3 is highly expressed in the SAN in a strictly complementary fashion compared with markers of atrial working myocardium.52 From a molecular standpoint, Tbx3 directly represses atrial myocardial genes (Cx40/Cx43/ANF) to prevent atrialization of the SAN.50 In fact, overexpression of Tbx3 in atrial working myocardium is sufficient to drive ectopic pacemaker formation and concomitant arrhythmias in mice.36 These findings have been further substantiated by recent studies in an allelic series of Tbx3 mutant mice, demonstrating that Tbx3 is a sensitive dosage-dependent regulator of SAN formation and function.53 Tbx3 has additional roles in development of other components of the cardiac conduction system, which are discussed below.
In addition to regulating left-right asymmetry in the developing embryo and heart,54,55 Pitx2 directly impacts pacemaker formation. In Pitx2 null embryos, a left-sided pacemaker is observed, suggesting that despite the competence of the LSH to form pacemaker tissue, Pitx2 normally acts to repress its formation.50 Additional studies have demonstrated that Pitx2 directly binds to a Shox2 enhancer element to mediate its transcriptional repression.56 These authors also provided a compelling link to a proposed genetic basis for atrial fibrillation57 by showing that Pitx2 heterozygous mice are susceptible to atrial tachycardia and fibrillation with programmed stimulation.56
In addition to its potential role in epicardial progenitor cells,58,59 Tbx18 is specifically expressed in SAN precursor cells within the sinus venosus myocardium.37,44,60 Similar to other SAN markers such as Tbx3 and Shox2, Tbx18 is expressed in a mutually exclusive manner with Nkx2.5.60 Anatomically, the SAN contains “head” and “tail” domains that together give rise to the normal comma-shape of the fully developed pacemaker. Christoffels et al demonstrated that Tbx18 is regionally expressed within the SAN “head,” and its function is required for “head” morphogenesis; on this initial field of SAN precursors, Tbx3 subsequently imposes the pacemaker gene regulatory program.61 Interestingly, recent experiments suggest that Tbx18 directly represses Cx43 without altering the levels of Cx40 or Cx45.62 Given that Tbx18 specifically affects Cx43 expression, it seems reasonable to surmise that Tbx18 and Tbx3 act in parallel during SAN development; nevertheless, the molecular mechanisms by which Tbx18 operates are largely unknown and merit further investigation.
Atrioventricular Node
Human linkage studies demonstrating an association between congenital heart defects (including AV conduction block) and familial mutations in the transcription factors NKX2-563 and TBX564 served as the initial entry point for subsequent analysis of the AVN gene regulatory network27,28,45,65 (Figure 4). Subsequent analysis of Nkx2.5-mutant mice confirmed that they exhibited phenotypes that closely resemble the nonsyndromic congenital heart defects caused by human mutations in NKX2-5.27,65 A genetically modified mouse was also generated to model Holt-Oram syndrome, a heart-hand syndrome caused by mutations in TBX5.45 Collectively, these studies demonstrated that Tbx5 and Nkx2.5 regulate downstream events that contribute to proper AVN and AVB formation by cooperatively regulating specific target genes such as Cx4027,45 which is required for rapid conduction through lower AVN cells and the proximal AVB.66 Thus, Tbx5 and Nkx2.5 appear to function synergistically to impact AVN morphogenesis, although their widespread expression within the embryonic heart implies that conduction system specificity is not likely to be achieved by these transcription factors acting alone.
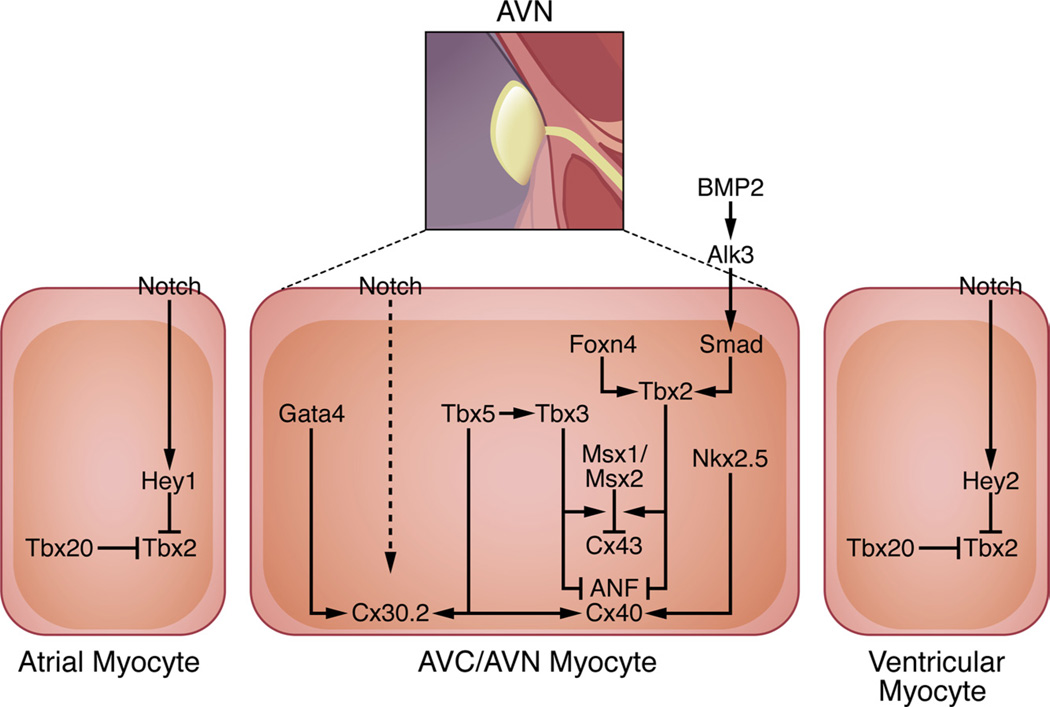
Figure 4. Gene networks in the AVN.
BMP and Notch signaling are known effectors of AVC and AVN development. BMP2 signals via the Alk3 receptor to activate Smad-dependent genes such as Tbx2; Foxn4 independently activates Tbx2 expression in zebrafish. Tbx5, which is expressed in the developing AVC, upregulates expression of Tbx3, which represses the chamber-specific genes ANF, Cx40, and Cx43 in conjunction with Tbx2, Msx1, and Msx2. While Tbx5 and Nkx2.5 coactivate expression of ANF and Cx40, Tbx5 functions in concert with Gata4 to activate Cx30.2 expression. Notch functions within the AVN to activate Cx30.2 expression either through a direct or indirect mechanism. Notch signaling also acts in a reciprocal manner in atrial and ventricular myocardium adjacent to the AVC by activating transcription of the Hey1 and Hey2 repressors, respectively, which inhibit Tbx2 expression. Moreover, Tbx20, which is expressed specifically in chamber myocardium, directly represses Tbx2 transcription.
