Abstract
Clathrin-mediated endocytosis (CME) is the best-studied pathway by which cells selectively internalize molecules from the plasma membrane and surrounding environment. Previous live-cell imaging studies using ectopically overexpressed fluorescent fusions of endocytic proteins indicated that mammalian CME is a highly dynamic but inefficient and heterogeneous process. In contrast, studies of endocytosis in budding yeast using fluorescent protein fusions expressed at physiological levels from native genomic loci have revealed a process that is very regular and efficient. To analyse endocytic dynamics in mammalian cells in which endogenous protein stoichiometry is preserved, we targeted zinc finger nucleases (ZFNs) to the clathrin light chain A and dynamin-2 genomic loci and generated cell lines expressing fluorescent protein fusions from each locus. The genome-edited cells exhibited enhanced endocytic function, dynamics and efficiency when compared with previously studied cells, indicating that CME is highly sensitive to the levels of its protein components. Our study establishes that ZFN-mediated genome editing is a robust tool for expressing protein fusions at endogenous levels to faithfully report subcellular localization and dynamics.
Clathrin-mediated endocytosis (CME) is characterized by recruitment of clathrin triskelia, composed of heavy and light chains, to the plasma membrane and their assembly into polygonal cages that mediate membrane invagination. During late stages of this process the GTPase dynamin is recruited to the necks of these invaginations1, where it functions in vesicle release through membrane scission and the subsequent internalization of plasma-membrane molecules, extracellular fluid and specific ligands from the environment2,3. Three decades of evidence directly connect perturbation of CME to a broad range of pathophysiological outcomes, including atherosclerosis4, disorders of the peripheral and central nervous system5 and infection by the hepatitis C virus6.
The study of CME has been greatly advanced through the use of fluorescent fusion proteins7. In yeast, direct genomic tagging of pairs of genes with different fluorescent tags has allowed researchers to define a very regular series of spatiotemporal events for CME in living cells8. Conversely, in mammalian cells these events have been described as being much more heterogeneous and inefficient9–12, but the inability to do precise genome editing has forced researchers to rely heavily on overexpression of fusion proteins1,9–14. Furthermore, these fusion proteins are often derived from a different cell type or species from the cell type being studied, and encode non-native splice variants. An accurate description of endocytic dynamics is the foundation for understanding the mechanism and regulation of this crucial process. Because evidence from fields ranging from developmental and cell biology to plant physiology has shown that overexpression can result in protein mislocalization, aggregation and altered signalling15–20, we sought to re-examine the highly dynamic process of CME using fluorescent fusion proteins expressed from their native loci.
RESULTS
ZFN-mediated editing as an efficient and accurate method for generating stable cell lines expressing fluorescent protein fusions from native loci
To engineer precise gene fusions at endogenous loci, we used zinc finger nucleases (ZFNs)21 designed to cleave their target genes near the stop codons. The resulting double-strand breaks were mended by homology-directed repair using an exogenously supplied DNA donor that encodes a fluorescent tag (Fig. 1a). In all cases, ZFNs and donor constructs were co-transfected into the cells. The high-editing efficiency and accuracy of the open reading frame (ORF) addition process, coupled with the expression of proteins bearing fluorescent markers, allowed us to obtain the desired cells simply by using fluorescence-activated cell sorting (FACS) without drug selection (Supplementary Fig. S1a and Table S1, top). Single-cell-derived clones were then generated by limiting dilution. We found that ZFN treatment had a negligible impact on cell survival (Supplementary Fig. S1b), suggesting that the editing strategy is an ideal method for generating stable cell lines. The complete panel of cells engineered in this study is described in Table 1.
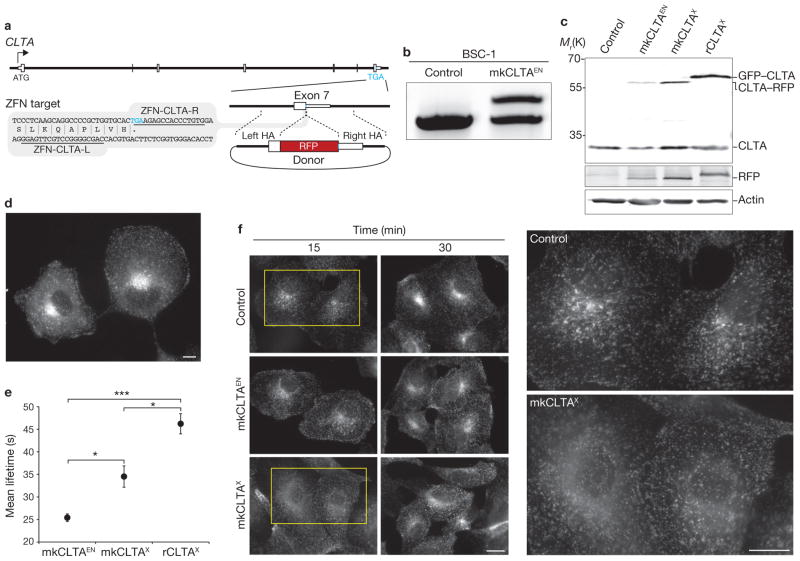
Figure 1.
Editing of CLTA using ZFNs in BSC-1 cells. (a) Schematic representation of the strategy for integration of RFP at the CLTA locus. White boxes, exons of CLTA; HA, donor plasmid region of homology to CLTA sequence; Blue letters, stop codon. The grey box indicates the region of CLTA exon 7 surrounding the translation stop codon; the underlined sequences indicate the recognition stretches of the individual zinc finger nucleases (ZFN-CLTA-L and ZFN-CLTA-R, respectively). (b) Out-out PCR showing targeted integration of RFP. Control, parental cell line; mkCLTAEN, single-allele CLTA–RFP tagged genome-edited line. (c) Western blot analysis of cell lysates immunoblotted for CLTA, RFP and actin. Note RFP antibody cross-reactivity with GFP. mkCLTAX, stable CLTA–RFP overexpression line; rCLTAX, stable GFP–CLTA (rat brain-derived) overexpression line. (d) Epifluorescence image of mkCLTAEN cells expressing endogenous CLTA–RFP. Scale bar, 10 μm. (e) CCP dynamics were assessed in the indicated BSC-1 cell lines by quantifying the lifetime of fluorescently tagged CLTA proteins at the plasma membrane. Data are means ± s.e.m. Tracks, 30,734–50,250; n = 5, 11 and 15 cells for the mkCLTAX, mkCLTAEN and rCLTAX lines, respectively. Triple asterisk indicates P < 0.0001 and single asterisk indicates P < 0.05. (f) Time course of Alexa Fluor 488-conjugated human transferrin uptake in the indicated BSC-1 cell lines. Areas enclosed by yellow boxes are enlarged (right) for better visualization. Scale bar, 10 μm. Uncropped images of gels and blots are shown in Supplementary Fig. S2a.
Table 1.
