Abstract
Background
We hypothesized that nitazoxanide (NTZ) added to pegylated inter-feron alfa-2a (PEG-IFN) and weight-based ribavirin (WBR) would improve hepatitis C virus (HCV) virologic responses in HCV treatment-naïve HIV-1/HCV genotype 1 coin-fected persons.
Methods
Prospective, single-arm study in which subjects received 4-week lead-in (NTZ 500 mg twice daily) followed by 48 weeks of NTZ, PEG-IFN, and WBR. We compared the HCV virologic responses of these subjects to historical controls from the completed ACTG study A5178 who received PEG-IFN and WBR and had similar subject characteristics. Primary endpoints were early virologic response and complete early virologic response (EVR and cEVR).
Results
Among 67 subjects (78% male; 48% Black; median age, 50 years), EVR was achieved in 65.7% (90% CI, 55.0%–75.3%), cEVR in 38.8% (28.8%–49.6%). and SVR in 32.8% (23.4%–43.5%). EVR was higher with NTZ (51.4% in A5178; P = .03), but the sustained virologic response (SVR) proportion was similar (27.3% in A5178; P = .24). In contrast to A5178, SVR was similar across IL28B genotypes. Overall, NTZ was safe and well-tolerated.
Conclusion
Whereas EVR proportion improved significantly in this pilot study, the addition of NTZ to PEG-IFN/WBR did not significantly improve SVR compared to historical controls. NTZ may be associated with an attenuation of the effect of IL28B on HCV treatment response.
Keywords: genotype 1, hepatitis C, HIV, nitazoxanide, pegylated interferon, ribavirin
Nitazoxanide (NTZ) has been shown to be a potent inhibitor of hepatitis C viral (HCV) replication in vitro, alone and in combina-tion.1 Clinical trials reported that the addition of NTZ to pegylated interferon alfa-2a (PEG-IFN) and ribavirin (RBV) increased sustained virologic response (SVR) proportions from 50% to 80% in genotype (GT)-4 HCV-infected Egyptian patients and from 32% to 44% in GT-1 HCV treatment-naïve subjects.2,3 These results prompted us to pursue NTZ for treatment of HCV in HIV-1/HCV genotype 1 (GT-1) coinfected subjects, particularly given the drug's minimal known drug interactions with antiretrovirals and favorable safety profile in HIV-1–infected patients treated for cryptosporidium.4
NTZ's putative mechanism of action is via intracellular upregulation of intrinsic antiviral responses within interferon-mediated pathways.5 Because NTZ sensitizes cells in vitro to the subsequent effect of interferon and acts in synergy with interferon,1 a treatment strategy was undertaken whereby an NTZ lead-in was followed by triple therapy with NTZ/PEG-IFN/RBV. The primary objective was to evaluate early virologic (EVR) and complete early virologic response (cEVR) proportions in HIV-1/HCV GT-1 coinfected subjects naïve to HCV therapy. SVR analysis was a secondary objective. Additional objectives were to explore associations between cellular IL28B alleles and HCV treatment response to assess whether NTZ would attenuate the negative effect of an unfavorable IL28B genotype status and to examine the association between insulin resistance and treatment response to NTZ-based therapy.
Methods
Study Design
The study was a prospective single-arm trial (National Institutes of Health Registration number NCT00991289) designed to evaluate NTZ added to PEG-IFN and weight-based ribavirin (WBR) at 1,000 mg per day for <75 kg and 1,200 mg per day for ≥75 kg. A completed AIDS Clinical Trials Group (ACTG) study (A5178) that was restricted to HCV treatment-naïve GT-1 subjects with HIV-1 coinfection provided historical controls on treatment with PEG-IFN and WBR alone.6 All eligible subjects received a 4-week lead-in course of oral NTZ 500 mg 2 times per day followed by 48 weeks of triple therapy with NTZ, PEG-IFN-alfa 2a 180 mcg subcutaneously per week, and WBR administered orally 2 times per day.