Development of the AVN involves at least 2 distinct steps evident even from early histological studies.12,13 Initially, the AVC is partitioned from atrial and ventricular myocardium as early as E8.5 to establish AV delay in the embryonic heart.52 The AV cushions then give rise to the AV valves, while a portion of the AVC myocardium differentiates into the AVN, the preferential site of conduction after electric isolation of the atria and ventricles takes place by E13.5.67 Tbx2 and Tbx3 are well-established markers of the AVC and its derivatives,52,68 and several recent studies have provided details about how these two transcription factors function within the AVN gene regulatory network.69–71 Both Tbx2 and Tbx3 play an instrumental role in ensuring that the AVC myocardium retains its primitive phenotype by repressing chamber-specific genes such as ANF and Cx4068,71; indeed, Tbx2 is sufficient to repress chamber morphogenesis even when it is expressed ectopically.70 At the molecular level, both Tbx2 and Tbx3 directly interact with Nkx2.5 and repress genes that are regulated by both Nkx2.5 and Tbx5 in working atrial and ventricular myocardium.52,68 Consistently, Tbx2 and Tbx3 are redundantly required for boundary formation between the AVC and working atrial and ventricular myocardium in vivo.69
More recently, 2 studies have further elucidated the in vivo functions of Tbx2 and Tbx3.53,72 Myocardial-specific Tbx2 knockout mice have persistent myocardial connections between the left atrium and ventricle with a malformed annulus fibrosus,72 providing the substrate for Wolff-Parkinson-White (WPW) syndrome.73 These mice ectopically upregulate chamber-specific genes in the left AVC and display electrophysiological properties consistent with ventricular preexcitation in the absence of any functional or morphological defects involving the AVN.72 Using an allelic series of mutant mice, Tbx3 was shown to specifically regulate cardiac conduction system function in vivo in a highly dosage-sensitive manner.53 Tbx3 mutant mice were found to manifest a variety of arrhythmias attributable to both SAN and AVN dysfunction. The authors also developed a method for assessing arrhythmias in utero to demonstrate that Tbx3 mutant mice have arrhythmias during development, suggesting that cardiac electric disturbances contribute to their fetal demise.53 As predicted from prior studies, Tbx3 downregulation was associated with upregulation and misexpression of the chamber-specific gene Cx43 in the AVN.53 Tbx3 affects AV conduction through cell-autonomous roles in both the AVC and AVB, as demonstrated by analyzing tissue-specific knockout mice using specific Cre-drivers.53 Furthermore, Tbx3 is necessary for AVN homeostasis, since deletion in adult mice also causes AV conduction abnormalities.53 Taken together, these studies demonstrate that Tbx2 and Tbx3 have potentially nonredundant roles in AVC and AVN formation. Whereas Tbx2 regulates AVC patterning, Tbx3 appears to have a more dominant role in the formation and function of the AVN per se.
Building on the critical role of repressive T-box transcription factors in AVN development, additional studies have shed new light on the transcriptional cascades involved in this process. Recently, a forward genetic screen for regulators of cardiac conduction in zebrafish led to the discovery of Foxn4 as an AVC-enriched gene that binds to a Tbx2b enhancer element necessary for its AVC-specific expression.74 Foxn4 mutant fish have severe cardiovascular abnormalities in large part stemming from defects in AV conduction.74 A Foxn4 ortholog exists in higher vertebrates, although cardiovascular defects have not been described in the knockout mice,75 suggesting that other Foxn family members may act redundantly with Foxn4 in higher vertebrates. In addition, a recent study showed that Tbx2 and Tbx3 collaborate with the muscle-segment homeobox transcription factors Msx1 and Msx2 to directly repress Cx43 expression in the AVC.76 Given that Tbx2 and Tbx3 function in boundary formation to repress genes expressed in working myocardium, it is perhaps not surprising that reciprocal repressive mechanisms exist in working atrial and ventricular myocytes.77–79 Notch-dependent signaling in atrial and ventricular myocytes upregulates Hey1 and Hey2, respectively, which prevent expansion of the AVC boundary by inhibiting Tbx2 expression. Moreover, Tbx20 delimits the AVC boundary by repressing BMP/Smad-dependent activation of Tbx2 transcription in working cardiomyocytes,79 although some of the mechanistic details of this process remain to be clarified.80
Studies correlating the anatomy and electric properties of the mature AVN have revealed an unanticipated complexity of its cellular makeup, suggesting the existence of at least 6 electrophysiologically distinct cell types.81 Whereas Cx45 and Cx40 mark lower AV nodal cells and the AVB (characterized by rapid conduction), Cx30.2 and Cx45 are the predominant gap junctions demarcating the compact AVN and posterior nodal extension (characterized by slow conduction).82 Given that Cx30.2 is necessary for proper AV delay in mice and its expression is confined to slower-conducting components of the AVN,83,84 we recently described an enhancer responsible for this expression pattern regulated by Tbx5 and Gata4.85 Furthermore, Gata4 heterozygous mice have shortened PR intervals, suggesting that Gata4-dependent regulation of Cx30.2 (and other genes) contributes to slow AV nodal conduction and normal AV delay.85 Thus, it appears that Tbx5 plays a global role in AVN morphogenesis and gene expression by collaborating with Nkx2.5 to regulate fast-conduction genes and with Gata4 to regulate slow-conduction genes. Although it is possible that tissue specificity can be achieved solely by the combinatorial action of these transcription factors, it is likely that additional cofactors (eg, Tbx2/Tbx3) are necessary.
Aside from roles in early cardiac and AVC morphogenesis, BMP signaling has also been implicated in definitive AVN formation. Mice with AVC-restricted deletion of the BMP receptor Alk3 have defects in AVN and annulus fibrosus morphogenesis; thus, these mice exhibit AV conduction disturbances consistent with ventricular preexcitation secondary to persistent AV myocardial connections.86,87 Interestingly, a human microdeletion that includes the human BMP2 gene has been associated with a familial case of WPW syndrome, although the fact that BMP2 mutant mice do not display similar abnormalities suggests that another BMP family member (eg, BMP4) may pattern the AVC in mice.88 Moreover, although Smad transcription factors (nuclear effectors of BMP signaling) have been shown to activate Tbx2 expression during AVC formation,79 it remains to be seen whether these transcription factors play a similar role during terminal differentiation of the AVN.