Cell lines used in this study
| Cell lines with marker ORFs added to the endogenous locus
| ||||
|---|---|---|---|---|
| Designation | Cell line | Species | Gene | Marker |
| mkCLTAEN | BSC-1 | Cercopithecus aethiops | monkey CLTA (clathrin light chain A) | RFP |
| hCLTAEN | SK-MEL-2 | Homo sapiens | human CLTA (clathrin light chain A) | RFP |
| hDNM2EN | SK-MEL-2 | Homo sapiens | human DNM2 (dynamin 2) | GFP |
| hCLTAEN/DNM2EN | SK-MEL-2 | Homo sapiens | human CLTA, DNM2 | RFP/GFP |
| Cell lines carrying overexpressed, randomly integrated marker ORFs
| ||||
|---|---|---|---|---|
| Designation | Cell line | Species | Gene | Marker |
| rCLTAX | BSC-1 | Cercopithecus aethiops | rat brain CLTA (clathrin light chain A) | GFP |
| mkCLTAX | BSC-1 | Cercopithecus aethiops | monkey CLTA (clathrin light chain A) | RFP |
| hCLTAX | SK-MEL-2 | Homo sapiens | human CLTA (clathrin light chain A) | RFP |
| hDNM2X | SK-MEL-2 | Homo sapiens | human DNM2 (dynamin 2) | GFP |
ZFN-mediated insertion of RFP at the CLTA locus in monkey cells
We designed ZFNs targeting the 3′-terminus of clathrin light chain A (CLTA; Fig. 1a and Supplementary Table S2) and first used them in African green monkey kidney epithelial cells (BSC-1), which have been used for many important studies of endocytic dynamics10,11,22,23. We generated a cell line, mkCLTAEN, in which one CLTA allele was tagged with red fluorescent protein (RFP; Table 1 and Supplementary Fig. S1c and Table S1, bottom). Genotyping and western blot analysis confirmed the accurate addition of the tag to the CLTA locus (Fig. 1b, c and Supplementary Fig. S2a). As expected, mkCLTAEN cells demonstrated punctate plasma membrane and perinuclear CLTA–RFP distribution (Fig. 1d and Supplementary Fig. S3a), and complete co-localization of CLTA–RFP with clathrin heavy chain (CHC; Supplementary Fig. S3b), indicating that the endogenously tagged CLTA is correctly incorporated into clathrin-coated pits (CCPs).
Genome-edited CLTA–RFP monkey cells exhibit increased CME dynamics and function relative to overexpression lines
Numerous reports have documented striking heterogeneity in the size, dynamics and productivity of CCPs10–12, yet the nature of this heterogeneity and its functional implications have not been established. Because determining the time required to form coated pits and vesicles is critical for gaining a mechanistic and regulatory understanding of CME11,22–24, we sought to quantitatively analyse clathrin dynamics in mkCLTAEN cells using total internal reflection fluorescence (TIRF) microscopy and semiautomated particle tracking. Analysis of a well-studied BSC-1-derived cell line (rCLTAX, Table 1), which overexpresses rat brain CLTA fused to green fluorescent protein (GFP) from a randomly integrated transgene12, yielded an overall mean lifetime for the fusion protein at the plasma membrane of 46 ± 2.2 s (Fig. 1e and Supplementary Fig. S4a, Table S3 and Movie S1), similar to previously reported values of 39 and 46 s (refs 11, 12). Strikingly, analysis of the endogenously tagged mkCLTAEN cell line revealed a significantly shorter mean lifetime (25 ± 0.9 s, P < 0.0001; Fig. 1e and Supplementary Fig. S4a, Table S3 and Movie S2).
Our single-allele tagged, genome-edited cell lines exhibited considerably dimmer fluorescent structures than the stable overexpression cell lines. As such, we were concerned that this decreased fluorescence might affect our ability to faithfully quantify endocytic protein dynamics. To address this concern, we conducted two tests of the effects of low fluorescence signal on our lifetime measurements. First, we determined whether the measured lifetime for an overexpression line is affected by decreasing the exposure time. Whether the rCLTAX cell line was analysed with an exposure time of 900 ms (as used in this study) or 30 ms (to simulate low fluorescence), similar mean lifetimes of 46 ± 2.2 s and 51 ± 6.3 s, respectively, were obtained. Second, we predicted that if we are faithfully measuring the lifetimes of low-fluorescence structures in the single-allele edited lines, then their arrival and disappearance at individual endocytic sites should be coincident with those of the same protein fused to a different tag, and expressed in the same cell. Indeed, when we transiently overexpressed human CLTA fused to GFP in the hCLTAEN–1 cell line (expressing CLTA–RFP), we found that, despite the low fluorescence of the endogenous CLTA–RFP, its arrival and disappearance were coincident with those of CLTA–GFP (data not shown). These data collectively demonstrate that the observed shorter mean lifetimes of our single-allele genome-edited lines, compared with all-allele edited and overexpression lines, were not the result of a decreased ability to detect clathrin for its entire membrane lifetime.
To address whether the species (rat versus monkey) and splice variant of rCLTAX could account for the discrepancy in CCP lifetimes, we generated a stable overexpression BSC-1 cell line (mkCLTAX; Table 1) in which the major endogenously expressed mkCLTA splice variant was fused to RFP. Relative to the endogenously tagged mkCLTAEN cell line, CLTA was overexpressed 3.5-fold in mkCLTAX and 5.7-fold in rCLTAX cells (Fig. 1c and Supplementary Fig. S2a). mkCLTAX cells had an average CLTA–RFP lifetime of 35 ± 2.4 s—a value significantly longer than that of the mkCLTAEN line (P < 0.05; Fig. 1e and Supplementary Fig. S4a, Table S3, and Movie S3)—indicating that protein overexpression increases CCP lifetime. Moreover, mkCLTAEN cells showed robust uptake of transferrin (a CME cargo), similar to that of control cells, with intense perinuclear accumulation. By contrast, the transferrin distribution was more diffuse in mkCLTAX cells, suggesting a diminished rate of internalization (Fig. 1f). These results indicate that overexpression of CLTA can also negatively impact endocytic function and are consistent with previous studies implicating clathrin light chains as negative regulators of CME7,23,25. Collectively, these findings demonstrate a more faithful representation of CME than was achieved using previous methods requiring overexpression and highlight the importance of studying the dynamics of proteins at physiological expression levels.
Genome-edited CLTA–RFP human cells exhibit increased CME dynamics and function
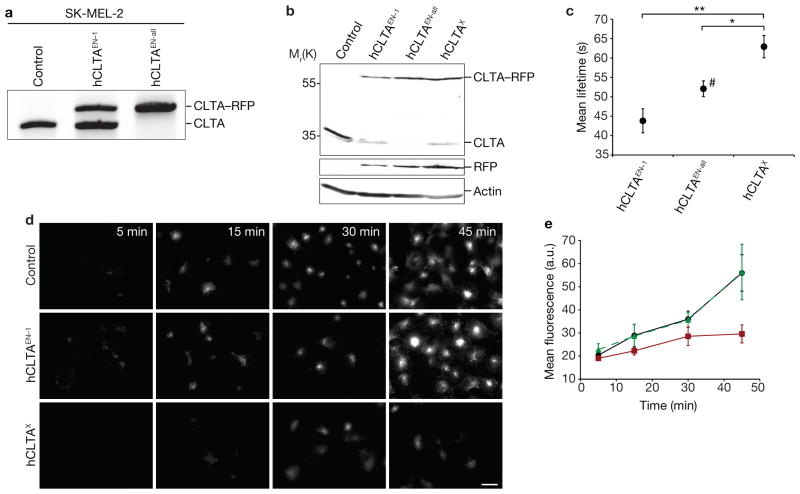
To test the generality of these results, we used the same ZFN pair to generate a human skin melanoma cell line, SK-MEL-2, in which CLTA was tagged with RFP at its native locus. Genotyping and western blot analysis confirmed the precise addition of the tag to the CLTA locus, yielding both single-allele tagged (hCLTAEN–1) and all-allele tagged (hCLTAEN–all) cell lines (Fig. 2a, b, Table 1 and Supplementary Fig. S2b). Importantly, hCLTAEN–all cells showed complete co-localization of CLTA–RFP with CHC (Supplementary Fig. S3c). Analysis of hCLTAEN–1 revealed a mean CLTA–RFP lifetime (44 ± 3.1 s) that was significantly shorter than the lifetime in a cell line overexpressing the same CLTA–RFP fusion (hCLTAX; 63 ± 2.9 s, P < 0.001; Fig. 2c and Supplementary Fig. S4b, Table S3 and Movies S4 and S5). Notably, analysis of the homozygous hCLTAEN–all cell line revealed large, stable clathrin-coated structures at the plasma membrane that were not seen in the isogenic heterozygous line. We also observed diffraction-limited CCPs in hCLTAEN–all cells, and their analysis yielded an intermediate CLTA–RFP mean lifetime of 52 ± 2.0 s (Fig. 2c and Supplementary Fig. S4b, Table S3 and Movie S6). Finally, we found that hCLTAEN cells maintain robust transferrin uptake, similar to control cells, whereas the overexpression hCLTAX cell line exhibited diminished endocytic function (Fig. 2d, e). Taken together, our data from isogenic cells in two different species support two major conclusions: CME in mammalian cells is a significantly more dynamic process than previously thought, and increasing fusion protein levels has a detrimental effect on CCP dynamics and endocytic function.