Study evaluations (safety and/or virologic) were performed every 4 weeks through the end of the treatment regimen at week 52 and at weeks 64 and 76. HCV RNA levels were obtained at baseline, every 4 weeks until week 16, and weeks 28, 40, 52, 64, and 76 for those who remained on therapy through week 52. For those who discontinued medication prematurely but did not meet virologic failure criteria, HCV RNA levels were obtained at baseline, every 4 weeks until week 16, and at 24 weeks following therapy. Virologic failures were defined as subjects who did not achieve at least a 2 log10 drop in HCV RNA level or undetectable HCV viral load (ie, <43 HCV RNA IU/mL) at 16 weeks of treatment (12 weeks of triple therapy) or in whom HCV viral load was detectable at week 28 (week 24 of triple therapy). Virologic failures discontinued study treatment, but evaluations continued for 4 weeks.
Adverse events (AEs) were graded according to the DAIDS criteria for the assessment of severity.7 Grade 2 and higher AEs and any AEs that led to changes in study treatment were collected. Subjects with anemia underwent RBV dose reductions, and those with neutropenia or thrombocytopenia underwent PEG-IFN dose reductions. The use of erythropoietin and granulocyte-colony stimulating factor was allowed at the discretion of the site investigators. Criteria for discontinuation included protocol-defined toxicity of severe thrombocytopenia, refractory anemia, neutropenia, and other clinical events thought to be life-threatening.
Historical controls were subjects from A5178 who were HCV GT-1 and naïve to HCV therapy; they were similar to the A5269 study population in distributions of sex, race/ethnicity, age, and baseline HCV RNA profiles. In A5178, subjects who achieved EVR (at least 2 log10 drop or undetectable HCV RNA level) at week 12 received a 72-week course of PEG-IFN-alfa-2a and WBR. After completion of treatment, subjects were monitored for 24 additional weeks to determine SVR. Subjects who failed to achieve EVR were considered non-SVR and were randomized to discontinue treatment or continue on PEG-IFN alone for 72 weeks in a subsequent study step. HCV RNA was determined using Roche Cobas Amplicor HCV Monitor with a lower limit of quantification (LLQ) of 600 IU/mL for determining EVR and with a lower limit of detection of 60 IU/mL for subsequent qualitative determinations of treatment response including SVR. Rapid virologic response (RVR) data at week 4 were not collected in A5178.
Study Population
Subjects were at least 18 years of age and coinfected with HCV GT-1 and HIV-1. Subjects were HCV treatment-naïve and without decompensated liver disease, active depression, or alcohol use. Liver biopsy was not required. Subjects not on HIV-1 antiretroviral therapy (ART) or on a stable ART regimen for at least 60 days prior to enrollment were eligible. Use of didanosine was prohibited. HIV-1 infection was documented by a positive ELISA assay confirmed by Western blot, HIV-1 culture, HIV-1 antigen, or HIV-1 RNA. All subjects were required to have >200 CD4+ T-lymphocytes/mm3.
The clinical protocol was reviewed and approved by National Institute of Allergy and Infectious Diseases, US Food and Drug Administration, and local institutional review boards at all clinical sites. All subjects provided informed consent. The study was monitored by an independent ACTG Study Monitoring Committee.
Virologic Testing
HCV RNA assay and virologic response definitions
HCV RNA was determined using the Roche TaqMan HCV, v1.0 (Roche Molecular Systems, Pleasanton, CA) with LLQ of 43 IU/mL.8 RVR was defined as undetectable HCV viral load at week 8 of the study (week 4 on triple therapy). EVR was defined as at least 2 log10 drop in HCV RNA from baseline or undetectable HCV RNA (<43 IU/mL) at week 16 (week 12 of triple therapy). cEVR was defined as undetectable HCV RNA at week 16, end of treatment response (ETR) as undetectable HCV RNA at the end of treatment, and SVR as undetectable HCV RNA at least 24 weeks after end of treatment.