Similar to BMP pathways, Notch signaling has also been implicated in AVN morphogenesis at multiple developmental time points. Mice expressing a dominant-negative Mastermind construct (DN-MAML) that inhibits Notch activity in the AVN have short PR intervals as a result of a hypoplastic Cx30.2+ compact AVN and inferior nodal extension; this phenotype is partially phenocopied by myocardium-restricted Notch1 knockout mice.89 Interestingly, mice that overexpress the transcriptionally active Notch intracellular domain have AVC patterning defects resulting in an abnormally large AVN and persistent AV connections. As a result, these mice display electrophysiological properties consistent with ventricular preexcitation and can potentially serve as a model for human WPW syndromes.89 Since the Notch intracellular domain does not appear to target the Cx30.2 enhancer directly (unpublished observations), dissecting the transcriptional regulatory networks downstream of Notch signaling in the AVN will be extremely informative. Furthermore, identification of the precise cell types involved in the Notch and BMP signaling processes could potentially provide valuable insights regarding the inductive influences of neighboring conduction and nonconduction cells during AVN development.
AVB, BB, and PF Network
Lineage tracing and cellular birth-dating studies suggest that the AVB and BB develop in parallel with the nodal structure of the heart and share a common progenitor with ventricular myocytes.35,36,39,90,91 Interestingly, distinct gene expression programs appear to regulate early versus late development of the AVB71 (Figure 5). During early AVB formation, Cx40 and the working myocardial genes Cx43 and ANF are repressed by Tbx3, whereas Cx40 and ANF expression increase (with maintenance of Cx43 repression) during late development. Taken together, these observations suggest that Tbx3 regulates early development of the AVB, whereas Tbx5 may play a more prominent role during the later stages of AVB maturation; given that some studies have demonstrated that Tbx3 expression is maintained in the adult AVB,37,53,71,92,93 the mechanisms by which this selective gene regulation is achieved remain to be elucidated. Furthermore, Tbx5 and Nkx2.5 act within the AVB and BB transcriptional networks by cooperatively activating expression of the transcriptional repressor Id2.90 Mice carrying compound heterozygous mutations in Tbx5 and Nkx2.5 phenocopy the conduction defects observed in Id2 null mice (prolonged AV conduction and left BB block), which can be explained by the fact that Tbx5 and Nkx2.5 directly activate Id2 expression through a common enhancer element.90 Although the downstream transcriptional targets of Id2 remain uncharacterized, cellular birth-dating studies along with known functions for this transcription factor in other systems suggest that Id2 directs cell-cycle exit in the AVB and BB.90
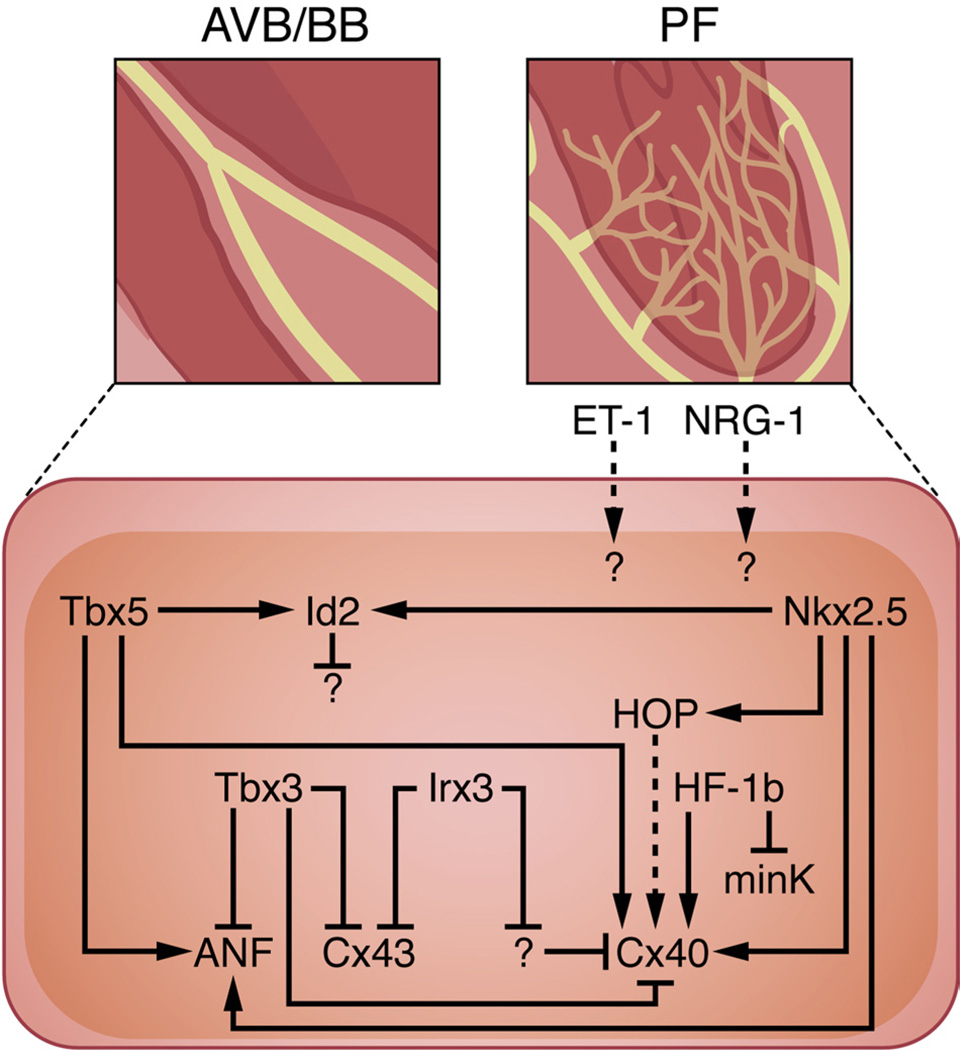
Figure 5. Gene networks in the AVB, BB, and PF network.
While Nkx2.5 orchestrates morphogenesis of the entire ventricular conduction system, Tbx5 regulates development of the AVB and BB. Both Nkx2.5 and Tbx5 synergistically activate Id2 expression in the developing AVB and BB to mediate cell-cycle exit. Whereas Tbx3 inhibits expression of Cx43, Cx40, and ANF during early AVB development, Tbx5 activates Cx40 and ANF expression during maturation of the AVB and BB. In addition, Nkx2.5 increases Cx40 expression directly by binding to the Cx40 promoter and indirectly by activating HOP in the AVB, BB, and PF network. HF-1b is also expressed within the ventricular conduction system and impacts its electrical properties by simultaneously upregulating Cx40 and downregulating minK levels. Similarly, Irx3 indirectly activates Cx40 expression while directly repressing Cx43 transcription in the developing AVB, BB, and PF network. ET-1 and NRG-1 have been shown to stimulate PF formation in avian and murine systems, respectively, although the downstream effectors of this process remain unknown.
