Preface
Advances in genome sequencing technologies have created new opportunities for comparative primate genomics. Genome assemblies have been published for several primates, with analyses of several others underway. Whole genome assemblies for the great apes provide remarkable new information about the evolutionary origins of the human genome and the processes involved. Genomic data for macaques and other nonhuman primates provide valuable insight into genetic similarities and differences among species used as models for disease-related research. This review summarizes current knowledge regarding primate genome content and dynamics and offers a series of goals for the near future.
Introduction
Current technologies for large-scale DNA sequencing have opened new avenues for the study of nonhuman primate genomes. While the major focus of genomics research is human genetics and its relationship to disease, investigators are also pursuing comparative primate genomics. Two basic motivations exist for the detailed study of nonhuman primate genomes: the application of this information in studies using primates as models for the analysis of human disease, and comparative evolutionary analyses that reconstruct the history and mechanisms of genomic change, with a particular focus on the origin of the human genome. One unexpected outcome from new genomic data for the great apes (chimpanzees, bonobos, gorillas, orangutans) and humans is a new perspective on the process of speciation and genetic divergence among these evolutionary lineages.
The first nonhuman primate genome sequenced and published was that of the chimpanzee, Pan troglodytes1, followed soon by the rhesus macaque, Macaca mulatta2. Both genomes were analysed using shotgun sequencing employing exclusively Sanger sequencing methods. As a result these projects entailed considerable cost and effort. A legacy of further primate sequencing projects that were initiated when Sanger sequencing was the only option is now reaching its end (Table 1). The genomes of all extant great ape species have been sequenced to draft quality. Analysis of a gibbon genome, the only remaining group of extant hominoids, is underway, and other nonhuman primate genome assemblies are at various stages of completion (see Supplementary information S1 (table)). Remarkably, researchers have also produced extensive sequence information for two extinct hominins, the Neanderthals3 and the Denisovans4 (Box 1). Investigators are also producing substantial information concerning primate transcriptomes and genetic variation within species.
Table 1.
Published primate genome sequences
| Common Name |
Species Name | Bases in contigs |
Contig N50 |
Scaffold N50 |
Ref. |
|---|---|---|---|---|---|
| Draft Genome Assemblies | |||||
| Chimpanzee | Pan troglodytes | 2.7 Gb | 15.7 kb | 8.6 Mb | 1 |
| Bonobo | Pan paniscus | 2.7 Gb | 67 kb | 9.6 Mb | 64 |
| Gorilla | Gorilla gorilla | 2.8 Gb | 11.8 kb | 914 kb | 5 |
| Orangutan | Pongo abelli | 3.1 Gb | 15.5 kb | 739 kb | 13 |
| Indian rhesus macaque | Macaca mulatta | 2.9 Gb | 25.7 kb | 24.3 Mb | 2 |
| Chinese rhesus macaque | Macaca mulatta | 2.8 Gb | 12.0 kb | 891 kb | 91 |
| Vietnamese cynomolgus macaque | Macaca fascicularis | 2.9 Gb | 12.5 kb | 652 kb | 91 |
| Aye-aye | Daubentonia madagascarensis | 3.0 Gb | na | 13.6 kb | 9 |
| 2× Sanger Genome Sequences* | |||||
| Mouse lemur Bushbaby Tarsier | Microcebus murinus Otolemur garnetti Tarsier syrichta | na | na | Na | 11 |
| Whole genome re-sequencing studies without assembly | |||||
| Indian rhesus macaque | Macaca mulatta | na | na | Na | 33 |
| Chinese rhesus macaque | Macaca mulatta | na | na | Na | 112 |
| Mauritian cynomolgus macaque | Macaca fascicularis | na | na | Na | 91 |
| Malaysian cynomolgus macaque | Macaca fascicularis | na | na | Na | 114 |
2× whole genome coverage.
na, not applicable. N50, weighted median statistic such that 50% of the entire assembly is contained in contigs or scaffolds equal to or larger than this value.
YES, OK.
Box 1. Genome analysis of ancient hominins.
The fossil record for recent human evolution (i.e. the last several hundred thousand years) is substantial. A great deal is known about morphology, biogeography and the archeological evidence for behavior concerning several extinct hominin species. Remarkably, through dramatic advances in techniques for investigating ancient DNA, we now have access to extensive genome sequence data for Neanderthals, an extinct hominin population from Europe and western Asia that diverged at least 250,000 years ago from the lineage leading to modern humans3. This work has shown that 1–4% of DNA sequences carried by modern humans outside Africa are derived from Neanderthals, the result of interbreeding and gene flow54. Another extinct hominin population (the Denisovans) were only recently recognized using genome sequence produced by extracting DNA from a finger bone found in the Altai Mountains4. The Denisovans diverged from human ancestors 170,000–700,000 years ago. Gene flow from the Denisovans into the modern human population has so far been detected only among aboriginal Australians and populations in Melanesia and southeast Asia4. These findings indicate ancestral human populations interbred to some biologically significant degree with other populations that were distinct in their genetics, and at least in the case of Neanderthals, distinct also in morphology. There is also evidence that introgression from Neanderthals into modern humans introduced alleles now associated with disease among modern humans, and that negative selection after this hybridization may have been driven by adverse effects of that hybridization on male fertility110.
The widespread availability of next-generation sequencing technology promises even more rapid progress. The amount of genomic information available for nonhuman primates is certain to grow at an accelerating pace. Our understanding of comparative primate genome content, diversity and evolution will necessarily change as new data appear. Conclusions based on current information may, therefore, soon be amended. Nevertheless, substantial progress has been made in the last several years, justifying an assessment of the insights gained to date.
This review begins by summarizing available information about the content of and differences among primate genomes. Next, we present some new insights regarding genomic differentiation and speciation, with particular reference to human evolution. Finally, we illustrate some of the ways genomic data are expanding and improving the use of nonhuman primates in studies concerning human health and disease.
Genomic differences among primates
Comparisons of annotated genome sequences across species allow researchers to directly identify genomic elements that are shared and that are species-specific. We have known for many years that protein-coding sequences exhibit a greater similarity between primate species than do intronic and inter-genic sequences. Past studies also indicated that a large proportion of human protein-coding genes are found in most or all primates. But detailed comparisons across all components of the genome have been impossible until recently. Investigators can now address fundamental questions concerning the content and function of genomic features across multiple species, thus providing new insight into the genetic basis of phenotypic similarity and differences across humans and other primates.
Differences in single-copy alignable sequence
With time, genomes accumulate mutations (single basepair substitutions) that may, through genetic drift or selection, become fixed differences distinguishing one species from its close relatives. Divergence in single-copy sequences occurs steadily among primate genomes but not at a uniform rate in all branches of the primate tree. Alignment of sequences across species demonstrates that pair-wise differences between species correlate fairly well with evolutionary divergence times inferred from other information. The human–chimpanzee sequence divergence is estimated at 1.1–1.4%1, 5. The time of separation of the human from the chimpanzee lineage remains somewhat controversial6, but is generally dated to 5–9 million years ago (Figure 1). The uncertainty concerning the date of the human-chimpanzee divergence results from several factors, including the lack of a reliable paleontological record for that event, and ambiguity concerning the appropriate mutation rate to use to infer the time of divergence from DNA sequences alone. The difference in single copy sequence between human and rhesus macaque is approximately 6.5%2, and the divergence of those two lineages is more confidently dated at 25–28 million years ago. Dates for the human-chimpanzee divergence calculated using estimates of mutation rate that are derived from other between-species differences (e.g. the human-macaque or human-orangutan divergence) differ from dates based on mutation rates obtained through pedigree-based analyses of current human mutation.