HCV genotype
HCV GT-1 documentation status was obtained for historical controls; if it was not available, it was tested centrally (HCV TRUGENE HCV 5′NC, version 4.0, research use only [RUO]; Siemens Healthcare Diagnostics Inc., Tarrytown, NY). HCV GT-1 status was confirmed for all enrolled subjects on banked study entry specimens by the VERSANT HCV Genotype assay (version 2.0 line probe assay, RUO; Siemens Healthcare Diagnostics Inc., Tarrytown, NY).
HIV-1 RNA assays
HIV-1 RNA was measured at local sites at entry and weeks 4, 8, and 16 (plus weeks 28, 40, and 52 among those remaining on study). The assays used were Roche COBAS AmpliPrep/Taqman HIV-1 (LLQ, 48 copies/mL), Abbot RealTime HIV-1 (LLQ, 40 copies/mL), and Versant HIV-1 RNA 3.0 assay (bDNA) (LLQ, 75 copies/mL).
Noninvasive liver fibrosis testing
Both the AST to platelet ratio index (APRI) and FIB-4 indices were used as surrogate indicators of fibrosis, using APRI >1.5 and FIB-4 >3.25 for advanced fibrosis or cirrhosis.9,10
IL28B Testing
Genotyping of the chromosome 19 IL28B single nucleotide polymorphism (SNP) at rs12979860 and at rs8099917 was performed by ABI TaqMan (for basepairs C/C, C/T, or T/T and for G/G, G/T, or T/T, respectively) using centrally banked frozen peripheral blood mononuclear cells (PBMCs). IL28B genotypic analysis was performed on 62 subjects co-enrolled in a genetic study (A5128) on frozen whole blood samples. IL28B genotype at rs12979860 in the historical comparison group from A5178 was assessed by MassARRAY iPLEX Gold.
Metabolic Testing
Fasting glucose and insulin measurements (Quest Diagnostics) were assessed at entry and at weeks 16, 28, 52 (end of treatment), and 76, and homeostatic model for insulin resistance (HOMA-IR) was calculated.11 Insulin resistance was defined as HOMA-IR >2.5.12
Statistical Analysis
The pilot study was designed to assess whether: (a)cEVR proportion was greater than 40%, and (b) EVR proportion was greater than 50%, without adjustment for multiple testing. One-sided tests at a significance level of 5% on the primary endpoints were planned to explore whether the proposed regimen was promising compared to the standard-of-care HCV treatment regimen at the time of A5269. Two-sided 90% Clopper-Pearson confidence intervals (CIs) around proportion estimates are provided for the HCV virologic endpoints. Early dropouts and missed data for HCV virologic endpoints were considered failures in the primary analysis. For all other analyses on secondary and exploratory endpoints, 2-sided tests at a significance level of 5% were conducted, and 2-sided 95% CIs are provided. For comparisons between groups, Wilcoxon rank-sum tests and Fisher exact tests were used, as appropriate. For trend assessments, Cochran-Armitage trend tests were used. Within a group, Wilcoxon signed rank tests were used to assess significant changes from baseline, and exact McNemar's tests were used for correlated proportions over time.
Results
Study Population
Sixty-seven subjects with confi rmed HCV GT-1 and HIV-1 coinfection were enrolled (1 subject with protocol eligibility violation was replaced shortly after enrollment) from January to July 2010 at 18 ACTG Clinical Research Sites in the United States including Puerto Rico. Baseline characteristics are presented in Table 1. Median age was 50 years, 77.6% of subjects were male, and 47.8% were African American. Most (91.0%) subjects were on ART, and 73.1% had undetectable HIV-1 viral loads (<LLQ) with median CD4+ count of 452 cells/mm3. APRI scores were >1.5 in 13.4%, suggesting the presence of advanced fibrosis or cirrhosis.9 Historical controls from A5178 had similar characteristics.