HF-1b, a transcription factor originally identified by its ability to activate ventricle-specific expression of myosin light-chain 2, also functions to regulate development of the AVB, BB, and PF network.29 HF-1b knockout mice survive to term, but they subsequently succumb to lethal ventricular tachyarrhythmias. Based on molecular phenotyping of HF-1b null animals, it appears that HF-1b activates expression of Cx40 while repressing expression of minK, an accessory potassium channel protein.29 Although these results suggest that HF-1b affects electrical excitability by coordinating the location and levels of specific membrane proteins within the ventricular conduction system, the molecular mechanisms that mediate this regulation remain to be elucidated.
Analysis of HOP-lacZ knock-in mice revealed its expression within the developing AVB, BBs, and PFs.94 Invasive electrophysiology studies demonstrated that HOP knockout animals have conduction defects distal to the AVN, consistent with the pattern of its expression. Furthermore, Cx40 levels were downregulated in HOP null mice, suggesting that HOP directly or indirectly affects Cx40 expression.94 Given that HOP has been shown to be an Nkx2.5-dependent repressor of SRF,95,96 one can envision that HOP inhibits the expression of a putative Cx40 inhibitor, although the exact mechanistic details require further investigation.
A recent study has provided a comprehensive analysis of the mechanisms by which Irx3 regulates formation and function of the His-Purkinje system.97 Consistent with the confinement of Irx3 expression to the AVB, BB, and PF network, Irx3 knockout mice have prolonged ventricular activation. Cellular and molecular studies suggest that Irx3 coordinates expression of the gap junction proteins Cx40 and Cx43 in a manner that is conceptually similar to the function of HF-1b (see above). Irx3 directly represses Cx43 expression in the ventricular conduction system by directly binding to a site which overlaps with an Nkx2.5 binding motif; in contrast, Irx3 simultaneously mediates Cx40 activation through an indirect mechanism.97 Taken together, these studies suggest that at least 5 separate transcription factors (Id2/Hop/Tbx3/HF- 1b/Irx3) function in parallel during formation of the ventricular conduction system to ensure deployment of the correct gene expression program and proper timing of cell-cycle exit.
The distal PF network remains the least-characterized component of the ventricular conduction system, which is required for rapid conduction and coordinated ventricular contraction. As mentioned earlier, ET-1 and NRG-1 have been shown to promote differentiation of PFs in avian21 and murine26 systems, respectively; however, the downstream transcriptional regulators that mediate these responses are completely unknown. One potential target of ET-1 and/or NRG-1 signaling is Nkx2.5, which plays a cell-autonomous role in PF network formation.98 Nkx2.5 heterozygous mice exhibit hypoplastic PFs, as assessed in Cx40-eGFP knock-in mice. In turn, these defects result from delayed cell-cycle withdrawal in PFs, leading to slowed ventricular conduction velocities in Nkx2.5+/− animals.98 Although our current knowledge of PF-specific gene expression programs is limited, the availability of recently developed lines that specifically mark this cell type should facilitate future experiments to expand our current level of understanding.99
Other Transcriptional Regulators of Cardiac Conduction
In addition to the transcriptional networks that orchestrate conduction system development, there exist additional regulators of cardiac conduction that are not easily categorized (Figure 6). For example, Irx5 is expressed in a gradient fashion within the ventricular myocardium from epicardium (lowest) to endocardium (highest); the potassium channel largely responsible for the major repolarization current (Kv4.2) is expressed in a gradient opposite to that of Irx5. Thus, Irx5 knockout mice are more susceptible to arrhythmias due to abnormal ventricular repolarization in the absence of a normal Kv4.2 gradient.100 Consistent with these observations, Irx5 recruits the mBop corepressor to a Kv4.2 silencer element to mediate transcriptional inhibition and establish the Kv4.2 gradient.100 It should be noted that a more recent study suggests that Irx4, rather than mBop, may be responsible for Irx5-dependent Kv4.2 repression.101 Taken together, Irx5 plays a crucial role in cardiac rhythm by establishing the ventricular repolarization gradient, although it is not specifically expressed within the conduction system.
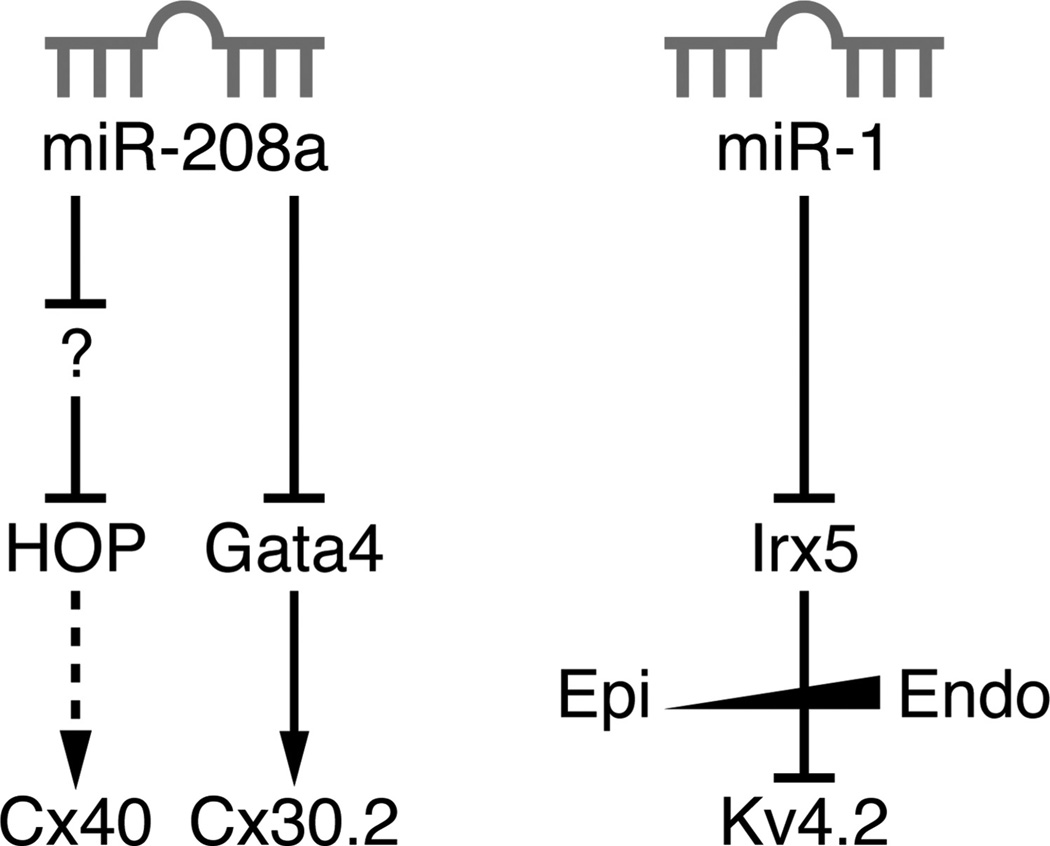
Figure 6. MicroRNA regulation of cardiac conduction.