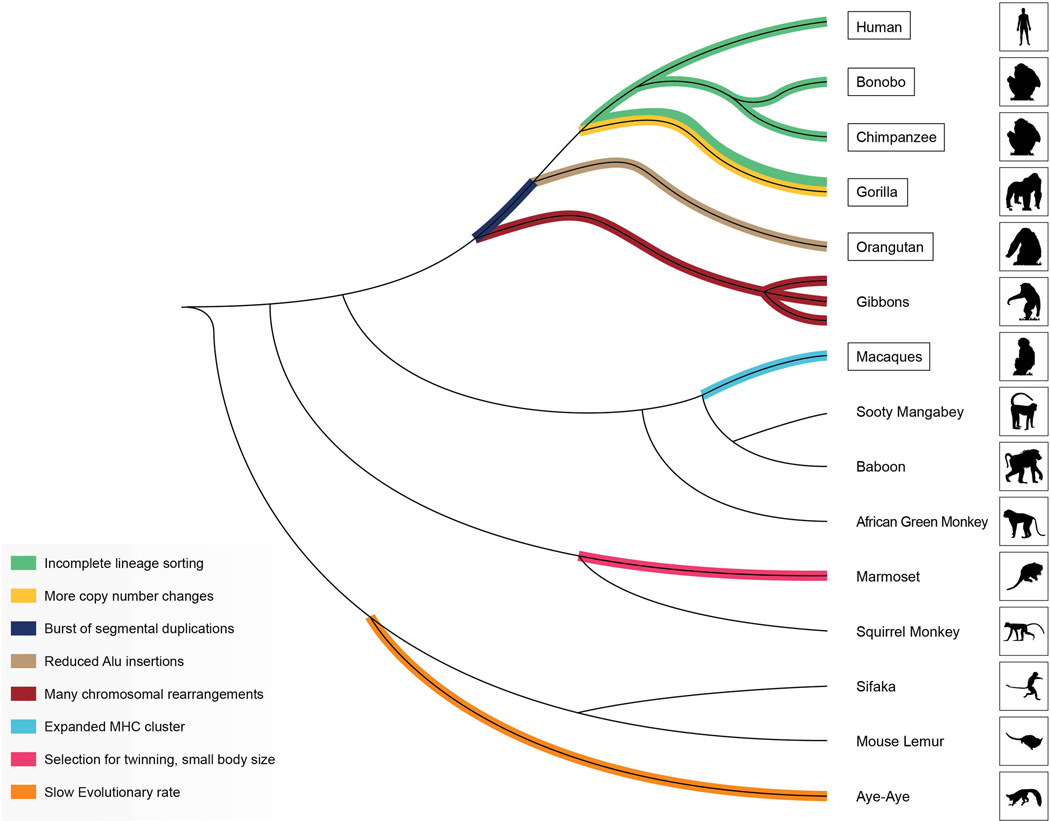
Figure 1. Primate phylogenetic tree.
This diagram presents the evolutionary relationships among species for which genome sequences are published, available or in progress. The genomes for species enclosed in boxes are already published. Among the macaques, the rhesus and cynomolgus macaque genomes are published2, 91, but sequencing of other macaque genomes is underway. Selected lineages are highlighted to indicate specific genomic features of interest, or unexpected genomic traits, such as reduced rate of Alu insertion in the orangutan genome13 or the lower evolutionary rate in the aye-aye9.
Despite the uncertainties, various analyses suggest that single copy DNA accumulates individual basepair substitutions more slowly through time among gorillas, chimpanzees, bonobos and humans than in other primates such as Old or New World monkeys. This is not entirely unexpected given differences in generation time7, 8. One exception may be the aye-aye (Daubentonia madagascarensis), a Malagasy lemur with extraordinarily unique morphology. Synonymous substitutions are reported to accumulate more slowly than expected in the aye-aye, based on comparisons with other species9. Additional sequencing projects may identify other primate lineages that do not fit current expectations.
Small insertions and deletions
While the difference in genome sequences between humans and chimpanzees is recently estimated at 1.4%5, this is correct only for nucleotide substitutions in regions where the two genomes can be directly aligned. As Roy Britten first noted10, small insertions and deletions (<100 bp) account for more total nucleotide differences among closely related species than do single base changes in alignable sequences. Both the human and chimpanzee genomes each contain about 1.5% unique sequence, not found in the other, primarily due to small indels. The rhesus macaque sequence alignable to human is 93.5% identical, but when including small indels it is only 90.8% identical to human2. These indel differences among species are found more frequently in intronic and inter-genic regions than in protein coding exons, primarily because indels in coding sequences will generally have negative consequences on protein function. For example, only indels involving a multiple of three nucleotides do not induce frameshifts that result in substantial changes in protein amino acid sequence. Available comparisons show that small indels are most common and presumably better tolerated in non-coding regions. However, as more non-coding regions with functional significance are identified11, 12, some indels in flanking or inter-genic segments will become more interesting, and potentially gain importance for understanding changes that affect enhancers and other regulatory sequences that influence gene expression and phenotypic differences among species.
Alu and other repetitive elements
The insertion of Alu repeats and other retroposons is an on-going process in primate genomes. Taken together, repetitive elements make up about 50% of the total genome in humans, apes and monkeys. But the number of species-specific insertions differs substantially across species, from about 5,000 in humans to 2,300 in chimpanzees, and only 250 in orangutans13. It is not entirely clear why the rate of accumulation differs. Nevertheless, de novo Alu insertions constitute a major source of genomic change, but have not affected all primates equally14. Retroposons also facilitate duplication or deletion events, which affect much larger DNA segments15 and thus can have broader effects on gene and genome content.
Copy number differences and gene family changes
The majority of protein-coding genes have 1:1 homologues among humans, the great apes and Old World monkeys sequenced to date, but gene content is not identical among species. Particular gene families have expanded or contracted in individual lineages. For example, 1,358 genes were identified as new duplications in the rhesus genome compared with human2. The HLA gene cluster, which is critical for response to pathogens as well as other immunological processes, is expanded in macaques relative to humans16. Other interesting cases are changes in zinc-finger transcription factor genes, which show gains and losses that distinguish humans, chimpanzees and orangutans17, as well as the marked expansion of genes containing DUF1220 protein domains in humans18–20, which might be related to the expansion of brain size in our species.
However, the draft quality of current nonhuman primate genome assemblies makes it difficult to define all copy number variations accurately. One can compare gene lists from different assemblies, but gaps and other issues in assemblies create ambiguity21, 22. Available evidence suggests that humans and chimpanzees experienced more rapid changes in gene copy number than did orangutans or rhesus macaques13. Among the great apes, gorillas exhibit more copy number variants than others5. Complete analyses await additional data, including better genome assemblies and information concerning copy number polymorphism within nonhuman primates.
Segmental duplications
Segmental duplications (that is, chromosomal regions >1 kb that are ≥90% identical to other segments in the same genome) are a significant aspect of primate genome structure and dynamics. Duplication and deletion of these segments is active in the human genome. Some of these mutations are apparently neutral, but many lead to adverse consequences and disease23. Just as segmental duplications create variation among humans, they are drivers of evolutionary change across primate genomes. About 5% of the human and chimpanzee genomes, and 3.8% of the orangutan genome, consist of segmental duplications13, 24. The human and great ape genomes are enriched with dispersed duplications, having experienced an interval after their divergence from Old World monkeys when the production of new duplications was particularly active25, 26. Many expansions of specific protein-coding gene families result from segmental duplications, which sometimes involve repeated expansions of a given sequence24, 27, 28. Some genes within segmental duplications show evidence of positive selection acting on coding sequence as well as copy number18, 20, 29, 30. Among the great apes, some of these expansion events have occurred as independent parallel events in different lineages, strengthening the interpretation that these genomic changes are often the result of positive selection for both gene copy number and gene sequences25, 31.