Table 1. Baseline demographics of subjects on NTZ/PEG-IFN/WBR triple therapy (A5269) and PEG-IFN/ WBR (A5178).
| Demographics | A5269 (n = 67) | A5178a (n = 183) |
|---|---|---|
| Race | ||
| White | 21 (31.3%) | 61 (33.3%) |
| Black | 32 (47.8%) | 89 (48.6%) |
| Hispanic | 12 (17.9%) | 25 (13.7%) |
| >1 race | 1 (1.5%) | 4 (2.2%) |
| Other/unknown | 1 (1.5%) | 4 (2.2%) |
| Sex | ||
| Male | 52 (77.6%) | 151 (82.5%) |
| Female | 15 (22.4%) | 32 (17.5%) |
| Median age, years (IQR) | 50 (44–54) | 48 (41–52) |
| <50 | 33 (49.3%) | 116 (63.4%) |
| IVDU history | ||
| Never | 30 (44.8%) | 73 (39.9%) |
| Currently | 1 (1.5%) | 4 (2.2%) |
| Previous | 36 (53.7%) | 106 (57.9%) |
| IL28B | ||
| C/C | 15 (24.2%) | 38 (33.9%) |
| C/T | 37 (59.7%) [n = 62] | 51 (45.5%) [n = 112] |
| T/T | 10 (16.1%) | 23 (20.5%) |
| HIV-1 | ||
| Receiving ART | 61 (91.0%) | 144 (78.7%) |
| Median CD4+, cells/mm3 (IQR) | 452 (323–738) | 495 (373–697) |
| HIV RNA (<LLQ) | 49 (73.1%) [n = 65] | 129 (70.5%) |
| HCV G-1 subtype 1b | 19 (28.4%) [n = 65b] | Not available |
| Median log10 HCV RNA (IQR) | 6.38 (6.03–6.73) | 6.55 (6.12–6.85) |
| <800,000 IU/mL | 12 (17.9%) | 33 (18.0%) |
| Baseline APRI >1.5 | 9 (13.4%) | 24 (13.3%) [n = 180] |
| Baseline FIB-4 >3.25 | 9 (13.4%) | 21 (11.7%) [n = 180] |
| Baseline insulin resistance: HOMA-IR >2.5 | 35 (56.5%) [n = 62] | 47 (54.7%) [n = 86c] |
Note: APRI = AST to platelet ratio index; ART = antiretroviral treatment; LLQ = lower limit of quantitation; IQR = interquartile range; IVDU = intravenous drug use; HOMA-IR = homeostatic model for insulin resistance; PEG-IFN = pegylated interferon alfa-2a; WBR = weight-based ribavirin; NTZ = nitazoxanide.
These subjects were a select subset from ACTG 5178 who were HCV G-1 and HCV treatment naïve.
Two of the confirmed genotype 1 samples were unable to be subtyped.
In A5178, metabolic testing was only performed in subjects who enrolled under protocol version 1.0.
HCV Virologic Response
Figure 1 demonstrates virologic outcomes at key time points in A5269 (A) and in comparison to historical data from A5178 (B). There was a minimal drop in HCV viral load during the initial 4-week NTZ lead-in (median decline 0.12 log10; P = .04). Seven subjects achieved RVR (10.4%; 90% CI, 5.0%–18.7%). EVR occurred in 65.7% (44 of 67) (90% CI, 55.0%–75.3%) of subjects compared to 51.4% of A5178 subjects, which was a significant difference (90% CI, 3.0%–25.6%; P = .03). Complete EVR was observed in 38.8% (26 of 67) (90% CI, 28.8%–49.6%) of subjects. Because the LLQ used at week 12 in A5178 for HCV RNA was 600 IU/mL, the proportion of subjects with HCV RNA <600 IU/mL at 12 weeks of NTZ/PEG-IFN/WBR was also assessed (W12R): It was 44.8% (30 of 67) versus 39.9% in A5178 (4.9% difference; 90% CI, –6.7%– 16.5%; P = .29). SVR was achieved in 32.8% (22) of subjects (90% CI, 23.4%–43.5%) compared to 27.3% in A5178 (5.5% difference; 90% CI, -5.4%–16.4%; P = .24). Thirty-four subjects (50.7%) had undetectable HCV RNA at week 28 (90% CI, 40.1%–61.4%) and 32 subjects (47.8%) had ETR. One subject had virologic breakthrough after week 28 and discontinued therapy. Of the 32 subjects with ETR, 6 (18.7%) had virologic relapse and 4 (12.5%) discontinued the study before determination of SVR.