Figure 2.
Editing of CLTA using ZFNs in SK-MEL-2 cells. (a) Out-out PCR showing targeted integration of RFP. Control, parental cell line; hCLTAEN–1, single-allele CLTA–RFP tagged genome-edited line; hCLTAEN–all, all-allele tagged genome-edited line. (b) Western blot analysis of cell lysates immunoblotted for CLTA, RFP, and actin. hCLTAX, stable CLTA–RFP overexpression line. (c) CCP dynamics were assessed in the indicated cell lines by quantifying the lifetime of CLTA–RFP at the plasma membrane. Data are means ± s.e.m. Tracks, 14,868–19,525; n = 8, 13 and 11 cells (hCLTAEN–1, hCLTAEN–all and hCLTAX, respectively). Double asterisks indicate P < 0.001, single asterisk indicates P < 0.05. #; only diffraction-limited CCPs were analysed, despite the presence of larger, stable clathrin-coated structures. (d) Time course of Alexa Fluor 488-conjugated human transferrin uptake in SK-MEL-2 cell lines. Scale bar, 10 μm. (e) Quantification of cell fluorescence of hCLTAEN–1 cells (green curve; n = 40), compared with parental (black curve; n = 45) and hCLTAX (red curve; n = 21) cell lines from experiment performed as in d. Data are means ± s.d. Uncropped images of gels and blots are shown in Supplementary Fig. S2b.
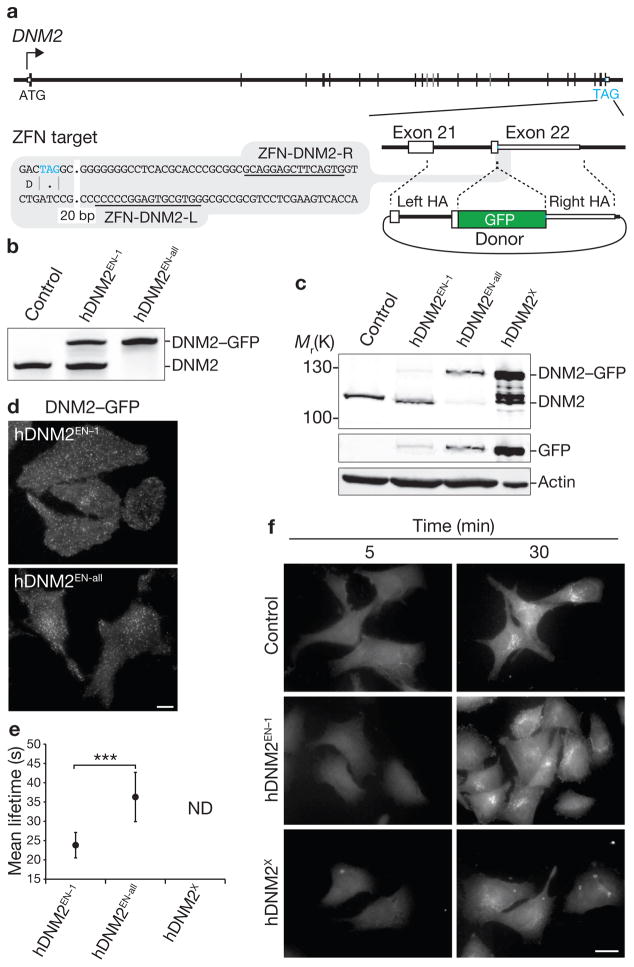
ZFN-mediated insertion of GFP at the DNM2 locus in human cells reveals unprecedented dynamics
We next sought to address whether overexpression might affect the native dynamics of another CME factor, dynamin-2 (DNM2). Previous studies on the dynamics of dynamin fluorescent protein fusions required examination of cells with a low level of overexpression, shortly after transfection (14–16 h), because high levels of overexpression at longer intervals resulted in the appearance of large, stationary structures12,24. We created SK-MEL-2 cell lines in which one (hDNM2EN–1) or all (hDNM2EN–all) endogenous DNM2 alleles were tagged with GFP (Fig. 3a, Table 1 and Supplementary Fig. S1a, d) and compared them with a cell line overexpressing the identical DNM2–GFP fusion that was generated from a randomly integrated transgene (hDNM2X; Table 1). The hDNM2EN lines were shown by genotyping and phenotypic analysis to express physiological levels of fusion protein (Fig. 3b, c and Supplementary Fig. S2c) that localized as punctate structures on the plasma membrane (Fig. 3d).
Figure 3.
Editing of DNM2 using ZFNs in SK-MEL-2 cells. (a) Schematic representation of the strategy for integration of GFP at the DNM2 locus. (b) Out-out PCR showing targeted integration of GFP. Control, parental cell line; hDNM2EN–1, single-allele tagged genome-edited line; hDNM2EN–all, all-allele tagged genome-edited line. (c) Western blot analysis of cell lysates immunoblotted for DNM2, GFP or actin. hDNM2X, stable overexpression DNM2–GFP cell line. (d) TIRF microscopy images of genome-edited hDNM2EN cell lines expressing DNM2–GFP. Scale bar, 10 μm. (e) Lifetime of DNM2–GFP at the plasma membrane in the indicated cell lines. Data are means ± s.e.m. Tracks, 14,726 or 29,681; n = 10 and 13 cells (hDNM2EN–1 and hDNM2EN–all, respectively). Asterisks indicate P < 0.0001. Not determined (ND) denotes an inability to perform lifetime analysis owing to the presence of stable, non-diffraction-limited dynamin structures. (f) Time course of Texas-Red-conjugated human transferrin uptake in SK-MEL-2 cell lines. Scale bar, 10 μm. Uncropped images of gels and blots are shown in Supplementary Fig. S2c.
Consistent with previous reports12,24, our attempts to generate stable overexpression hDNM2X cell lines by random transgene insertion resulted in cells that exhibited bright, irregular and highly stable dynamin structures. This made it impossible, as previously reported11–12,22–24, to implement a global and unbiased analysis of DNM2 plasma membrane dynamics in these lines. In striking contrast, the genome-edited lines exhibited highly dynamic DNM2–GFP localization, allowing us to globally quantify their lifetimes. The single-allele-tagged hDNM2EN–1 cell line exhibited significantly shorter average lifetimes than isogenic hDNM2EN–all cells: 24 ± 1.0 s versus 36 ± 2.1 s (P < 0.0001; Fig. 3e and Supplementary Fig. S4c, Table S3, and Movies S7 and S8). Furthermore, hDNM2X-overexpressing cells had diminished endocytic function when compared with the endogenously tagged cell lines (Fig. 3f). Thus, similarly to CLTA, DNM2 dynamics and function are highly sensitive to protein expression level and tagging.