Genetic variation within primate species
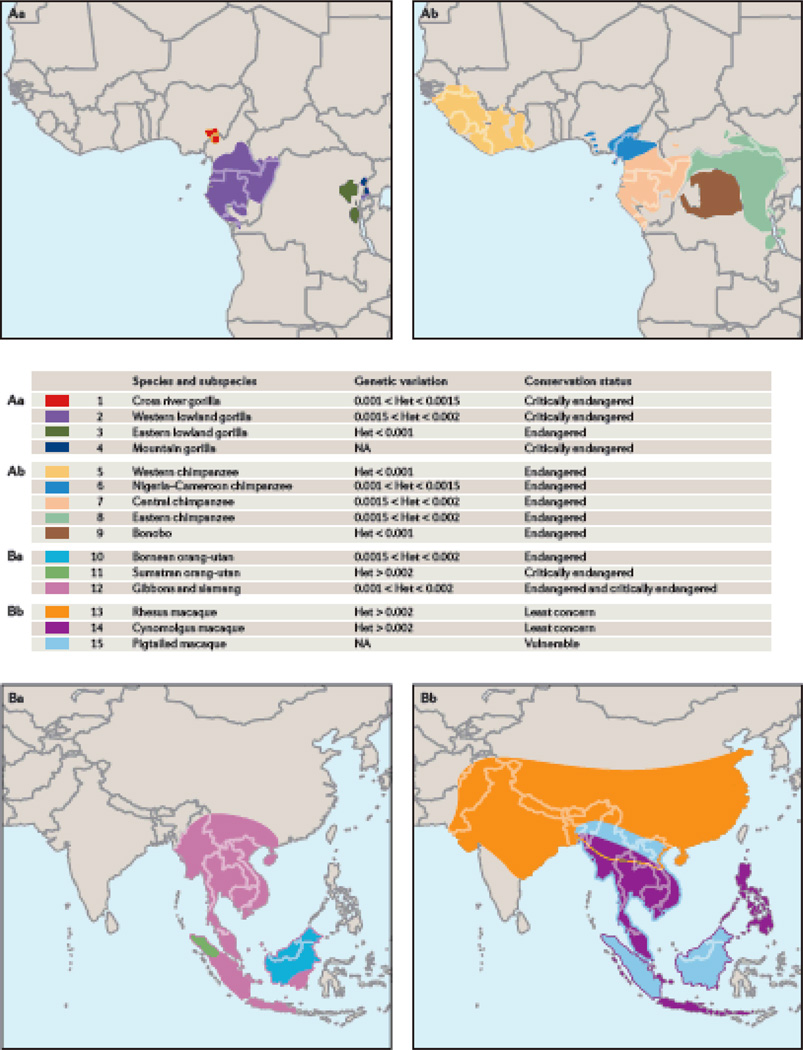
Individual primate genome projects have assessed within-species genetic variation in different ways, with a broad range of sample sizes and population genetic parameters used to quantify variation. Earlier studies that analysed small samples or only small fractions of the genome had suggested that, for the most part, nonhuman primate species exhibit higher levels of intra-species genetic variation than humans (e.g.32–35 and others), and this pattern holds in the larger datasets published more recently. Great ape species have all been reduced to low total population census numbers, but studies using whole-genome data indicate that genetic diversity within great ape species is consistent with effective population sizes as large or even larger than humans13, 24. The Great Ape Genome Project investigated genome-wide variation within and between all six great ape species36 and found that some subspecies and species show levels of intra-specific diversity roughly equivalent to non-African humans (Figure 2). Chimpanzees from east and central Africa, and the Nigeria-Cameroon subspecies, as well as western lowland gorillas and both orangutan species, exhibit significantly higher variation than is found among humans. Application of coalescent and incomplete lineage-sorting models allowed the researchers to re-estimate effective population size for different ape species and subspecies. Each species has a unique history of population expansion and decline, while each chimpanzee subspecies has an independent history36. This observation of separate unique demographic histories for regional populations or subspecies within a species is likely true for most or all primates34.
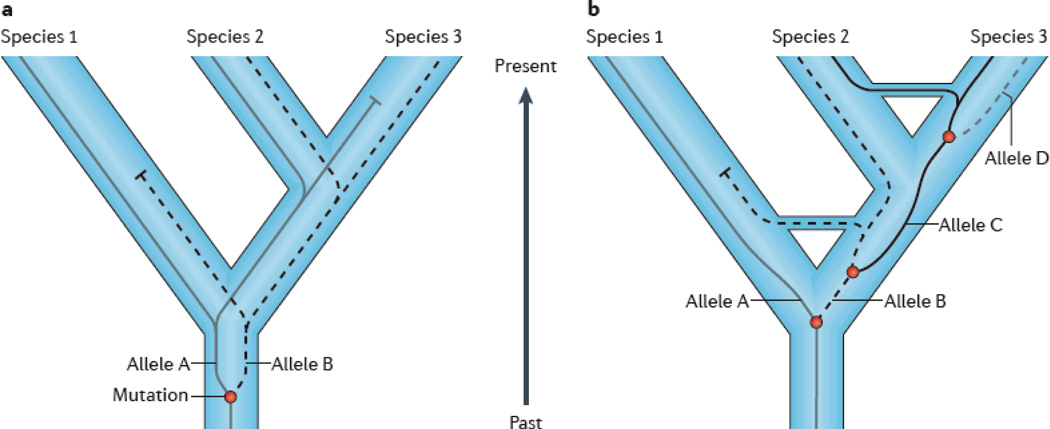
Figure 3. Incomplete lineage sorting.
Incomplete lineage sorting can produce discrepancy between the phylogenetic tree for a specific gene or genomic segment and the overall species-level phylogenetic tree. If an ancestral species is polymorphic, segregating alleles A and B, and divides into two descendent lineages, both alleles can be retained in both daughter lineages. If one of those lineages then divides again relatively soon, all three species lineages may carry both alleles. Over time, each lineage will lose one or the other allele due to drift or selection. In this case, assume that Species 1 retains allele A and Species 3 retains B. Species 2 will, for this genomic segment, appear more closely related to either Species 1 or Species 3 depending on whether it retains allele A or B. Retention of allele B would mean this genomic segment matches the overall species-level phylogenetic tree, but retention of allele A would lead to discrepancy. Analysis of whole genome sequences for human, chimpanzee and gorilla indicate that “gene trees” for a significant fraction of the genome do not match the overall species-level phylogeny, which places chimpanzees are more closely related to humans than to gorillas19.
While the great apes have high intra-species diversity despite low present day census sizes, the rhesus macaque is much more widely distributed geographically, with larger extant population numbers. As part of the rhesus genome sequencing project, DNA from Chinese- and Indian-origin rhesus macaques was sequenced for 150 kb from five genomic regions34. The density of SNPs was significantly higher than is found in human populations. Only about one-third of SNPs were shared between the two geographic populations, which indicates that most variation is region-specific. Similar results were obtained in a survey of 3’ UTR sequences in a small number of macaques37. A study of whole genome sequences for three Indian-origin rhesus macaques found >3 million variants present in at least two of the datasets examined33. Including singletons, about 14 million single nucleotide variants were found33, a density substantially greater than that found in humans. For example, the 1000Genomes Project estimated the average number of SNPs (including singletons) per human individual at 3.6 million38.
Overall levels of variation are high in nonhuman primates, including additional non-hominoid species that have very low population numbers and are in serious danger of extinction39. However, the amount of functionally significant variation within particular species is not yet clear. Significant numbers of non-synonymous substitutions that are predicted to be possibly damaging have been identified in even small numbers of macaques33, 40. But a recent comparison of protein-coding variation between humans and rhesus macaques found little difference between species41. It is possible that macaques and other nonhuman primates are segregating greater total within-species variation compared with humans but equivalent levels of damaging or adverse variation.
Differences in gene expression
Alan Wilson and his colleagues predicted years ago that much of the adaptively significant phenotypic change that distinguishes species results from changes in gene expression, rather than mutations in protein-coding sequences42, 43. Recent information concerning comparative primate gene expression is consistent with this prediction. It is likely that a large proportion of adaptive evolution involves changes in transcription factor binding, possibly rivaling adaptation through protein evolution44. Overall, the description of nonhuman primate transcriptomes lags behind knowledge for human and mouse, but researchers are now developing larger information resources, such as the Allen Institute for Brain Science atlas of gene expression for the rhesus macaque brain (http://www.blueprintnhpatlas.org) and the Nonhuman Primate Reference Transcriptome Resource45.