Two miRs have been implicated in regulating cardiac conduction: miR-208a and miR-1. miR-208a directly targets the 3′UTR of Gata4 (a direct activator of Cx30.2), but it activates HOP expression by inhibiting a putative HOP inhibitor. miR-1 directly targets the 3′UTR of Irx5, which is responsible for the normal ventricular repolarization sequence by establishing an epicardial-to-endocardial gradient of Kv4.2 expression.
MicroRNAs (miRs) are 19- to 25-nucleotide short RNA molecules that fine-tune gene expression by posttranscriptional gene silencing.102 Several miRs are enriched in the heart and have been shown to modulate gene expression during development and on specific stressful stimuli.103 Among these, miR-208a and miR-1 also have roles in regulating certain aspects of cardiac conduction.103 miR-208a plays an instrumental role in stress-dependent myocardial remodeling by regulating genes involved in the process of myosin heavy-chain isotype switching.104 Subsequently, an independent study characterized additional miR-208a targets to explain the prolongation in AV conduction that they observed in both miR-208a knockout mice and transgenic mice with myocyte-specific miR-208a overexpression.105 These experiments revealed that whereas HOP and Cx40 were indirect targets of miR-208a, Gata4 contains a miR binding site in its 3′UTR that mediates miR-208a-dependent posttranscriptional repression.105 miR-1 knockout mice have several conduction abnormalities, including a reduced heart rate, short PR interval, and prolonged QRS interval.106 Irx5 was identified as a miR-1 target by both bioinformatic and biochemical experiments, suggesting that disruption of the Kv4.2 ventricular gradient is at least partially responsible for the observed conduction phenotypes.106
Conclusions and Perspectives
Building on descriptive anatomic studies that made bold conclusions about the existence of a specialized cardiac conduction system,7–15 the field has rapidly adapted with the availability of cutting-edge techniques to provide detailed mechanistic perspectives on the transcriptional networks involved in conduction system development. One feature of the conduction system that particularly hampered early investigators was the fact that it does not encompass a single cell type but rather a collection of cell types that are individually distinct from surrounding working myocardial cells. Studies aimed at dissecting the developmental mechanisms of each of these components have added tremendously to our working knowledge of the ontogeny and development of the cardiac conduction system and how it functions as an integrated syncytium.
Although our knowledge of the transcriptional networks orchestrating cardiac conduction system development has expanded exponentially in recent years, several important questions remain. Fate-mapping experiments have suggested that whereas the SAN and AVN are specified early, the AVB, BB, and PF network undergo at least some degree of ongoing recruitment, implying that the proximal and distal components of the conduction system possess different modes of development. From an evolutionary standpoint, what caused the fusion of the two systems during ventricular septation, since this step would also require coordinated insulation of the atria from the ventricles? Furthermore, how do these two components of the conduction system communicate with one another to facilitate their connection during development?
Despite extensive analysis of the mechanisms responsible for AVN development, several interesting avenues remain for future investigation. BMP and Notch signaling pathways clearly play reiterative roles during AVC and AVN development, but what are the cells of origin for these signaling molecules, and which cells receive and transduce their signals? Is there cross-talk between the BMP and Notch pathways and are additional signaling networks involved in this process? Given that multiple core transcription factors, such as Tbx5, Nkx2.5, and Gata4, are involved in so many aspects of cardiac development, how is their specificity achieved within the conduction system? One strategy for which there is ample evidence is the restriction of developmental potential mediated by active transcriptional repressors, such as Tbx2 and Tbx3. Another possibility remains that BMP and Notch pathways, for example, are required in concert with core transcription factors to coregulate downstream gene expression. The concept of dominant repression, however, raises its own set of questions. For example, although Tbx2, Tbx3, and Tbx5 have similar binding specificities and are coexpressed in the AVC, how is the overall transcriptional effect (activation versus repression) on a particular target gene determined? In the case where T-box and Nkx2.5 binding sites coexist, this can be explained by Tbx2/3-mediated repression via direct interaction with Nkx2.5; however, not all genes expressed in the AVC or AVN are regulated by Nkx2.5.
Given the state of our current understanding of ventricular conduction system development, perhaps the most unresolved issues pertain to its developmental gene network. Although several studies have demonstrated the expression and function of specific transcription factors (eg, HF-1b, HOP, Id2) in the AVB, BB, and PF network, their mechanistic impact on downstream gene regulation remains to be clarified. While NRG-1 plays an instructive role in PF formation based on CCS-lacZ staining, does ET-1 have a similar role in conduction system development using alternative methods of assessment? What pathways are involved in transducing the NRG-1 and/or ET-1 signals from the cell surface to the nucleus, and how do they specifically impact gene expression in the developing ventricular conduction system? Based on previous studies demonstrating that mechanical forces can shape the developing conduction system in chick107 and zebrafish,108,109 how are these mechanisms incorporated into PF development in higher vertebrates? Also, the ventricular conduction system is the only component that exhibits persistent left/right asymmetry; thus, how is this accomplished in 4-chambered vertebrates, and are there hints of these mechanisms operating in 2- and 3-chambered organisms?
Although in-depth studies of the mechanisms underlying cardiac conduction system development have been facilitated by using model organisms, the ultimate goal of these collective efforts is to gain a better understanding of normal and pathological cardiac rhythms in humans. Indeed, the seminal studies leading to the identification of TBX5 and NKX2.5 as key players in conduction system morphogenesis were performed by linkage analysis in well-phenotyped human pedigrees.63,64 In addition, TBX3 has been linked to Ulnar- Mammary syndrome, and genotype-phenotype correlations suggest that certain rare mutations may involve disease of the conduction system.110 To complement human linkage analysis, human geneticists have recently turned to genome-wide association studies (GWAS) to uncover common alleles with relatively weak effects that impact a broader segment of the general population.111 Thus, several GWAS conducted on large cohorts have revealed a number of common variants in loci near transcription factor genes known to play roles in conduction system formation and maintenance112–121 (Table). Interestingly, several transcription factors have been identified with potential roles in conduction system function that were unanticipated. Taken together, the combination of linkage studies and GWAS has reinforced the notion that mechanistic analyses in model organisms is directly applicable to humans with potential ramifications on both risk stratification and novel pharmaceutical development.
Table.