Simultaneous tagging of CLTA and DNM2 reveals that mammalian CME is highly efficient
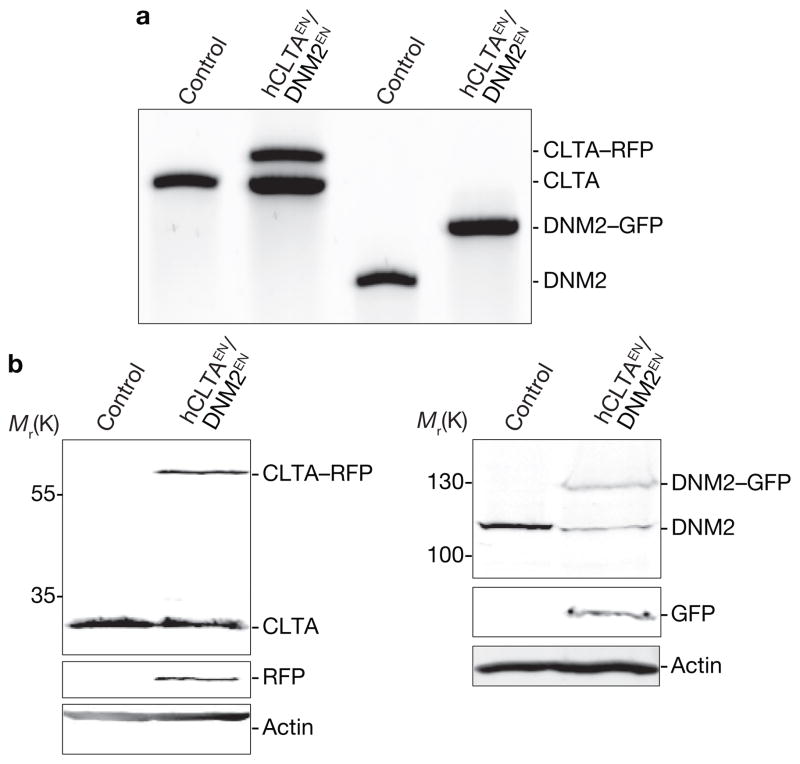
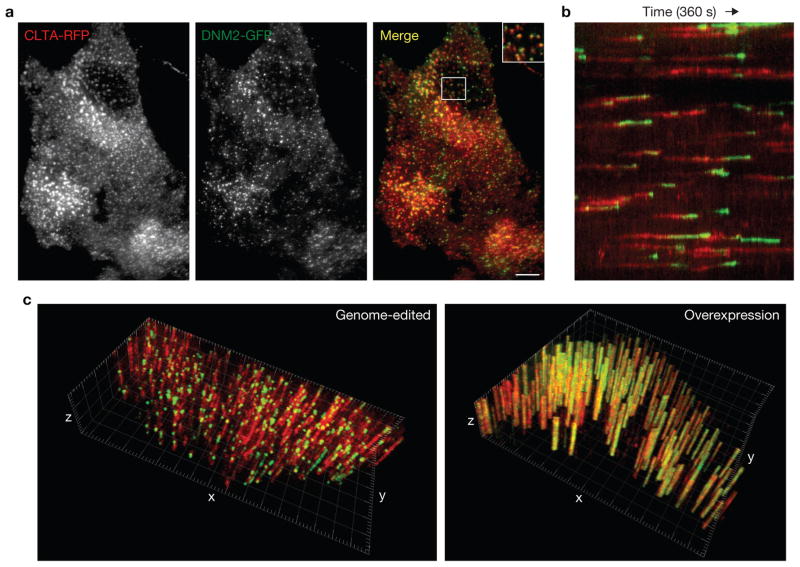
In budding yeast, direct genomic tagging of pairs of genes encoding endocytic proteins with different fluorescent tags has implicated > 60 proteins in CME and has allowed researchers to define, in detail, a very regular series of spatiotemporal events8. To generate mammalian cells possessing tagged forms of CLTA and DNM2 in a single cell, we co-delivered ZFNs and donors for both CLTA and DNM2 and used FACS to isolate a pool of double-marker-positive cells. We isolated a single-cell-derived clone with one copy of CLTA tagged with RFP and all copies of DNM2 tagged with GFP (hCLTAEN/DNM2EN; Fig. 4a, b, Table 1 and Supplementary Fig. S2d). To our knowledge, this represents the first example in human somatic cell genetics of a cell carrying tags at two distinct endogenous non-allelic loci. Similarly to the individually tagged cell lines (Supplementary Table S3), the measured lifetimes of CLTA and DNM2 in this dual-tagged cell line were 45 ± 1.0 s and 32 ± 3.3 s, respectively. As expected1, DNM2 was robustly recruited to clathrin-intense plasma membrane puncta in the final moments of CCP maturation (Fig. 5a, b and Supplementary Movie S9).
Figure 4.
Simultaneous editing of both CLTA and DNM2 using ZFNs in SK-MEL-2 cells. (a) Out-out PCR of a single-cell-derived clone showing targeted integration of RFP and GFP into the CLTA and DNM2 loci, respectively. Control, parental cell line; hCLTAEN/DNM2EN, single-allele tagged CLTA–RFP and all-allele tagged DNM2–GFP genome-edited line. (b) Western blot analysis of cell lysates immunoblotted for CLTA, DNM2, GFP, RFP or actin. Uncropped images of gels and blots are shown in Supplementary Fig. S2d.
Figure 5.
Fluorescence microscopy analysis of the hCLTAEN/DNM2EN cell line. (a) TIRF microscopy image of the hCLTAEN/DNM2EN cell line expressing both CLTA–RFP and DNM2–GFP. The top right of merged image is a higher magnification view of indicated area. Scale bar, 10 μm. (b) Kymograph analysis of CLTA–RFP and DNM2–GFP fluorescence over time. (c) Three-dimensional kymograph analysis of the genome-edited hCLTAEN/DNM2EN cell line (left), compared with a SK-MEL-2 cell transiently overexpressing CLTA–RFP and DNM2–GFP (right). x–y plane (2.5 μm2 grid), cell plasma membrane; z axis, time (240 s, 2 s per slice).
Clathrin-coated structures have been classified as either abortive, in which clathrin assembly does not lead to an endocytic event, or productive, in which clathrin assembly leads to scission and internalization. Previous efforts to define the percentage of productive clathrin-coated structures have relied on inference-based statistical analysis of population lifetimes in several overexpression cell lines, yielding CME efficiency estimates of 40% (refs 11,22,23). As dynamin is required for clathrin-coated vesicle scission at the plasma membrane, the hCLTAEN/DNM2EN line allowed us to monitor clathrin and dynamin dynamics simultaneously in a global and unbiased manner and to use dynamin as a marker to directly measure CCP productivity. To our surprise, 91% (n = 276 CCPs) of disappearing clathrin puncta in our genome-edited cells displayed bright peaks of dynamin recruitment, suggesting that most CME events are productive (Fig. 5c, left panel). By contrast, cells overexpressing both CLTA–RFP and DNM2–GFP displayed extensive, large stationary dynamin-containing clathrin-coated structures (Fig. 5c, right panel). This result suggests that CME efficiency in mammals and yeast is more similar than was previously appreciated.
DISCUSSION
Significant advances in our understanding of endocytosis have been made by generating cell lines carrying overexpressed transgenes encoding fluorescent protein fusions1,2,9–14,22–29. Although such studies have provided unique and powerful insights into understanding endocytic mechanisms and regulation, the impact of fusion protein overexpression on proper endocytic function was unclear. We found that the dynamics and function of both clathrin and dynamin in two different species (monkey and human) are significantly impaired by overexpression. We expect the same phenomenon to be true for many other proteins, including those that have multiple interaction partners and/or regulate rate-limiting steps in biosynthetic or signalling pathways.
In this study, we performed ZFN-mediated genome editing in mammalian cells to create precise, in-frame fluorescent protein fusions at the genomic loci of endocytic proteins. In yeast cells, where construction of precise protein fusions at genomic loci has long been routine for expression of fluorescent protein fusions at endogenous levels, endocytosis has been described as a very regular process. The predictable timing and ordering of endocytic protein recruitment in yeast have allowed for detailed descriptions of the timing of events in the endocytic pathway and for extensive mutant analysis of the contributions of individual proteins to CME dynamics8. By contrast, reports of variability in the size and dynamic behaviour of clathrin structures in mammalian cells have raised questions about the underlying biological significance of the different populations and about the regulatory mechanisms responsible for this heterogeneity. Furthermore, this heterogeneity has limited the ability to perform a global analysis of protein dynamics to detect the phenotypes that are caused when endocytic protein functions are impaired. Our data suggest that conventional overexpression-based methods in mammalian cells contribute to heterogeneity in the size and dynamics of clathrin-coated structures, thus warranting the use of genome-edited cells for re-examination of the effects of cargo, cell physiology and function-perturbing manipulations on endocytic dynamics and productivity.