Nevertheless, comparisons of gene expression across primates have already proven valuable. Differences in gene expression among humans, chimpanzees and rhesus macaques are influenced by natural selection46–48, and include substantial differences in alternative splicing47, 49. RNA sequencing from the livers of humans and other mammals, including 11 primates, found strong evidence for positive selection in a number of the genes expressed39. The results of this study show enrichment for changes affecting genes involved in peroxisome function, such as GGH, PEX7 and HACL139. Patterns of DNA methylation in the prefrontal cortex differ between humans and chimpanzees and correlate with differences in gene expression50. Three-way comparisons find greater overall similarity in gene expression between chimpanzees and humans as compared to gorillas, which matches the overall phylogeny (Figure 1). However, and notably, genes falling in specific regions of the genome (i.e. chromosomal segments exhibiting incomplete lineage sorting, see text below and Figure 3) show a contrary pattern5. Studies of gene expression have recently been extended to wild populations of baboons51, an approach with outstanding potential for future discoveries.
Primate evolutionary dynamics
One of the primary motivations for comparative primate genomics is the desire to understand the origin of the human genome. Whole genome information now available for our closest relatives is altering and refining ideas about the processes of speciation, diversification and genome evolution for this clade. This new picture, though still incomplete, reveals previously unappreciated complexity in the processes that produced the modern human genome. The theory and modeling of speciation (Box 2) is a complex topic with a long history and extensive literature, and consequently is outside the scope of this article. However, the remarkable insights concerning gene exchange among the early human and chimpanzee ancestors52, as well as among ancient hominins3, 4, 53, 54, are dramatic indications that this history is of greater interest than previously recognized (Box 1). In parallel, these between-species comparisons are greatly increasing our ability to identify genes or genomic regions that have undergone positive selection during recent human evolution, thereby pointing us to genetic changes and phenotypes that have been important in human and nonhuman primate adaptation. Comparisons also demonstrate that fundamental genetic processes such as recombination can undergo rapid change, as local hotspots of recombination are not conserved in humans and chimpanzees despite the overall high sequence similarity and the general conservation of large-scale patterns of recombination55.
Box 2. Initial genomic divergence and incipient speciation.
The theory and modeling of speciation is a complex topic that has generated a large amount of discussion. Historically, the founders of the modern evolutionary synthesis (e.g. Ernst Mayr, Theodosius Dobzhansky) argued that genetic and reproductive isolation among populations precedes phenotypic and/or genetic differentiation that is significant enough to justify recognizing those populations as distinct species111. Ernst Mayr’s ‘biological species concept’ and the allopatric speciation model long dominated discussion111. By contrast, the model of punctuated equilibrium112 posited that most adaptively important genetic differentiation occurs during or immediately after initial divergence and isolation of incipient species (see also113). More recently, other models and theories have addressed the greater complexity now known to be inherent in speciation and the genetic differentiation of many lineages86–89. For various types of species, the process of genetic divergence and incipient speciation appears to be more complex than the traditional allopatric speciation model proposed.
Signatures of selection
There is substantial evidence for positive selection on protein-coding genes in various nonhuman primate species. Two classes of genes provide consistent evidence of positive selection, those involved in the immune system and pathogen resistance and genes involved in reproductive biology and gametogenesis2, 5, 13, 56. These results are quite reasonable, as the constant pressure of infectious disease is a plausible driver of selection on the primate immune system, while reproductive competition within species likely accounts for the evidence of selection on those systems. Within individual species, positive selection has been detected for a wide array of phenotypes. The whole genome comparisons among human, chimpanzee and gorilla suggest that these three species have experienced approximately equal levels of positive selection5. This analysis also indicates shared positive selection in hominoids related to neurodevelopment and brain morphology. The orangutan genome provides evidence of selection on glycolipid metabolism and hearing acuity that is specific to that lineage13. Studies in marmosets and other callitrichine primates provide evidence for selection on multiple genes related to phyletic reduction in body size and the development of a unique form of dizygotic twinning in which co-twins exchange hematopoietic stem cells early in gestation, and consequently become life-long hematopoietic chimeras57.
Genes involved in the evolution of unique human traits have received much attention, and been reviewed elsewhere58. Detailed description of the genetic basis of human-specific traits has obvious interest to evolutionary biologists, anthropologists and the broader public. Studies have now attributed human-specific adaptations to deletions of gene regulatory elements (enhancers)59, rapid lineage-specific evolution of such elements60, changes in gene copy number19, 61, and other types of genetic changes58.
Initial genomic divergence and incipient speciation
Whole genome sequence data from humans, chimpanzees and gorillas are not consistent with simple models of reproductive isolation, allopatric genetic divergence or the rapid development of species boundaries (Box 2). Two processes that are known to shape genome evolution in other groups of animals, incomplete lineage sorting and gene flow, are now critical elements in discussions concerning mechanisms of human and nonhuman primate genome differentiation.
Incomplete lineage sorting
Incomplete lineage sorting (ILS) occurs when a polymorphic ancestral species, with two or more alleles (haplotypes) at a given locus divides into two lineages. Both alleles can be retained in the descendant branches, and when one of those lineages divides again, the phylogenetic tree for that locus (the gene tree) may or may not match the branching order for the species-level evolutionary tree (Figure 3). The likelihood of discrepancy between the species-level phylogeny and any one particular gene tree increases as either the time between the two successive branching events decreases or effective population size increases62. Prior analyses of a few genes suggested that different regions within the human, chimpanzee and gorilla genomes exhibit different evolutionary relationships, i.e. different gene trees63, 64. Following assembly of the gorilla genome5, researchers determined the evolutionary relationships for arbitrary segments across the human-chimpanzee-gorilla genomes. They found, as expected, that for most of the genome, chimpanzees are more closely related to humans than to gorillas. But for ~15% of the genome, chimpanzee DNA sequences share a more recent common ancestor with the homologous sequences in the gorilla genome, not the human. For another 15%, gorillas and humans are most closely related. ILS from a polymorphic common ancestor is a likely contributing factor, but not the only possibility. Gene flow among differentiating lineages may also be a factor. This developing picture of evolutionary process complexity also applies in other cases. Bonobos and chimpanzees are undoubtedly sister taxa, more closely related to each other than to any other species. Nevertheless, for 1.6% of the genome, sequences in bonobos are more similar to homologues in humans than chimpanzees, whereas 1.7% places bonobos as the outlying group65. ILS is the likely explanation and may be quite common in primates66.
Gene flow among incipient lineages
Analysis of the human, chimpanzee and gorilla genomes indicates that genetic exchange between divergent lineages is not restricted to recent periods (Box 1). The common ancestor of humans and chimpanzees began differentiating 5–12 million years ago, depending on the assumed mutation rate1, 52. Using coalescent models, Mailund et al. estimated that those diverging lineages experienced reciprocal gene flow for about 3 million years. In other words, the separation of the last common ancestor of humans and chimpanzees into two independent lineages was not a rapid event but included an extended period of progressive genetic divergence simultaneous with gene flow52. Such divergence with continuing gene flow is also true for Bornean (Pongo pygmaeus) and Sumatran (P. abelli) orangutans13, 52.
Whole genome data are not yet available for enough species of Old or New World monkeys, or strepsirrhine primates, to support similar analyses of evolutionary divergence and exchange across partially isolated lineages. However, smaller datasets suggest that the complexity documented for humans and apes may be common across primates. The number of documented hybrid zones between morphologically and/or behaviorally distinct primate populations is increasing67. Active hybrid zones facilitate the study of the process of genetic differentiation, including demographic and phenotypic correlates of hybridization. Various reviews of primate hybridization are available67, 68, but some examples will illustrate general principles.