Transcription Factors Implicated in Regulating Human Cardiac Rhythm
| TF Gene | Phenotypic Association |
Evidence | Function |
|---|---|---|---|
| HAND1 | QRS | GWAS | Unknown |
| MEIS1 | PR | GWAS | Unknown |
| NFIA | QRS | GWAS | Unknown |
| NKX2-5 | PR CHD/AV block |
GWAS Linkage |
Formation of AVN/AVB/BB/PF |
| PITX2 | AF | GWAS | Inhibits ectopic sinus node formation |
| SOX5 | PR | GWAS | Unknown |
| TBX3 | PR/QRS UMS/WPW |
GWAS Linkage |
Formation of SAN/AVN/AVB |
| TBX5 | PR/QRS/QT HOS/AV block |
GWAS Linkage |
Formation of SAN/AVN/AVB/BB |
| TBX20 | QRS | GWAS | Inhibits ectopic conduction tissue |
Summary of specific transcription factor loci associated with particular human cardiac electrical phenotypes. The type of evidence supporting these associations is provided along with the function of individual transcription factors in cardiac conduction system development (if known). CHD indicates congenital heart disease; UMS, Ulnar-Mammary Syndrome; HOS, Holt-Oram Syndrome.
Acknowledgments
I apologize to the many investigators whose work could not be cited due to space constraints. I thank Eric Olson for critical reading of the manuscript and Jose Cabrera for providing expert graphical assistance. I also wish to thank the anonymous reviewers for their constructive comments on the manuscript.
Sources of Funding
N.V.M. is supported by a K08 award from the National Institutes of Health/National Heat, Lung, and Blood Institute, a Career Award for Medical Scientists from the Burroughs Wellcome Foundation, and a Disease-Oriented Clinical Scholars Award from University of Texas Southwestern Medical Center.
Non-standard Abbreviations and Acronyms
- ANF
atrial natriuretic factor
- AV
atrioventricular
- AVB
atrioventricular bundle
- AVC
atrioventricular canal
- AVCS
atrioventricular conduction system
- AVN
atrioventricular node
- BB
bundle branch
- CreERT2
Cre recombinase fused with estrogen receptor T2
- eGFP
enhanced green fluorescent protein
- ET-1
endothelin-1
- LSH
left sinus horn
- miR
microRNA
- NRG-1
neuregulin-1
- PF
Purkinje fiber
- SAN
sinuatrial node
- WPW
Wolff-Parkinson-White
Footnotes
Disclosures
None.
References
- 1.Rubart M, Zipes DP. Mechanisms of sudden cardiac death. J Clin Invest. 2005;115:2305–2315. doi: 10.1172/JCI26381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echt DS, Liebson PR, Mitchell LB, et al. Mortality and morbidity in patients receiving encainide, flecainide, or placebo: the cardiac arrhythmia suppression trial. N Engl J Med. 1991;324:781–788. doi: 10.1056/NEJM199103213241201. [DOI] [PubMed] [Google Scholar]
- 3.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hmink gene cause long QT syndrome and suppress Iks function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 4.Keating MT, Sanguinetti MC. Pathophysiology of ion channel mutations. Curr Opin Genet Dev. 1996;6:326–333. doi: 10.1016/s0959-437x(96)80010-4. [DOI] [PubMed] [Google Scholar]
- 5.Keating MT, Sanguinetti MC. Molecular genetic insights into cardiovascular disease. Science. 1996;272:681–685. doi: 10.1126/science.272.5262.681. [DOI] [PubMed] [Google Scholar]
- 6.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: Herg encodes the Ikr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 7.His W. Die thatigkeit des embryonalen herzens und deren bedeutung fur die lehre von der herzbewegung beim erwachsenen. Arb Aus d Med Klinik Zu Leipzig. 1893;1:14–49. [Google Scholar]
- 8.Tawara S. Das reizleitungs system des saugetierherzens: eine anatomisch-histologische studie uber das atrioventrikularbundel und die purkinjeeschen faden. Jena: Gustav Fischer; 1906. [Google Scholar]
- 9.Keith A, Flack M. The form and nature of the muscular connections between the primary divisions of the vertebrate heart. J Anat Physiol. 1907;41:172–189. [PMC free article] [PubMed] [Google Scholar]
- 10.Aschoff L. Referat uber die herzstorungen in ihren beziehungen zu den spezifischen muskelsystem des herzens. Verh Dtsch Pathol Ges. 1910;14:3–35. [Google Scholar]
- 11.Monckeberg JG. Beitrage zur normalen und pathologischen anatomie des herzens. Verh Dtsc Pathol Ges. 1910;14:64–71. [Google Scholar]
- 12.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart, II: histogenesis of the atrioventricular node and bundle. Dev Biol. 1977;56:397–411. doi: 10.1016/0012-1606(77)90279-2. [DOI] [PubMed] [Google Scholar]
- 13.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart, I: the first embryonic a-v conduction pathway. Dev Biol. 1977;56:382–396. doi: 10.1016/0012-1606(77)90278-0. [DOI] [PubMed] [Google Scholar]
- 14.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol. 1980;80:28–45. doi: 10.1016/0012-1606(80)90496-0. [DOI] [PubMed] [Google Scholar]
- 15.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol. 1982;89:25–40. doi: 10.1016/0012-1606(82)90290-1. [DOI] [PubMed] [Google Scholar]
- 16.Cheng G, Litchenberg WH, Cole GJ, Mikawa T, Thompson RP, Gourdie RG. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126:5041–5049. doi: 10.1242/dev.126.22.5041. [DOI] [PubMed] [Google Scholar]
- 17.Gourdie RG, Mima T, Thompson RP, Mikawa T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development. 1995;121:1423–1431. doi: 10.1242/dev.121.5.1423. [DOI] [PubMed] [Google Scholar]
- 18.Kanzawa N, Poma CP, Takebayashi-Suzuki K, Diaz KG, Layliev J, Mikawa T. Competency of embryonic cardiomyocytes to undergo Purkinje fiber differentiation is regulated by endothelin receptor expression. Development. 2002;129:3185–3194. doi: 10.1242/dev.129.13.3185. [DOI] [PubMed] [Google Scholar]
- 19.Takebayashi-Suzuki K, Yanagisawa M, Gourdie RG, Kanzawa N, Mikawa T. In vivo induction of cardiac Purkinje fiber differentiation by coexpression of preproendothelin-1 and endothelin converting enzyme-1. Development. 2000;127:3523–3532. doi: 10.1242/dev.127.16.3523. [DOI] [PubMed] [Google Scholar]
- 20.Hyer J, Johansen M, Prasad A, Wessels A, Kirby ML, Gourdie RG, Mikawa T. Induction of Purkinje fiber differentiation by coronary arterialization. Proc Natl Acad Sci U S A. 1999;96:13214–13218. doi: 10.1073/pnas.96.23.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gourdie RG, Wei Y, Kim D, Klatt SC, Mikawa T. Endothelin-induced conversion of embryonic heart muscle cells into impulse-conducting Purkinje fibers. Proc Natl Acad Sci U S A. 1998;95:6815–6818. doi: 10.1073/pnas.95.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE, Jalife J, Fishman GI. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JB. A gata-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–119. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- 24.Kondo RP, Anderson RH, Kupershmidt S, Roden DM, Evans SM. Development of the cardiac conduction system as delineated by mink-lacz. J Cardiovasc Electrophysiol. 2003;14:383–391. doi: 10.1046/j.1540-8167.2003.02467.x. [DOI] [PubMed] [Google Scholar]