ZFN-mediated genome editing has several advantages over conventional methods for generating cell lines that stably express fluorescent fusion proteins. First, conventional methods require selection of a single splice variant for study and, consequently, dominant expression of a single isoform. Moreover, these methods result in transgene overexpression, leading to total protein levels (comprising both tagged and untagged forms) that are significantly higher than in wild-type cells. Although transient overexpression methods can generate cells that express a fusion protein closer to physiological levels, they often result in heterogeneous cell-to-cell protein expression levels, necessitating experimental bias in the selection of ‘low-expression’ cells for study. By contrast, a ZFN-mediated strategy allows for creation of single- or multiple-allele tagged cell lines for greater control of a tagged protein’s expression level. Furthermore, our tagging strategy preserves native RNA splicing, enabling the analysis of protein isoforms at physiological stoichiometry and levels. Although such an approach precludes an analysis of individual splice variants, alternative ZFNs and donors could be readily designed to achieve exclusive expression of a desired isoform at endogenous levels.
Although the need to perturb a biological process to observe it is a general shortcoming of experimental research, we believe that our studies strike a balance between modifying a cell to faithfully report on its biology and retaining its natural state. In particular, the ability to engineer isogenic human cells using ZFN technology21,30,31 in a manner that has been the gold standard in budding and fission yeast for decades has the potential to transform the way that researchers study dynamic processes.
METHODS
ZFN design, screening and characterization
ZFNs designed to cleave precisely at or in the immediate vicinity of the stop codons of CLTA and DNM2 were assembled using an archive of pre-validated zinc finger modules32–34. All ZFNs carried enhanced obligate heterodimer (eHF) alleles of the FokI endonuclease35: Q486E/I499L/N496D and E490K/I538K/H537R. ZFNs were pre-screened for activity in K562 and SK-MEL-2 cells using Surveyor nuclease (Cel-1, Transgenomic)33 to detect insertions and deletions of DNA segments (indels). Briefly, Accuprime Taq HiFi (Invitrogen) was used with the Cel-1 primers listed in Supplementary Table S4 to amplify a short region surrounding the ZFN-target site in a 28-cycle PCR. Resulting amplicons were denatured, slowly re-annealed, and treated with Cel-1 for 20 min at 42 °C, then separated on 10% criterion gels (Bio-Rad) and visualized by ethidium-bromide staining.
Out-out PCR to detect targeted integration at CLTA and DNM2
PCR was used to screen BSC-1 and SK-MEL-2 clones that were RFP- (CLTA) and/or GFP- (DNM2) positive after FACS enrichment. For the initial pre-screening of clones, 2 μl of genomic DNA prepared using QuickExtract (Epicentre Biotechnologies) was amplified in a 35-cycle Accuprime Taq HiFi (Invitrogen) PCR with the TI primers listed in Supplementary Table S4. An extension time of 10 min was used to minimize bias against the tagged allele(s) of the targeted gene. PCR products were analysed on 5% criterion gels (Bio-Rad) that were run for 3–4 h at 150 V and visualized with ethidium bromide. Positive clones were then re-analysed independently using genomic DNA prepared with the Masterpure kit (Epicentre Biotechnologies) in 30-cycle PCRs with 100 ng template DNA. The resulting amplicons were analysed as described above. Clones suitable for further analysis were identified by cloning the out-out PCR products into pCR2.1 TOPO (Invitrogen) and conventional sequencing.
Cell culture
SK-MEL-2 cells and BSC-1 cells were purchased from the ATCC. BSC-1 GFP–LCa (rCLTAX) cells were provided by T. Kirchhausen (Harvard Medical School, USA)12. All cells were maintained under 5% CO2 at 37 °C in DMEM/F-12 (Invitrogen) supplemented with 10% FBS (fetal bovine serum; HyClone), except for BSC-1 GFP–CLTA cells, for which the media was supplemented with 0.5 mg ml−1 G418 (Invitrogen) for selection.
Plasmid construction
Human clathrin light chain A (CLTA) was amplified by PCR from a plasmid that was provided by L. Greene (NIH, USA)36. This CLTA sequence is identical to the CLTA expressed in SK-MEL-2 cells. The African green monkey CLTA gene was amplified by reverse transcription PCR (RT–PCR) from poly-A mRNA of BSC-1 cells and subcloned into a pEGFP-N1 vector (Clontech) in which GFP was replaced with TagRFP-T (RFP)37. TagRFP-T was a gift from R. Tsien (UC, San Diego, USA). A GTSGGS linker was placed between CLTA and RFP. Dynamin-2 (DNM2) was amplified by RT–PCR from poly-A mRNA of SK-MEL-2 cells and subcloned into a pEGFP-N1 vector with a GTSGGS linker between DNM2 and GFP.
Donor Design
Approximately 1.5 kb of genomic DNA sequence surrounding the CLTA (monkey or human for BSC-1 or SK-MEL-2, respectively) and DNM2 stop codons was amplified using the ‘HA’ primers listed in Supplementary Table S4, and cloned into pCR8-TOPO (Invitrogen). Next, a unique KpnI site was introduced through Quikchange (Agilent Technologies) site-directed mutagenesis with the ‘QC’ primers. Finally, RFP or GFP was amplified using the ‘KpnI’ primers, which encode an amino-terminal GTSGGS linker, and cloned into the KpnI site of the CLTA or DNM2 homology arm constructs, respectively.
Generation of stable overexpression cell lines
Human CLTA–RFP and DNM2–GFP plasmids were transfected into SK-MEL-2 cells using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. After 72 h, transfected SK-MEL-2 cells were selected in DMEM/F-12 containing 10% FBS and 1 mg ml−1 G-418. African green monkey CLTA–RFP plasmid was transfected into BSC-1 cells using Fugene 6 (Roche), following a standard protocol. After 72 h, transfected BSC-1 cells were selected in DMEM/F-12 containing 10% FBS and 0.4 mg ml−1 G418 (Geneticin). Single-cell clones were selected and amplified by dilution cloning in 96-well plates.
Generation of genome-edited cell lines
ZFN and donor plasmids were transfected into cells using a single cuvette Amaxa Nucleofector device (Lonza), according to manufacturer’s instructions. In brief, cells grown to ~80% confluency were harvested by trypsinization, and 1–1.5 × 106 BSC-1 or SK-MEL-2 cells were resuspended in Nucleofector solution L or R and transfected using Nucleofector program A-020 or T-020, respectively. After transfection, cells were subjected to cold-shock treatment38 by incubation at 30 °C for 48 h before being transferred to 37 °C, 5% CO2. Depending on the experiment, recovered cells were sorted for RFP-positive, GFP-positive or RFP- and GFP-positive signals using a DAKO-Cytomation MoFlo High Speed Sorter, either directly into 96-well plates, or followed by growth for 1 week and cloning by limiting dilution.