Baboons (genus Papio) exhibit unusual phenotypic diversity and evolutionary complexity69, 70. Baboon taxonomy has been controversial, but six morphologically distinct species with parapatric geographic ranges are now widely recognized69, 71–73. Hybridization occurs where distinct baboon ‘types’ meet73–75, despite morphological differences and an increased frequency of developmental abnormalities in hybrids76. The evidence suggests a long history of gene flow71, 77, 78. Among baboon species the gene trees from different non-recombining genetic elements (mtDNA, Y-chromosomes, etc.) do not necessarily match observable phenotypic similarity among populations69, 71, 77.
Rhesus macaques and cynomolgus macaques (Macaca fascicularis) are closely related but universally regarded as separate species72. Across mainland Indochina, somewhat similar to the African baboons, these macaque species form a hybrid zone with apparently substantial gene flow. Y-chromosomes from rhesus macaques are found in animals that are phenotypically cynomolgus macaques79. Autosomal gene flow occurs from rhesus into cynomolgus macaque populations, affecting mainland but not other populations of cynomolgus that are isolated on Indonesian islands and in the Philippines80. Thus, these two species present clear evidence for gene flow between well-differentiated species.
One recently discovered species of African monkey (Rungwacebus kipunji) exhibits evidence of ancient hybridization. Of the two geographically isolated populations of kipunji81, 82, one carries mtDNA sequences associated with Cercocebus mangabeys while the other carries mtDNA more closely related to Papio baboons. Despite this apparent genetic introgression from baboons, the second population retains morphological and nuclear DNA similarities with its conspecific sister population. In another case of phylogenetic complexity, the genus Cercopithecus, commonly known as guenons, contains about 24 species72, 83. But their phylogeny has proved difficult to resolve. Novel Alu insertions generate a phylogeny84 with multiple inconsistencies suggesting either ILS or ancient hybridization among differentiated species. Chromosome painting analysis also indicates inter-species hybridization85, and field studies document active hybrid zones68.
Thus, simple allopatric speciation models and associated ideas positing the rapid origin of species boundaries do not generally hold either for humans or other primates. (The bonobo-chimpanzee speciation may be one notable exception52, possibly related to a rapid shift in the Congo River that may have created a robust barrier to gene flow). Newer models of speciation address these complexities86–89 and provide frameworks for future studies. Unresolved questions regarding primate genome evolution include: what are the demographic circumstances associated with extended periods of progressive genetic differentiation despite continuing genetic exchange; what types of genes are able to transfer between lineages and what genes or genetic pathways are first to develop significant differences between diverging lineages; and finally, what changes correlate with the cessation of gene flow between differentiating lineages?
Biomedical relevance
The two most commonly used nonhuman primates in biomedical research are the rhesus macaque (M. mulatta) and the cynomolgus or long-tailed macaque (M. fascicularis). Their importance as models for studies of human health and disease justifies extensive analysis of these genomes (see Supplemental Information S1 (table)). These two species are members of the genus Macaca, a very successful radiation of Old World monkeys that contains 18 extant species72 and is distributed across Asia from Afghanistan to Japan and the Philippines, with relict populations in Morocco. Other important primate model organisms are now also receiving attention. The genomes of the marmoset (Callithrix jacchus), sooty mangabey (Cercocebus atys), African green monkey (Chlorocebus aethiops) and olive baboon (Papio anubis) have been sequenced and assembled. Genome assemblies for mouse lemur (Microcebus murinus) and pig-tailed macaque (M. nemestrina) are in progress. These species are all used as animal models in disease-related research, and therefore whole genome assemblies, transcriptome data and other information is valuable. For example, both the sooty mangabey and African green monkey are important model species for Simian immunodeficiency virus (SIV) research, because these animals are natural hosts that tolerate long-term infection with specific SIV viruses without developing disease90. Development of sequence data and related tools facilitates analyses of how these species tolerate SIV infection that is pathogenic in other primates.
Differences between species in disease-relevant variants
Comparisons of the cynomolgus, rhesus and human genomes are producing information directly relevant to specific biomedical questions. The rhesus and cynomolgus genomes are <1% different in single copy sequence, but the two species do carry specific differences in cytochrome p450 genes involved in drug metabolism91. Although most p450 genes are expressed at similar levels in humans, rhesus and cynomolgus, particular loci (e.g. CYP17A1) are not. Knowledge of genomic differences should improve the interpretation of pharmacological studies using these species.
Humans and the two macaque species also exhibit differences in other genetic pathways relevant to disease, such as melanocortin receptor activity, methyltransferase activity and the parathyroid hormone receptor 192, Moreover, importantly, macaques have an expanded array of MHC Class I genes that are central to their response to infectious agents and other immune system processes16.
Several nonhuman primates carry sequences for coding genes that are associated with increased risk of specific diseases in humans. Rhesus macaques carry variants in OTC, PAH and NAGLU that predispose some humans to disease (i.e. OTC deficiency, a potentially severe disruption of the urea cycle, and phenylketonuria, a relatively common metabolic disorder affecting amino acid levels)2. Chimpanzees carry ‘disease’ alleles in genes related to cancer (MLH1), diabetes mellitus (PPARG) and Alzheimer disease (e.g. APOE)1. Gorillas exhibit alleles at PGRN that in humans are associated with dementia, and variants at TCAP that are associated with hypertrophic cardiomyopathy in humans5.
Polymorphism within species and disease phenotypes
Macaques and other primate species generally have higher levels of within-species genetic variation than humans (see above). Thousands of non-synonymous and splice site variants have been identified in rhesus macaques33, 41, and such variability may influence the response of individual monkeys to experimental protocols. This naturally occurring genetic variation can be exploited to identify novel relationships between specific genes and disease-related phenotypes93, 94 or to study the phenotypic consequences of variation in genes already implicated in human disease risk95–97. Variation in OPRM1, the gene encoding mu-opioid receptor, illustrates the parallels in monkeys and humans, as naturally occurring non-synonymous variation in rhesus OPRM1 influences both the behavioral response of animals to alcohol consumption and the pharmacogenetic response to treatment, similar to non-synonymous variation in humans96. Large-scale DNA re-sequencing of macaques, baboons, African green monkeys, marmosets and other laboratory primates will undoubtedly identify many functionally important genetic variants useful for investigating genetic mechanisms of disease in experimentally controlled primate models33, 41.
Transcriptomics
Analysis of gene expression in primate models of disease will be fundamental to future studies. Primates are, for example, critical for the development and testing of new drugs. Expression of drug-metabolizing p450 genes, as well as some amino acid sequences, differ between cynomolgus and rhesus macaques98, which has implications for pharmacokinetics. Using linkage analysis and quantified differences in gene expression among pedigreed African green monkeys, researchers have mapped eQTLs99. Analyses of rhesus show that gene expression in the immune system is sensitive to differences in social dominance rank, a fundamental aspect of macaque behavior100, thus indicating that common social interactions can influence gene function related to immunology and disease. Primate microRNAs will be critical to understanding disease models101 as well as evolutionary adaptation102, 103. Species differences in microRNA expression may affect expression of transcription factors104, with important consequences for multiple pathways.
Future directions
The initial draft assemblies for nonhuman primates all provide much useful information, but are not complete or reliable enough to support all current scientific goals21, 22. One limitation of draft genomes is the presence of gaps in chromosomal sequences, resulting in missing exons or genes. Although growing use of RNA sequencing to identify transcribed genes is improving the completeness and annotation of nonhuman genome assemblies, the available assemblies still contain gaps. For example, the recent assembly of the gorilla genome incorporates 2.8 gigabases of sequence into contigs5. However, when Scally et al. aligned the human, chimpanzee, gorilla and orangutan genome assemblies in order to conduct “whole genome” analyses of sequence differences, they were only able to produce a four-way “great ape plus human” alignment that included 2.0 gigabases5, due in part to gaps and other problems among the ape assemblies. Another significant issue that affects the ability of researchers to perform comprehensive analyses is problems with identifying and properly assembling segmental duplications and gene copy number differences among species21. Mis-assemblies are also a recurrent problem among draft assemblies produced using next-gen short read technologies only22.