Figure 1.
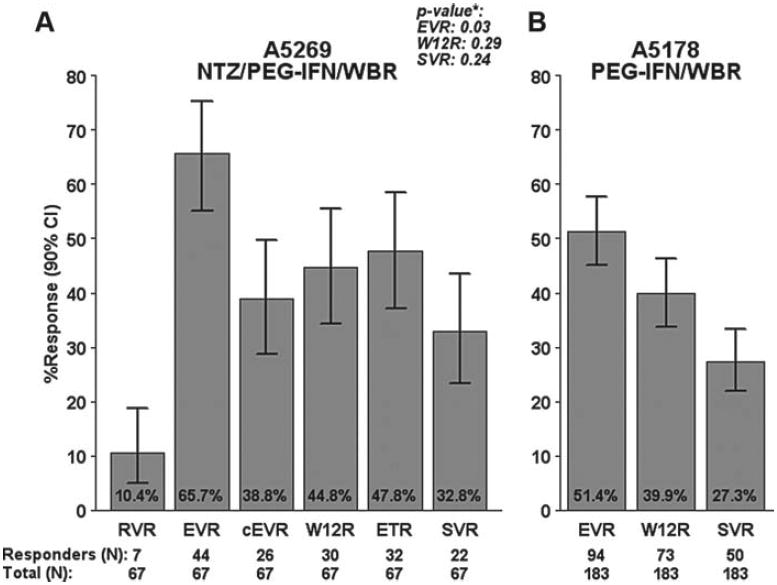
Proportion of subjects with virologic response in A5269 and A5178. *P values for A5269 in (A) are from one-sided Fisher exact test for comparison with A5178 results presented in (B). cEVR = complete early virologic response; EVR = early virologic response; RVR = rapid virologic response; SVR = sustained virologic response; W12R = week-12 response.
Predictors of SVR
Fifty percent (22 of 44) of subjects with EVR achieved SVR, and 76.9% (20 of 26) of those with cEVR achieved SVR. Only 5.7% (2 of 35) of subjects who did not achieve cEVR achieved SVR. Six of the 7 (85.7%) subjects with RVR achieved SVR, and the remaining subject withdrew from the study prematurely. SVR was observed more often in younger subjects (<50 years), men, non-Black, HCV GT-1b, baseline HCV RNA <800,000 IU/mL, APRI ≤ 1.5, and FIB-4 ≤3.25; however, with limited sample size, none of these reached statistical significance (P > .25, for each).
Safety
Among the 67 subjects, 18 (26.9%) discontinued treatment prematurely for AE-related reasons. Five discontinued due to protocol-defined toxicities (4 for cytopenias and 1 for diarrhea), 1 due to a hyper-sensitivity reaction to NTZ with lip and perioral swelling after a single dose that resolved over 3 days, and 1 due to grade 3 rash. Five discontinued for AEs that did not meet protocol-defined criteria for discontinuation (but subjects refused to continue): 1 for intolerance, and 4 for noncompliance. Among the clinical events noted were 4 cases of bacterial pneumonia (6% overall) and 1 case each of esophageal candidiasis and oral hairy leukoplakia.