Western immunoblotting
For the preparation of lysates, cells were briefly washed in PBS, incubated with 0.5 mM EDTA for 5 min, and pelleted at 110g for 2 min. The supernatant was subsequently removed, and hot 2 × protein sample buffer (125 mM Tris–HCl at pH 6.8, 10% glycerol, 10% SDS, 130 mM dithiothreitol; DTT, 0.05% bromophenol blue and12.5% β-mercaptoethanol) was immediately added. After pellet resuspension by pipetting, samples were heated to 95 °C, for 3 min, separated by 8% or 12% SDS–PAGE, and transferred to Immobilon-FL PVDF transfer membrane (IPFL-00010, Millipore) or nitrocellulose membrane in transfer buffer (25 mM Trizma base, 200 mM glycine, 20% methanol and 0.025% SDS) at 50 V and 4 °C, for 1 h. The membrane was subsequently rinsed in TBS (Tris-buffered saline) and incubated in Odyssey blocking buffer (diluted 1:1 in PBS) at room temperature for 1 h. Primary antibodies were diluted in blocking buffer and incubated with the membrane at 4 °C overnight. The membrane was incubated in the following primary antibodies: anti-CLTA (H-55; 1:100; Santa Cruz Biotechnology), anti-DNM2 (C-18; 1:1000; Santa Cruz Biotechnology), anti-actin (C4; 1:20,000; MP Biomedicals), anti-GFP (B2; 1:2000; Santa Cruz Biotechnology) or anti-tRFP (AB233; 1:500; Evrogen). Blots were subsequently incubated in the dark for 1 h at room temperature with secondary IRDye 680 or 800CW antibodies (LI-COR Biosciences) diluted to 1:5000 in Odyssey blocking buffer/PBS solution containing 0.1% Tween-20. After washing with TBST (Tris-Buffered Saline Tween-20), the membrane was incubated in TBS and scanned on an Odyssey infrared imager (LI-COR Biosciences). Quantification of protein bands was performed using the Odyssey Infrared Imaging System (version 3.0).
Total internal reflection fluorescence (TIRF) microscopy and live-cell imaging
TIRF microscopy images were captured using Metamorph software on an Olympus IX-81 microscope using a ×60/NA 1.49 objective and an ORCA-R2 camera (Hamamatsu). The system was maintained at 37 °C using a WeatherStation chamber and temperature controller (PrecisionControl). At 16–24 h before imaging, cells were seeded onto uncoated glass coverslips in growth medium. During imaging, cells were maintained in DMEM without phenol red that was supplemented with 5% FBS and 10 mM HEPES. A 488 nm solid-state laser (Melles Griot) and a 561 nm diode-pumped solid-state laser (Melles Griot) were used to excite GFP and RFP fluorophores, respectively. For lifetime analysis, images were acquired every 2 s for 6 min, with an exposure time of 900 ms. Simultaneous two-colour TIRF images were obtained using a DV2 image splitter (MAG Biosystems) to separate GFP and RFP emission signals.
Transferrin uptake assay
Cells were grown on glass coverslips in 6-well plates overnight. The cells were serum-starved at 37 °C for 1 h in starving medium (DMEM containing 20 mM HEPES at pH 7.4 and 5 mg ml−1 BSA; bovine serum albumin). They were then incubated in starving medium containing 25 mg ml−1 Alexa Fluor 488- or Texas Red-conjugated human transferrin (Invitrogen) at 37 °C for 0–45 min before fixation in 4% paraformaldehyde at room temperature for 20 min. After three washes with PBS, the coverslips were mounted onto glass slides using ProLong Gold Antifade Reagent (Invitrogen). The degree of transferrin uptake was quantified using ImageJ software (NIH, V1.44e). In brief, cell outlines were traced by using polygon or freehand selection tools, and the mean fluorescence intensity (arbitrary units) was measured for individual cells.
Immunofluorescence staining
Cells were grown on glass coverslips in 6-well plates overnight before fixation in 4% paraformaldehyde at room temperature for 20 min. After three washes with PBS, the coverslips were quenched with 1 mg ml−1 NaBH4 for 15 min twice, and the cells were permeabilized in 0.1% saponin/PBS or 0.1% Triton X-100/PBS. After brief washing with PBS, the coverslips were incubated with anti-clathrin HC antibody (MA1-065, Thermo Scientific) at 1:500 overnight at 4 °C. The coverslips were then washed with PBS for 15 min before incubation with anti-mouse secondary antibody conjugated to Alexa Fluor 488 (A21202, Invitrogen) or Alexa Fluor 568 (A10037, Invitrogen) at 1:1000 dilution for 2 h at room temperature. After three washes with PBS, the coverslips were mounted on glass slides using ProLong Gold Antifade Reagent (Invitrogen).
Particle detection, tracking and image analysis
Particle detection and tracking were performed using Imaris 7.1 software (Bitplane Scientific). Before particle detection, images were first subjected to background subtraction. The Spots module of Imaris was then used to automatically detect point-like particles with an estimated spot diameter of 500 nm (~5 pixels). After automatic detection, detected spots were filtered based on satisfying minimum ‘intensity standard deviation’ and ‘quality’ control parameters. Only spots with values higher than the set threshold values were considered for analyses. Quality is defined as the intensity at the centre of the spot, Gaussian filtered by ¾ of the spot radius. Appropriate threshold values were confirmed by visual inspection of correct particle detection. To trace objects through sequential frames of time data, a Brownian motion particle-tracking algorithm was applied. A maximum search distance of 350 nm was defined to disallow connections between a spot and a candidate match if the distance between the predicted future position of the spot and the candidate position exceeded the maximum distance. A gap-closing algorithm was also implemented to link track segment ends to track segment starts to recover tracks that were interrupted by the temporary disappearance of particles. The maximum permissible gap length was set equal to 7 frames. Track outputs were then visually inspected and, as necessary, edited to correct for tracking errors. Only tracks that appeared and disappeared during the lifetime of the acquisition (that is, 181 frames, 6 min) were subjected to lifetime analyses.
Statistical analysis
One-way ANOVAs were used to assess the effect of genome editing on the mean lifetime of endocytic proteins at the plasma membrane. Significant results were followed by post-hoc Tukey–Kramer HSD (honestly significant difference) tests. All tests were performed using JMP 8.0.2 (SAS Institute). Data that had undergone statistical analysis are presented as means ± s.e.m.
Supplementary Material
Figure S1 Genotypic analysis of genome-edited cell lines. (a) Representative editing efficiency using FACS enrichment. RFP-positive (i) BSC-1 or (ii and iii) SK-MEL-2 cells were enriched by FACS and individual clones were selected from colonies in 96-well plates. After ~4 weeks, clones were analyzed by out-out PCR for CLTA and DNM2 using the oligos listed in Supplementary Information, Table S4 as “TI.” (i) 25 randomly chosen clones of the total 26, or (ii) 50 randomly chosen clones of the total 73 that were genotyped are shown. (iii) GFP-positive SK-MEL-2 clones were analyzed by out-out PCR for DNM2. (b) Stability of ZFN-modified BSC-1 and SK-MEL-2 cells. (i) BSC-1 cells were transfected with CLTA ZFNs, harvested at the indicated days post transfection, and processed for PCR and the Cel-1 cleavage assay using primers as described in Supplementary Information, Table S4. (ii to iv) SK-MEL-2 cells were transfected with (ii) CLTA ZFNs, (iii) DNM2 ZFNs, or (iv) both (top panel, CLTA; lower panel, DNM2), and processed similarly. GFP, ZFN transfection control. Arrows indicate the expected position of the Cel-1 cleavage products. % indels was calculated using the fraction of the cleaved signal. (c and d) Representative sequence genotyping for two genome-edited clones. (c) BSC-1 CLTA-RFP clone mkCLTAEN was processed by out-out PCR (as in Fig. 1b) and the resulting amplicons were cloned into pCR2.1. 24 colonies were sequenced using CLTA-SQ-R (Supplementary Information, Table S4). Of the 23 clones that produced legible sequence, 17 perfectly matched WT CLTA near the ZFN cleavage site, and six matched the expected sequence of the RFP donor. “Stop” denotes the position of the endogenous or donor-provided stop codons. An asterisk (*) marks the location of SNPs in the ZFN-binding site of the donor. Red shading indicates the RFP coding sequence provided by the donor. (d) SK-MEL-2 DNM2-GFP clone hDNM2EN-1 was processed as in (c). 24 colonies were sequenced using DNM2-SQ-R (Supplementary Information, Table S4). Of the 22 clones that produced legible sequence, 19 perfectly matched WT DNM2 near the ZFN cleavage and donor insertion sites, and three matched the expected sequence of the GFP donor (note that the transgenic allele is a less efficient substrate both for cloning and for amplification). “Stop” denotes the position of the endogenous or donor-provided stop codons. Green shading indicates the GFP coding sequence provided by the donor.