Improved assemblies with longer contigs and more complete coverage in high-quality sequence data (i.e. comprehensive delineation of segmental duplications and fewer genes with gaps and errors) are needed. Deeper sequence coverage will improve some assemblies, but new technologies that provide longer reads will yield better assemblies by filling remaining gaps. The Pacific Biosciences RS II platform is one plausible option for upgrading primate genomes105.
In addition, annotation of functional elements can improve with contiguity and quality of the reference genome as well as access to transcript data. Identification and validation of transcripts for both protein-coding and other transcribed sequences is a high priority. Long non-coding RNAs, microRNAs and other genome features are today poorly annotated for most nonhuman primates. Experimental study of those genomic elements in primate model systems is likely to produce significant dividends for both biomedical and evolutionary studies.
With the sequencing technologies now available, researchers are able to generate large amounts of DNA and RNA sequence data rapidly. This is creating an increasing need for software tools to process comparative data and speed interpretation. The natural emphasis among researchers in human genetics has been the development of computational tools that are specifically designed to analyze human genomes, some of which are not easily applicable to nonhuman species. However, some new tools are readily useful in analyses of nonhuman primates106, 107, and several online databases are collecting, organizing and synthesizing comparative genomic data. [http://biologiaevolutiva.org/greatape/index.html; http://www.rhesusbase.org; http://www.genome.uscs.org; http://www.ensembl.org] However, the speed with which comparative data is being generated creates an ever-growing need for additional computational tools designed to meet the needs of comparative analysis.
Most efforts in primate sequencing to date have been directed toward the great apes, as is natural given their phylogenetic relationships to humans. The sequencing of species from other branches of the primate evolutionary tree, in particular New World monkeys and strepsirrhine primates, will provide increased power to identify conserved genomic segments unique to primates, or to subsets of primates (e.g. catarrhines). Each new species sequenced adds evolutionary perspective and generates new potential models of human genetic disorders.
Little is known about genetic variation in most primate species, although they generally display as much or more variation than do humans. Re-sequencing in commonly used laboratory primates will discover new variants of interest for biomedical research. Furthermore, there is substantial opportunity to use this naturally occurring functional variation to explore gene-gene or gene-environment interactions108, 109.
Finally, nonhuman primates can facilitate investigation of epigenetic control of genome function. Experimental manipulation of environmental factors influencing the human epigenome will be feasible in better characterized primate genomes. Detailed analysis and manipulation of the primate microbiome may also have a substantial impact.
Conclusions
Comparative primate genomics is in a phase of rapid growth, as information about transcriptomes, intra-species polymorphism and other aspects of genomics is being generated at a rapid pace. The major impact to date has been to provide novel information concerning the history and mechanisms of human genome evolution including evidence for a complex history of genetic divergence and exchange among ancestral evolutionary lineages (Figure 1). Nonhuman primate genomics is also expanding the scope of biomedical research with innovative analyses of primate models of human disease. Despite recent progress, both evolutionary and biomedical studies would benefit significantly from additional information. There is real opportunity to examine the continuum from microevolutionary processes controlling within-species variation (e.g. positive and negative fitness effects of segregating polymorphisms within species) to macroevolutionary processes affecting between-species differences.
Either from the perspective of understanding the origin of humans or elucidating the genetic basis of human disease, nonhuman primates are indispensable resources for comparative and experimental study. Genomics is now central to all of biology and so it is both sensible and timely that comparative primate genomics is receiving increased attention. Analyses to date have provided valuable and sometimes unexpected results. There will be many further advances, including a few more surprises, and ultimately a much richer understanding of genome structure, function and dynamics as investigators with a wide range of interests continue to generate new information concerning the genomes of nonhuman primates.
Supplementary Material
Key Points.
Whole genome assemblies are now available for all the great apes and several other nonhuman primate species. The published analyses document between-species differences in gene content, segmental duplications, retroposon insertions and other genomic features.
Next-gen sequencing has made whole genome sequencing and draft assembly more practical, and consequently additional nonhuman primate genome assemblies, with detailed annotation and other associated analyses, are in progress.
The available data concerning nonhuman primate population genomics indicate that these species exhibit as much or more within-species genetic variation than is found among humans, with some species showing substantially higher rates of polymorphism.
Current information indicates that differences between species in patterns of gene expression are common, have been influenced by natural selection and are likely to contribute to phenotypic differences among species.
Analyses suggest that the evolutionary radiation that produced the extant human, chimpanzee and gorilla lineages (i.e. the speciation events) resulted from a complex process characterized by incomplete lineage sorting and/or gene flow among partially differentiated lineages.
Whole genome analysis as well as more targeted sequencing in nonhuman primate species used in disease-related research has identified specific variants relevant to human disease risk, and finds differences among primate model species that are directly relevant to disease mechanisms and other biomedically significant phenotypes such as drug metabolism.
Acknowledgments
The authors thank Kim Worley, Stephen Richards, Muthuswamy Raveendran, Gloria Fawcett, David Rio Deiros and Fuli Yu for valuable discussion, and three anonymous reviewers for their comments. This work was supported by NIH grants U54-HG006484 and R24-OD011173.
Glossary
- catarrhine
any member of the primate evolutionary lineage that includes Old World monkeys (superfamily Cercopithecoidea) or hominoids (superfamily Hominoidea). The catarrhines include all extant apes, anthropoid monkeys native to Asia and Africa, and humans
- coalescent models
an approach used in population genetics to investigate various aspects of population history and dynamics. These models are based on the genealogy or relationships within a gene tree among alleles of a specific DNA sequence. All alleles found in a population or set of related populations can be traced back to a common ancestral sequence, and the statistical properties of those allelic relationships are exploited to investigate questions of population genetics and history.
- effective population size
A basic concept from population genetics that describes the number of individuals required in an ideal breeding population (equal numbers of breeding males and females, with equal reproductive success among them) of constant size to sustain a given amount of within-population genetic variation. Because genetic variation in a given population is affected by current and past demographic factors, estimation of effective population size allows researchers to infer aspects of population history.
- hominins
members of the evolutionary lineage leading to humans after divergence from the ancestors of chimpanzees. Hominins include species directly ancestral to modern humans, and related species such as Neanderthals or older branches such as australopithecines. Tribe Hominini.
- Hybrid zones
Geographic areas, often but not always elongated and narrow in shape, where two distinct primate species occur together, mate and produce hybrid offspring that are fertile and reproductively successful themselves.
- Incomplete lineage sorting
The process by which, as a result of segregation of an ancestral polymorphism, the evolutionary relationships among a series of homologous DNA sequences present in a set of distinct populations do not match the phylogenetic relationships among those overall populations, i.e. that gene trees do not match population trees. See Figure 2.
- Old World monkeys
members of the branch of primates that includes extant anthropoid primates (monkeys) native to Asia and Africa, superfamily Cercopithecoidea
- New World monkeys
members of the branch of primates that includes extant anthropoid primates (monkeys) native to South and Central America, parvorder Platyrrhini
- positive selection
natural selection acting on phenotypes and the relevant DNA sequences that results in directional change toward a new sequence and phenotype. Contrasted with negative selection that acts to eliminate deleterious traits, and therefore acts against any new mutations that generate them
- strepsirrhine primates
members of the branch of primates that includes lemurs, lorises, galagoes and cheirogaleids, suborder Strepsirrhini
- allopatric
having separate, non-overlapping geographic distributions
- parapatric
having geographic distributions that are adjoining but do not overlap
Figure 2. Geographic distribution and genetic variation in selected primates.