- 25.Kupershmidt S, Yang T, Anderson ME, Wessels A, Niswender KD, Magnuson MA, Roden DM. Replacement by homologous recombination of the mink gene with lacz reveals restriction of mink expression to the mouse cardiac conduction system. Circ Res. 1999;84:146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- 26.Rentschler S, Zander J, Meyers K, France D, Levine R, Porter G, Rivkees SA, Morley GE, Fishman GI. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A. 2002;99:10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O’Brien TX, Gourdie RG, Berul CI, Izumo S. NKX2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moskowitz IP, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, Roden D, Berul CI, Seidman CE, Seidman JG. The t-box transcription factor tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen-Tran VT, Kubalak SW, Minamisawa S, et al. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell. 2000;102:671–682. doi: 10.1016/s0092-8674(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 30.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1 + progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 31.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of isl1 and gata factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multi-potential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- 33.Poelmann RE, Jongbloed MR, Molin DG, Fekkes ML, Wang Z, Fishman GI, Doetschman T, Azhar M, Gittenberger-de Groot AC. The neural crest is contiguous with the cardiac conduction system in the mouse embryo: a role in induction? Anat Embryol (Berl) 2004;208:389–393. doi: 10.1007/s00429-004-0401-6. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 35.Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, Moorman AF, Christoffels VM. The tbx2 + primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res. 2009;104:1267–1274. doi: 10.1161/CIRCRESAHA.108.192450. [DOI] [PubMed] [Google Scholar]
- 36.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aanhaanen WT, Mommersteeg MT, Norden J, Wakker V, de Gier-de Vries C, Anderson RH, Kispert A, Moorman AF, Christoffels VM. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- 38.Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- 39.Miquerol L, Moreno-Rascon N, Beyer S, Dupays L, Meilhac SM, Buckingham ME, Franco D, Kelly RG. Biphasic development of the mammalian ventricular conduction system. Circ Res. 2010;107:153–161. doi: 10.1161/CIRCRESAHA.110.218156. [DOI] [PubMed] [Google Scholar]
- 40.Arnolds DE, Moskowitz IP. Inducible recombination in the cardiac conduction system of mink: Creert(2) bac transgenic mice. Genesis. 2011;49:878–884. doi: 10.1002/dvg.20759. [DOI] [PubMed] [Google Scholar]
- 41.Hoesl E, Stieber J, Herrmann S, Feil S, Tybl E, Hofmann F, Feil R, Ludwig A. Tamoxifen-inducible gene deletion in the cardiac conduction system. J Mol Cell Cardiol. 2008;45:62–69. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Flames N, Hobert O. Transcriptional control of the terminal fate of monoaminergic neurons. Annu Rev Neurosci. 2011;34:153–184. doi: 10.1146/annurev-neuro-061010-113824. [DOI] [PubMed] [Google Scholar]
- 43.Howard ML, Davidson EH. Cis-regulatory control circuits in development. Dev Biol. 2004;271:109–118. doi: 10.1016/j.ydbio.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 44.Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010;87:92–101. doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 45.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the t-box transcription factor tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 46.Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, Henkelman RM, Bruneau BG. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 47.Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G. Shox2 mediates tbx5 activity by regulating bmp4 in the pacemaker region of the developing heart. Hum Mol Genet. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing NKX2-5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaschke RJ, Hahurij ND, Kuijper S, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 50.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 51.Espinoza-Lewis RA, Liu H, Sun C, Chen C, Jiao K, Chen Y. Ectopic expression of nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev Biol. 2011;356:359–369. doi: 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 53.Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM. Lethal arrhythmias in tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci U S A. 2012;109:E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- 55.Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, Black BL, Brown NA, Martin JF. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Damani SB, Topol EJ. Molecular genetics of atrial fibrillation. Genome Med. 2009;1:54. doi: 10.1186/gm54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–E9. doi: 10.1038/nature07916. [DOI] [PubMed] [Google Scholar]
- 60.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an NKX2-5-negative precursor population requires tbx18. Circ Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 61.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by tbx18 and tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 62.Kapoor N, Galang G, Marban E, Cho HC. Transcriptional suppression of connexin43 by tbx18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J Biol Chem. 2011;286:14073–14079. doi: 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 64.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human tbx5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 65.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR. NKX2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 66.Tamaddon HS, Vaidya D, Simon AM, Paul DL, Jalife J, Morley GE. High-resolution optical mapping of the right bundle branch in connexin40 knockout mice reveals slow conduction in the specialized conduction system. Circ Res. 2000;87:929–936. doi: 10.1161/01.res.87.10.929. [DOI] [PubMed] [Google Scholar]
- 67.Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, Kotlikoff MI. Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator gcamp2. Proc Natl Acad Sci U S A. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of tbx2 and nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh R, Hoogaars WM, Barnett P, Grieskamp T, Rana MS, Buermans H, Farin HF, Petry M, Heallen T, Martin JF, Moorman AF, t Hoen PA, Kispert A, Christoffels VM. Tbx2 and tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci. 2012;69:1377–1389. doi: 10.1007/s00018-011-0884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 71.Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 72.Aanhaanen WT, Boukens BJ, Sizarov A, Wakker V, de Gier-de Vries C, van Ginneken AC, Moorman AF, Coronel R, Christoffels VM. Defective tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. J Clin Invest. 2011;121:534–544. doi: 10.1172/JCI44350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson RH, Ho SY. Anatomy of the atrioventricular junctions with regard to ventricular preexcitation. Pacing Clin Electrophysiol. 1997;20:2072–2076. doi: 10.1111/j.1540-8159.1997.tb03631.x. [DOI] [PubMed] [Google Scholar]
- 74.Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 76.Boogerd KJ, Wong LY, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AF, Barnett P. Msx1 and msx2 are functional interacting partners of t-box factors in the regulation of connexin43. Cardiovasc Res. 2008;78:485–493. doi: 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- 77.Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 78.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by notch and hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh R, Horsthuis T, Farin HF, Grieskamp T, Norden J, Petry M, Wakker V, Moorman AF, Christoffels VM, Kispert A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- 80.Cai X, Nomura-Kitabayashi A, Cai W, Yan J, Christoffels VM, Cai CL. Myocardial tbx20 regulates early atrioventricular canal formation and endocardial epithelial-mesenchymal transition via bmp2. Dev Biol. 2011;360:381–390. doi: 10.1016/j.ydbio.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meijler FL, Janse MJ. Morphology and electrophysiology of the mammalian atrioventricular node. Physiol Rev. 1988;68:608–647. doi: 10.1152/physrev.1988.68.2.608. [DOI] [PubMed] [Google Scholar]