Figure S2 Western immunoblot analysis of genome-edited cell lines. Lysates of (a) BSC-1 or (b to d) SK-MEL-2 cells were immunoblotted against CLTA, DNM2, RFP, GFP, or actin. Control, parental cell line. (a) BSC-1 cell lines: mkCLTAEN, single-allele CLTA-RFP tagged genome-edited line; mkCLTAX, stable CLTA-RFP overexpression line; rCLTAX, stable GFP-CLTA (rat brain-derived) overexpression line. (b) SK-MEL-2 CLTA-RFP cell lines: hCLTAEN-1, single-allele CLTA-RFP tagged genome-edited line; hCLTAEN-all, all-allele tagged genome-edited line; stable CLTA-RFP overexpression line. (c) SK-MEL-2 DNM2-GFP cell lines: hDNM2EN-1, single-allele tagged genome-edited line; hDNM2EN-all, all-allele tagged genome-edited line; hDNM2X, stable overexpression DNM2-EGFP cell line. (d) SK-MEL-2 hCLTAEN/hDNM2EN, single-allele tagged CLTA-RFP and all-allele tagged DNM2-GFP genome-edited line. Note RFP antibody cross-reactivity with GFP.
Figure S3 Fluorescence microscopy analysis of genome-edited cell lines. (a) Epifluorescence images of BSC-1 cell lines. mkCLTAEN, single-allele tagged CLTA-RFP genome-edited cell line; mkCLTAX, stable CLTA-RFP (monkey-derived) overexpression cell line; rCLTAX, stable GFP-CLTA (rat brain-derived) overexpression cell line. Scale bar, 10 μm. (b and c) Immunostaining analysis of genome edited cell lines. Fixed (b) BSC-1 mkCLTAEN and (c) SK-MEL-2 hCLTAEN-all cell lines were stained for clathrin heavy chain (CHC) and visualized by epi-fluorescence and TIRF microscopy, respectively. Scale bar, 10 μm.
Figure S4 Endocytic protein lifetime distribution of genome-edited cell lines. Lifetime distribution for endocytic proteins was determined by the elapsed time between the appearance and disappearance of fluorescent structures present in the time series (6 min) from TIRF microscopy time-lapse videos of (a) BSC-1, (b) SK-MEL-2 CLTA-RFP and (c) SK-MEL-2 DNM2-GFP cell lines. Exposure time: 900 ms; Acquisition: 2 s/frame.
Table S1 Summary of genome-editing statistics. Top panel: The number of genotyped clones is given for each target in each cell type, along with the number (percentage) of correctly tagged clones having at least one (but less than all) allele tagged, or all alleles tagged. Clones that had smaller, larger, or unexpected combinations of out-out PCR products are classified as “Imprecise or Ambiguous”. Note that in all cases, cells were enriched by FACS for the cognate fluorescent marker prior to limiting dilution and genotyping. Lower panel: Summary of genotypes for genome-edited clones. Each clone was processed by out-out PCR (as in Figs. 1b, 2a, 3b, and 4a), and the number of topo clones representing tagged or untagged alleles is given. The genotype of these alleles is shown in parenthesis. An asterisk (*) denotes alleles containing the indicated short insertion/deletion (number in parentheses) at the predicted cleavage site for DNM2 ZFN pair, which is located 43 bp downstream of the DNM2 stop codon in the 3′ UTR.
Table S2 ZFN target sites and designed zinc finger helix sequences (top panel). Bases in lower-case are skipped from a DNA recognition perspective by the zinc finger proteins (ZFPs). The amino acid sequences of the ZFPs are in the bottom panel, with the recognition alpha-helices underlined.
Table S3 Lifetime analysis of endocytic proteins in various cell lines.
Table S4 Sequences of oligos used in the PCR, Cel-1, and sequencing reactions.
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 cell stably overexpressing rat brain-derived CLTA-GFP (rCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 single-allele tagged genome-edited cell expressing monkey-derived CLTA-RFP (mkCLTAEN). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 cell stably overexpressing monkey-derived CLTA-RFP (mkCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele tagged genome-edited cell expressing human-derived CLTA-RFP (hCLTAEN-1). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 cell stably overexpressing human-derived CLTA-RFP (hCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 all-allele tagged genome-edited cell expressing human-derived CLTA-RFP (hCLTAEN-all). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele tagged genome-edited cell (hDNM2EN-1) expressing human-derived DNM2-GFP. Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 all-allele tagged genome-edited cell (hDNM2EN-all) expressing human-derived DNM2-GFP. Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele CLTA-RFP and all-allele DNM2-GFP tagged genome-edited cell (hCLTAEN/DNM2EN). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 6.7 MB)
Acknowledgments
We thank H. Nolla and A. Valeros for their cell-sorting expertise, A. Fischer and M. Yasukawa for help with cell culture, the Biological Imaging Facility for use of the Imaris software, the Sangamo Production group for technical assistance and members of the Drubin/Barnes lab for critical reading of this manuscript. We also thank T. Kirchhausen, L. Greene, and R. Tsien for providing the BSC-1 GFP–CLTA cell line, human CLTA plasmid, and TagRFP-T plasmid, respectively. J.B.D. and J.C. were supported by postdoctoral fellowships from the Jane Coffin Childs Memorial Fund and The Croucher Foundation, respectively. This work was supported by NIH grant R01 GM65462 to D.G.D.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
AUTHOR CONTRIBUTIONS
J.B.D, B.Z., J.C., A.T.C., T.D.V., Y.D., J.C.M., D.E.P., L.Z., E.J.R., P.D.G., F.D.U. and D.G.D designed the study and experiments. J.B.D, B.Z., J.C., A.T.C., J.M.C., Y.S. and A.H.L. performed the experiments. J.B.D., B.Z., J.C., A.T.C., J.M.C. and F.D.U. analysed the data. J.B.D., B.Z., J.C., A.T.C., F.D.U. and D.G.D. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
B.Z., J.M.C., Y.S., A.H.L., T.D.V., Y.D., J.C.M., D.E.P., L.Z., E.J.R., P.D.G. and F.D.U are full-time employees of Sangamo BioSciences, Incorporated.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 2.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein JL, Brown MS. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu Rev Biochem. 1977;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- 5.Zuchner S, et al. Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet. 2005;37:289–294. doi: 10.1038/ng1514. [DOI] [PubMed] [Google Scholar]
- 6.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 8.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 9.Rappoport JZ, Kemal S, Benmerah A, Simon SM. Dynamics of clathrin and adaptor proteins during endocytosis. Am J Physiol. 2006;291:C1072–C1081. doi: 10.1152/ajpcell.00160.2006. [DOI] [PubMed] [Google Scholar]
- 10.Saffarian S, Cocucci E, Kirchhausen T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. Plos Biol. 2009;7:e1000191. doi: 10.1371/journal.pbio.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loerke D, et al. Cargo and dynamin regulate clathrin-coated pit maturation. Plos Biol. 2009;7:628–639. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur J Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- 14.Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoops L, Hornakova T, Royer Y, Constantinescu SN, Renauld JC. JAK kinases overexpression promotes in vitro cell transformation. Oncogene. 2008;27:1511–1519. doi: 10.1038/sj.onc.1210800. [DOI] [PubMed] [Google Scholar]
- 16.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 17.Luo T, Matsuo-Takasaki M, Sargent TD. Distinct roles for Distal-less genes Dlx3 and Dlx5 in regulating ectodermal development in Xenopus. Mol Reprod Dev. 2001;60:331–337. doi: 10.1002/mrd.1095. [DOI] [PubMed] [Google Scholar]