Despite having modest or small current population sizes, and in most cases being either endangered or critically endangered (www.iucnredlist.org), most nonhuman primate species investigated to date have substantial levels of within-species genetic variability. Panel A: The approximate geographic distributions of African ape species, although actual distributions are generally discontinuous isolated populations within these areas. Hatching indicates the level of genetic variation estimated through the Great Ape Genome Project36. The IUCN conservation status is indicated by legend font. Panel B: Approximate geographic distributions for gibbons, orangutans and selected macaque species. As for the African species, conservation status is indicated by the legend font and the estimated level of genetic variation is shown by hatching. Information on genetic variation from references #33, 34 and 66.
Biographies
Jeffrey Rogers, Ph.D.: Dr. Rogers is an Associate Professor in the Human Genome Sequencing Center and Dept. of Molecular and Human Genetics. His research focuses on the genomics of nonhuman primates, analysis of new genome assemblies, investigation of genetic variation in primate species used as models of disease, and targeted analysis of primate models of psychiatric disorders.
Richard A. Gibbs, Ph.D.: Dr. Gibbs is Director of the Human Genome Sequencing Center and holds the Wofford Cain Chair of Molecular and Human Genetics. His work includes many aspects of human and comparative genomics, with particular emphasis on application of new technologies to research concerning Mendelian disorders, cancer and common metabolic disease, as well as the introduction of genomics into diagnostic and clinical practice.
References
- 1.Consortium TCSaA. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 2.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 3.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci U S A. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiper ME, Seiffert ER. Evidence for a convergent slowdown in primate molecular rates and its implications for the timing of early primate evolution. Proc Natl Acad Sci U S A. 2012;109:6006–6011. doi: 10.1073/pnas.1119506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li WH, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 9.Perry GH, et al. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 2012;4:126–135. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Britten RJ, Rowen L, Williams J, Cameron RA. Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci U S A. 2003;100:4661–4665. doi: 10.1073/pnas.0330964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. This paper presents an outstanding example of the power of comparative genomics to identify novel conserved genomic regions rhat evolve slowly across species as a result of shared functional significance.
- 12.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gokcumen O, et al. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A. 2013;110:15764–15769. doi: 10.1073/pnas.1305904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowick K, et al. Gain, loss and divergence in primate zinc-finger genes: a rich resource for evolution of gene regulatory differences between species. PLoS One. 2011;6:e21553. doi: 10.1371/journal.pone.0021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 19.Dumas LJ, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Bleness MS, et al. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2012;2:977–986. doi: 10.1534/g3.112.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Goodsell J, Norgren RB., Jr Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics. 2012;13:206. doi: 10.1186/1471-2164-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 24.Marques-Bonet T, Ryder OA, Eichler EE. Sequencing primate genomes: what have we learned? Annu Rev Genomics Hum Genet. 2009;10:355–386. doi: 10.1146/annurev.genom.9.081307.164420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 27.Dumas L, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazave E, et al. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res. 2011;21:1626–1339. doi: 10.1101/gr.117242.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. This is one of the first studies to identify significant changes in gene copy number among humans and the great apes that appear to be driven by positive selection, with evidence for adaptive changes in both copy number and nucleotide sequences.
- 30.Lorente-Galdos B, et al. Accelerated exon evolution within primate segmental duplications. Genome Biol. 2013;14:R9. doi: 10.1186/gb-2013-14-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortna A, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen-Seaman MI, Deinard AS, Kidd KK. Modern African ape populations as genetic and demographic models of the last common ancestor of humans, chimpanzees, and gorillas. J Hered. 2001;92:475–480. doi: 10.1093/jhered/92.6.475. [DOI] [PubMed] [Google Scholar]
- 33.Fawcett GL, et al. Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2011;12:311. doi: 10.1186/1471-2164-12-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 35.Smith DG, McDonough JW, George DA. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol. 2007;69:182–198. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- 36. Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. This paper presents a substantial amount of genomic information concerning species and subspecies of great apes. This analysis provides important new insight into the evolution of these lineages.
- 37.Ferguson B, et al. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. This paper reports the first genome-scale analysis of several threatened or endangered primates, documenting unexpectd patterns of intra-specific variability and ancient selection on protein-coding genes.
- 40.Vallender EJ. Expanding whole exome resequencing into non-human primates. Genome Biol. 2011;12:R87. doi: 10.1186/gb-2011-12-9-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan Q, et al. The rhesus macaque is three times as diverse but more closely equivalent in damaging coding variation as compared to the human. BMC Genet. 2012;13:52. doi: 10.1186/1471-2156-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. In this classic and prescient paper, written long before researchers had access to substantial amounts of DNA sequence data, the authors used information about protein sequence differences and dissociation temperatures for hybrid human-chimpanzee DNA molecules to correctly infer that much of the anatomical and physiological difference between humans and chimpanzees are due to changes in gene regulation rather than changes in protein sequence.
- 43.Wilson AC, Maxson LR, Sarich VM. Two types of molecular evolution. Evidence from studies of interspecific hybridization. Proc Natl Acad Sci U S A. 1974;71:2843–2847. doi: 10.1073/pnas.71.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arbiza L, et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat Genet. 2013;45:723–972. doi: 10.1038/ng.2658. This paper describes an innovative analysis of evolutionary changes in transcription factor binding sites, finding that natural selection has exerted significant effects of these regulatory sequences during recent human evolution.
- 45.Pipes L, et al. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic Acids Res. 2013;41:D906–D914. doi: 10.1093/nar/gks1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sexspecific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. This report describes significant data concerning differences in gene splicing among nonhuman primates and humans, a potentially important mechanism for rapid evolutionary change.
- 48.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 49.Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng J, et al. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet. 2012;91:455–465. doi: 10.1016/j.ajhg.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babbitt CC, Tung J, Wray GA, Alberts SC. Changes in gene expression associated with reproductive maturation in wild female baboons. Genome Biol Evol. 2012;4:102–109. doi: 10.1093/gbe/evr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mailund T, Halager AE, Scally A. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLOS Genetics. 2012;8:e1003125. doi: 10.1371/journal.pgen.1003125. This paper reports a highly informative analysis of genomic differentiation among ancestral populations of hominoids, including analysis of the process of divergence that produced the evolutionary separation of the ancestors of humans, chimpanzees and gorillas.
- 53.Reich SPLP. The date of interbreeding between Neanderthals and modern humans. PLOS Genetics. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sankararaman S, Patterson N, Li H, Paabo S, Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.George RD, et al. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Res. 2011;21:1686–1694. doi: 10.1101/gr.121327.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harris RA, et al. Evolutionary genetics and implications of small size and twinning in callitrichine primates. Proceedings of the National Academy of Sciences USA. doi: 10.1073/pnas.1316037111. (in press). This report describes genetic evolution in an unusual group of nonhuman primates that exhibit unique adaptations for reproduction, identifying specific sequence changes that may contribute to those adaptations.
- 58.O'Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 2012;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charrier C, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pamilo P, Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 63.Rogers J. Levels of the genealogical hierarchy and the problem of homonoid phylogeny. Amer. J. Phys. Anthropol. 1994;94:81–88. doi: 10.1002/ajpa.1330940107. [DOI] [PubMed] [Google Scholar]
- 64.Ruvolo M. Molecular phylogeny of the hominoids: inferences from multiple independent DNA sequence data sets. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 65.Prufer K, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wall JD, et al. Incomplete lineage sorting is common in extant gibbon genera. PLoS One. 2013;8:e53682. doi: 10.1371/journal.pone.0053682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zinner D, Arnold ML, Roos C. The strange blood: natural hybridization in primates. Evol Anthropol. 2011;20:96–103. doi: 10.1002/evan.20301. [DOI] [PubMed] [Google Scholar]
- 68.Detwiler KM, Burrell AS, Jolly CJ. Conservation implications of hybridization in African cercopithecine monkeys. International Journal of Primatology. 2005;26:661–684. [Google Scholar]
- 69. Jolly CJ. A proper study for mankind: Analogies from the Papionin monkeys and their implications for human evolution. Am J Phys Anthropol. 2001;(Suppl 33):177–204. doi: 10.1002/ajpa.10021. This wide-ranging review by a highly influential biological anthropologist discusses issues related to the complexity of reconstructing ancient evolutionary processes and the value of information concerning the population biology of and hybridization among nonhuman primate species for understanding aspects of human evolution.