- 82.Kreuzberg MM, Willecke K, Bukauskas FF. Connexin-mediated cardiac impulse propagation: connexin 30.2 slows atrioventricular conduction in mouse heart. Trends Cardiovasc Med. 2006;16:266–272. doi: 10.1016/j.tcm.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kreuzberg MM, Schrickel JW, Ghanem A, Kim JS, Degen J, Janssen-Bienhold U, Lewalter T, Tiemann K, Willecke K. Connexin30.2 containing gap junction channels decelerate impulse propagation through the atrioventricular node. Proc Natl Acad Sci U S A. 2006;103:5959–5964. doi: 10.1073/pnas.0508512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kreuzberg MM, Sohl G, Kim JS, Verselis VK, Willecke K, Bukauskas FF. Functional properties of mouse connexin30.2 expressed in the conduction system of the heart. Circ Res. 2005;96:1169–1177. doi: 10.1161/01.RES.0000169271.33675.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munshi NV, McAnally J, Bezprozvannaya S, Berry JM, Richardson JA, Hill JA, Olson EN. Cx30.2 enhancer analysis identifies gata4 as a novel regulator of atrioventricular delay. Development. 2009;136:2665–2674. doi: 10.1242/dev.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stroud DM, Gaussin V, Burch JB, Yu C, Mishina Y, Schneider MD, Fishman GI, Morley GE. Abnormal conduction and morphology in the atrioventricular node of mice with atrioventricular canal targeted deletion of alk3/bmpr1a receptor. Circulation. 2007;116:2535–2543. doi: 10.1161/CIRCULATIONAHA.107.696583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaussin V, Morley GE, Cox L, et al. Alk3/bmpr1a receptor is required for development of the atrioventricular canal into valves and annulus fibrosus. Circ Res. 2005;97:219–226. doi: 10.1161/01.RES.0000177862.85474.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lalani SR, Thakuria JV, Cox GF, et al. 20p12.3 microdeletion predisposes to Wolff-Parkinson-White syndrome with variable neurocognitive deficits. J Med Genet. 2009;46:168–175. doi: 10.1136/jmg.2008.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, Morley GE, Patel VV, Epstein JA. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–533. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including id2, tbx5, and NKX2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 91.Sedmera D, Reckova M, DeAlmeida A, Coppen SR, Kubalak SW, Gourdie RG, Thompson RP. Spatiotemporal pattern of commitment to slowed proliferation in the embryonic mouse heart indicates progressive differentiation of the cardiac conduction system. Anat Rec A Discov Mol Cell Evol Biol. 2003;274:773–777. doi: 10.1002/ar.a.10085. [DOI] [PubMed] [Google Scholar]
- 92.Horsthuis T, Buermans HP, Brons JF, Verkerk AO, Bakker ML, Wakker V, Clout DE, Moorman AF, t Hoen PA, Christoffels VM. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ Res. 2009;105:61–69. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- 93.Greener ID, Monfredi O, Inada S, Chandler NJ, Tellez JO, Atkinson A, Taube MA, Billeter R, Anderson RH, Efimov IR, Molenaar P, Sigg DC, Sharma V, Boyett MR, Dobrzynski H. Molecular architecture of the human specialised atrioventricular conduction axis. J Mol Cell Cardiol. 2011;50:642–651. doi: 10.1016/j.yjmcc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 94.Ismat FA, Zhang M, Kook H, Huang B, Zhou R, Ferrari VA, Epstein JA, Patel VV. Homeobox protein hop functions in the adult cardiac conduction system. Circ Res. 2005;96:898–903. doi: 10.1161/01.RES.0000163108.47258.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by hop, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 96.Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 97.Zhang SS, Kim KH, Rosen A, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci U S A. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meysen S, Marger L, Hewett KW, Jarry-Guichard T, Agarkova I, Chauvin JP, Perriard JC, Izumo S, Gourdie RG, Mangoni ME, Nargeot J, Gros D, Miquerol L. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev Biol. 2007;303:740–753. doi: 10.1016/j.ydbio.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 99.Pallante BA, Giovannone S, Fang-Yu L, Zhang J, Liu N, Kang G, Dun W, Boyden PA, Fishman GI. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Costantini DL, Arruda EP, Agarwal P, et al. The homeodomain transcription factor irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He W, Jia Y, Takimoto K. Interaction between transcription factors Iroquois proteins 4 and 5 controls cardiac potassium channel kv4.2 gene transcription. Cardiovasc Res. 2009;81:64–71. doi: 10.1093/cvr/cvn259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 103.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 105.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 107.Reckova M, Rosengarten C, deAlmeida A, Stanley CP, Wessels A, Gourdie RG, Thompson RP, Sedmera D. Hemodynamics is a key epigenetic factor in development of the cardiac conduction system. Circ Res. 2003;93:77–85. doi: 10.1161/01.RES.0000079488.91342.B7. [DOI] [PubMed] [Google Scholar]
- 108.Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, Scott I, Beis D, Xiao T, Baier H, Jan LY, Tristani-Firouzi M, Stainier DY. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chi NC, Bussen M, Brand-Arzamendi K, Ding C, Olgin JE, Shaw RM, Martin GR, Stainier DY. Cardiac conduction is required to preserve cardiac chamber morphology. Proc Natl Acad Sci U S A. 2010;107:14662–14667. doi: 10.1073/pnas.0909432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Linden H, Williams R, King J, Blair E, Kini U. Ulnar mammary syndrome and tbx3: expanding the phenotype. Am J Med Genet A. 2009;149A:2809–2812. doi: 10.1002/ajmg.a.33096. [DOI] [PubMed] [Google Scholar]
- 111.O’Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011;365:2098–2109. doi: 10.1056/NEJMra1105239. [DOI] [PubMed] [Google Scholar]
- 112.Chambers JC, Zhao J, Terracciano CM, et al. Genetic variation in scn10a influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 113.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, Basford MA, Masys DR, Haines JL, Roden DM. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 115.Newton-Cheh C, Guo CY, Wang TJ, O’Donnell CJ, Levy D, Larson MG. Genome-wide association study of electrocardiographic and heart rate variability traits: the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S7. doi: 10.1186/1471-2350-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the qt interval duration in the QTSCD study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of pr interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sotoodehnia N, Isaacs A, de Bakker PI, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the nos1 regulator nos1ap modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 120.Nolte IM, Wallace C, Newhouse SJ, et al. Common genetic variation near the phospholamban gene is associated with cardiac repolarisation: Meta-analysis of three genome-wide association studies. PLoS One. 2009;4:e6138. doi: 10.1371/journal.pone.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]