- 18.Miyama K, et al. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev Biol. 1999;208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 19.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 20.Jensen TJ, et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 21.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 22.Mettlen M, Loerke D, Yarar D, Danuser G, Schmid SL. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J Cell Biol. 2010;188:919–933. doi: 10.1083/jcb.200908078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettlen M, et al. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol Biol Cell. 2009;20:3251–3260. doi: 10.1091/mbc.E09-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knockout cells. Mol Biol Cell. 2008;19:5347–5359. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang F, Khvorova A, Marshall W, Sorking A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 26.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchhausen T. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Clainche C, et al. A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis. EMBO J. 2007;26:1199–1210. doi: 10.1038/sj.emboj.7601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 30.DeKelver RC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promotor. Nat Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 34.DeKelver RC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, et al. Clathrin exchange during clathrin-mediated endocytosis. J Cell Biol. 2001;155:291–300. doi: 10.1083/jcb.200104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyon Y, et al. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Genotypic analysis of genome-edited cell lines. (a) Representative editing efficiency using FACS enrichment. RFP-positive (i) BSC-1 or (ii and iii) SK-MEL-2 cells were enriched by FACS and individual clones were selected from colonies in 96-well plates. After ~4 weeks, clones were analyzed by out-out PCR for CLTA and DNM2 using the oligos listed in Supplementary Information, Table S4 as “TI.” (i) 25 randomly chosen clones of the total 26, or (ii) 50 randomly chosen clones of the total 73 that were genotyped are shown. (iii) GFP-positive SK-MEL-2 clones were analyzed by out-out PCR for DNM2. (b) Stability of ZFN-modified BSC-1 and SK-MEL-2 cells. (i) BSC-1 cells were transfected with CLTA ZFNs, harvested at the indicated days post transfection, and processed for PCR and the Cel-1 cleavage assay using primers as described in Supplementary Information, Table S4. (ii to iv) SK-MEL-2 cells were transfected with (ii) CLTA ZFNs, (iii) DNM2 ZFNs, or (iv) both (top panel, CLTA; lower panel, DNM2), and processed similarly. GFP, ZFN transfection control. Arrows indicate the expected position of the Cel-1 cleavage products. % indels was calculated using the fraction of the cleaved signal. (c and d) Representative sequence genotyping for two genome-edited clones. (c) BSC-1 CLTA-RFP clone mkCLTAEN was processed by out-out PCR (as in Fig. 1b) and the resulting amplicons were cloned into pCR2.1. 24 colonies were sequenced using CLTA-SQ-R (Supplementary Information, Table S4). Of the 23 clones that produced legible sequence, 17 perfectly matched WT CLTA near the ZFN cleavage site, and six matched the expected sequence of the RFP donor. “Stop” denotes the position of the endogenous or donor-provided stop codons. An asterisk (*) marks the location of SNPs in the ZFN-binding site of the donor. Red shading indicates the RFP coding sequence provided by the donor. (d) SK-MEL-2 DNM2-GFP clone hDNM2EN-1 was processed as in (c). 24 colonies were sequenced using DNM2-SQ-R (Supplementary Information, Table S4). Of the 22 clones that produced legible sequence, 19 perfectly matched WT DNM2 near the ZFN cleavage and donor insertion sites, and three matched the expected sequence of the GFP donor (note that the transgenic allele is a less efficient substrate both for cloning and for amplification). “Stop” denotes the position of the endogenous or donor-provided stop codons. Green shading indicates the GFP coding sequence provided by the donor.
Figure S2 Western immunoblot analysis of genome-edited cell lines. Lysates of (a) BSC-1 or (b to d) SK-MEL-2 cells were immunoblotted against CLTA, DNM2, RFP, GFP, or actin. Control, parental cell line. (a) BSC-1 cell lines: mkCLTAEN, single-allele CLTA-RFP tagged genome-edited line; mkCLTAX, stable CLTA-RFP overexpression line; rCLTAX, stable GFP-CLTA (rat brain-derived) overexpression line. (b) SK-MEL-2 CLTA-RFP cell lines: hCLTAEN-1, single-allele CLTA-RFP tagged genome-edited line; hCLTAEN-all, all-allele tagged genome-edited line; stable CLTA-RFP overexpression line. (c) SK-MEL-2 DNM2-GFP cell lines: hDNM2EN-1, single-allele tagged genome-edited line; hDNM2EN-all, all-allele tagged genome-edited line; hDNM2X, stable overexpression DNM2-EGFP cell line. (d) SK-MEL-2 hCLTAEN/hDNM2EN, single-allele tagged CLTA-RFP and all-allele tagged DNM2-GFP genome-edited line. Note RFP antibody cross-reactivity with GFP.
Figure S3 Fluorescence microscopy analysis of genome-edited cell lines. (a) Epifluorescence images of BSC-1 cell lines. mkCLTAEN, single-allele tagged CLTA-RFP genome-edited cell line; mkCLTAX, stable CLTA-RFP (monkey-derived) overexpression cell line; rCLTAX, stable GFP-CLTA (rat brain-derived) overexpression cell line. Scale bar, 10 μm. (b and c) Immunostaining analysis of genome edited cell lines. Fixed (b) BSC-1 mkCLTAEN and (c) SK-MEL-2 hCLTAEN-all cell lines were stained for clathrin heavy chain (CHC) and visualized by epi-fluorescence and TIRF microscopy, respectively. Scale bar, 10 μm.
Figure S4 Endocytic protein lifetime distribution of genome-edited cell lines. Lifetime distribution for endocytic proteins was determined by the elapsed time between the appearance and disappearance of fluorescent structures present in the time series (6 min) from TIRF microscopy time-lapse videos of (a) BSC-1, (b) SK-MEL-2 CLTA-RFP and (c) SK-MEL-2 DNM2-GFP cell lines. Exposure time: 900 ms; Acquisition: 2 s/frame.
Table S1 Summary of genome-editing statistics. Top panel: The number of genotyped clones is given for each target in each cell type, along with the number (percentage) of correctly tagged clones having at least one (but less than all) allele tagged, or all alleles tagged. Clones that had smaller, larger, or unexpected combinations of out-out PCR products are classified as “Imprecise or Ambiguous”. Note that in all cases, cells were enriched by FACS for the cognate fluorescent marker prior to limiting dilution and genotyping. Lower panel: Summary of genotypes for genome-edited clones. Each clone was processed by out-out PCR (as in Figs. 1b, 2a, 3b, and 4a), and the number of topo clones representing tagged or untagged alleles is given. The genotype of these alleles is shown in parenthesis. An asterisk (*) denotes alleles containing the indicated short insertion/deletion (number in parentheses) at the predicted cleavage site for DNM2 ZFN pair, which is located 43 bp downstream of the DNM2 stop codon in the 3′ UTR.
Table S2 ZFN target sites and designed zinc finger helix sequences (top panel). Bases in lower-case are skipped from a DNA recognition perspective by the zinc finger proteins (ZFPs). The amino acid sequences of the ZFPs are in the bottom panel, with the recognition alpha-helices underlined.
Table S3 Lifetime analysis of endocytic proteins in various cell lines.
Table S4 Sequences of oligos used in the PCR, Cel-1, and sequencing reactions.
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 cell stably overexpressing rat brain-derived CLTA-GFP (rCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 single-allele tagged genome-edited cell expressing monkey-derived CLTA-RFP (mkCLTAEN). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative BSC-1 cell stably overexpressing monkey-derived CLTA-RFP (mkCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele tagged genome-edited cell expressing human-derived CLTA-RFP (hCLTAEN-1). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 cell stably overexpressing human-derived CLTA-RFP (hCLTAx). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 all-allele tagged genome-edited cell expressing human-derived CLTA-RFP (hCLTAEN-all). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele tagged genome-edited cell (hDNM2EN-1) expressing human-derived DNM2-GFP. Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 all-allele tagged genome-edited cell (hDNM2EN-all) expressing human-derived DNM2-GFP. Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 3.3 MB)
This movie shows a TIRF microscopy time-lapse video (6 min) of a representative SK-MEL-2 single-allele CLTA-RFP and all-allele DNM2-GFP tagged genome-edited cell (hCLTAEN/DNM2EN). Exposure time= 900 ms; Acquisition= 2 s/frame. (AVI, 6.7 MB)