- 70.Jolly CJ. In: Species, Species Concepts and Primate Evolution. Kimbel WH, Martin LB, editors. New York, NY: Plenum Press; 1993. pp. 67–107. [Google Scholar]
- 71.Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol. 2013;150:133–140. doi: 10.1002/ajpa.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groves C. Primate Taxonomy. Washington, DC: Smithsonian Institution Press; 2001. [Google Scholar]
- 73.Jolly CJ, Burrell AS, Phillips-Conroy JE, Bergey C, Rogers J. Kinda baboons (Papio kindae) and grayfoot chacma baboons (P. ursinus griseipes) hybridize in the Kafue river valley, Zambia. Am J Primatol. 2011;73:291–303. doi: 10.1002/ajp.20896. [DOI] [PubMed] [Google Scholar]
- 74.Alberts SC, Altmann J. Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. Am J Primatol. 2001;53:139–154. doi: 10.1002/ajp.1. [DOI] [PubMed] [Google Scholar]
- 75.Bergman TJ, Phillips-Conroy JE, Jolly CJ. Behavioral variation and reproductive success of male baboons (Papio anubis × Papio hamadryas) in a hybrid social group. Am J Primatol. 2008;70:136–147. doi: 10.1002/ajp.20467. [DOI] [PubMed] [Google Scholar]
- 76.Ackermann RR, Rogers J, Cheverud JM. Identifying the morphological signatures of hybridization in primate and human evolution. J Hum Evol. 2006;51:632–645. doi: 10.1016/j.jhevol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 77.Burrell AS. Dept. of Anthropology. New York, NY: New York University; 2009. p. 323. [Google Scholar]
- 78.Charpentier MJ, et al. Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Mol Ecol. 2012;21:715–7131. doi: 10.1111/j.1365-294X.2011.05302.x. [DOI] [PubMed] [Google Scholar]
- 79.Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 80.Stevison LS, Kohn MH. Divergence population genetic analysis of hybridization between rhesus and cynomolgus macaques. Mol Ecol. 2009;18:2457–2475. doi: 10.1111/j.1365-294X.2009.04212.x. [DOI] [PubMed] [Google Scholar]
- 81.Burrell AS, Jolly CJ, Tosi AJ, Disotell TR. Mitochondrial evidence for the hybrid origin of the kipunji, Rungwecebus kipunji (Primates: Papionini) Mol Phylogenet Evol. 2009;51:340–348. doi: 10.1016/j.ympev.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 82.Jones T, et al. The highland mangabey Lophocebus kipunji: a new species of African monkey. Science. 2005;308:1161–1164. doi: 10.1126/science.1109191. [DOI] [PubMed] [Google Scholar]
- 83.Jaffe KE, Isbell LA. In: Primates in Perspective. 2nd. edition. Campbell CJ, Fuentes A, MacKinnon KC, Bearder SK, Stumpf RM, editors. Oxford, UK: Oxford University Press; 2011. [Google Scholar]
- 84.Xing J, et al. A mobile element-based evolutionary history of guenons (tribe Cercopithecini) BMC Biol. 2007;5:5. doi: 10.1186/1741-7007-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moulin S, Gerbault-Seureau M, Dutrillaux B, Richard FA. Phylogenomics of African guenons. Chromosome Res. 2008;16:783–799. doi: 10.1007/s10577-008-1226-6. [DOI] [PubMed] [Google Scholar]
- 86.Gombert Z, Parchman TL, Buerkle CA. Genomics of isolation in hybrids. Philos Trans R Soc Lond B Biol Sci. 2012;367:439–450. doi: 10.1098/rstb.2011.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Network MCS. What do we need to know about speciation ? Trends Ecol Evol. 2012;27:27–39. doi: 10.1016/j.tree.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Nosil P, Feder JL. Genomic divergence during speciation: causes and consequences. Philos Trans R Soc Lond B Biol Sci. 2012;367:332–342. doi: 10.1098/rstb.2011.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 90.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science. 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ebeling M, et al. Genome-based analysis of the nonhuman primate Macaca fascicularis as a model for drug safety assessment. Genome Res. 2011;21:1746–1756. doi: 10.1101/gr.123117.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan G, et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 93.Cox LA, et al. Identification of promoter variants in baboon endothelial lipase that regulate high-density lipoprotein cholesterol levels. Circulation. 2007;116:1185–1195. doi: 10.1161/CIRCULATIONAHA.107.704346. [DOI] [PubMed] [Google Scholar]
- 94.Rogers J, et al. CRHR1 genotypes, neural circuits and the diathesis for anxiety and depression. Mol Psychiatry. 2013;18:700–707. doi: 10.1038/mp.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francis PJ, et al. Rhesus monkeys and humans share common susceptibility genes for age-related macular disease. Hum Mol Genet. 2008;17:2673–2680. doi: 10.1093/hmg/ddn167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vallender EJ, Ruedi-Bettschen D, Miller GM, Platt DM. A pharmacogenetic model of naltrexone-induced attenuation of alcohol consumption in rhesus monkeys. Drug Alcohol Depend. 2010;109:252–256. doi: 10.1016/j.drugalcdep.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barr CS, et al. Rearing condition and rh5-HTTLPR interact to influence limbic-hypothalamic-pituitary-adrenal axis response to stress in infant macaques. Biol Psychiatry. 2004;55:733–738. doi: 10.1016/j.biopsych.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 98.Ise R, et al. Expression profile of hepatic genes in cynomolgus macaques bred in Cambodia, China, and Indonesia: implications for cytochrome P450 genes. Drug Metab Pharmacokinet. 2012;27:307–316. doi: 10.2133/dmpk.dmpk-11-rg-133. [DOI] [PubMed] [Google Scholar]
- 99.Jasinska AJ, et al. A non-human primate system for large-scale genetic studies of complex traits. Hum Mol Genet. 2012;21:3307–3316. doi: 10.1093/hmg/dds160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tung J, et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci U S A. 2012;109:6490–6495. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karere GM, Glenn JP, VandeBerg JL, Cox LA. Differential microRNA response to a high-cholesterol, high-fat diet in livers of low and high LDL-C baboons. BMC Genomics. 2012;13:320. doi: 10.1186/1471-2164-13-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu HY, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Somel M, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dannemann M, et al. Transcription factors are targeted by differentially expressed miRNAs in primates. Genome Biol Evol. 2012;4:552–564. doi: 10.1093/gbe/evs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.English AC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, Lu J, Yu J, Gibbs RA, Yu F. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013;23:833–842. doi: 10.1101/gr.146084.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barr CS, et al. The utility of the non-human primate; model for studying gene by environment interactions in behavioral research. Genes Brain Behav. 2003;2:336–340. doi: 10.1046/j.1601-1848.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 109.Barr CS, et al. Interaction between serotonin transporter gene variation and rearing condition in alcohol preference and consumption in female primates. Arch Gen Psychiatry. 2004;61:1146–1152. doi: 10.1001/archpsyc.61.11.1146. [DOI] [PubMed] [Google Scholar]
- 110.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014 doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mayr E. Systematics and the Origin of Species. New York, NY: Columbia Univ. Press; 1942. [Google Scholar]
- 112.Gould SJ, Eldridge N. Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology. 1977;3:115–151. [Google Scholar]
- 113.Pagel M, Venditti C, Meade A. Large punctuational contribution of speciation to evolutionary divergence at the molecular level. Science. 2006;314:119–121. doi: 10.1126/science.1129647. [DOI] [PubMed] [Google Scholar]
- 114.Higashino A, et al. Whole-genome sequencing and analysis of the Malaysian cynomolgus macaque (Macaca fascicularis) genome. Genome Biol. 2012;13:R58. doi: 10.1186/gb-2012-13-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.