Table 2 summarizes common grade 2 or higher AEs. The predominant AEs were those attributable to PEG-IFN such as neutropenia, thrombocytopenia, pain, and weight loss. Although overall gastrointestinal (GI) AEs were common, with 31 subjects (46.2%) experiencing such events, no grade 3 GI toxicities were noted during the 4-week NTZ lead-in. Overall, 7.5% of GI toxicities were grade 3 versus 10.4% in A5178.
Table 2. Selected grade 2 or higher adverse events in A5269.
| Sign/symptom/lab abnormality | No. (%) with grade 2 or higher events |
|---|---|
| Neutropenia | 39 (58.2) |
| Fatigue | 23 (34.3) |
| Pain | 23 (34.3) |
| Thrombocytopenia | 22 (32.8) |
| Weight loss | 20 (29.9) |
| Diarrhea | 18 (26.9) |
| Nausea | 13 (19.4) |
| Appetite decrease, loss or anorexia | 13 (19.4) |
| Dizziness/lightheadedness | 9 (13.4) |
| Anemia | 9 (13.4) |
| Shortness of breath | 9 (13.4) |
| Swelling | 9 (13.4) |
| Headache | 8 (11.9) |
| Cough | 8 (11.9) |
| Vomiting | 8 (11.9) |
| Depression | 8 (11.9) |
| Pruritus | 6 (8.9) |
| Ache | 5 (7.5) |
After the NTZ 4-week lead-in, there was no significant drop in hemoglobin or CD4+ T-cell counts. After the addition of PEG-IFN and WBR, there was a median decline in CD4+ T-cell count of 145 cells/ mm3 at week 16 (interquartile range [IQR], 104 to 296 cells/mm3) and a median decline of 173 cells/ mm3 at end of treatment (IQR, -59 to 276 cells/ mm3). Overall CD4% increased while on therapy by 6.5% at week 52 (IQR, 2.5% to 11.0%). Of the 49 subjects on ART with undetectable HIV-1 RNA levels at baseline, 4 had detectable viremia at any time during the study: 3 had HIV-1 RNA <200 copies/ mL with no further HIV RNA data available due to study discontinuation, and 1 had week 4 HIV-1 RNA level of 362 copies/mL that returned to undetectable by week 8.
IL28B Analysis
Of the 62 subjects with available data, 24.2% had rs12979860 polymorphism C/C, 59.7% C/T, and 16.1% T/T. The unfavorable T/T polymorphism at rs12979860 was observed in 23.3% of Blacks and 7.4% of non-Blacks (P = .15). There was a nonsignificant trend of higher EVR and cEVR proportions among subjects with the favorable C/C polymorphism at rs12979860 (P = .45 for EVR; P = .08 for cEVR). Those subjects with the favorable C/C IL28B polymorphism at rs12979860 also had a greater median HCV viral load drop after 4 weeks of triple therapy (-2.80 log10 IU/mL [IQR, -3.67 to -1.88] vs -1.41 log10 IU/mL [-2.14 to -0.95]; P = .004). However, these subjects did not have a statistically significant higher SVR proportion (33.3% for C/C, 29.7% for C/T, 30.0% for T/T; P = .84). This was in contrast with A5178 in which 42.1% (95% CI, 26.3%-59.2%) of those with the C/C polymorphism at rs12979860 achieved SVR compared to 33.3% (20.8%-47.9%) of those with C/T and 4.3% (0.1%-21.9%) of those with T/T (P = .003). Dropout rates were similar across IL28B genotypes. Figure 2 demonstrates SVR results by IL28B genotype at rs12979860 in comparison to available data from 112 of 183 A5178 subjects. At the rs8099917 polymorphism, A5269 EVR and cEVR proportions were actually lower among those with the favorable T/T genotype (61% of T/T with EVR vs 71% of non-T/T, P = .57; 30% of T/T with cEVR vs 53% of non-T/T, P = .14).
Figure 2.
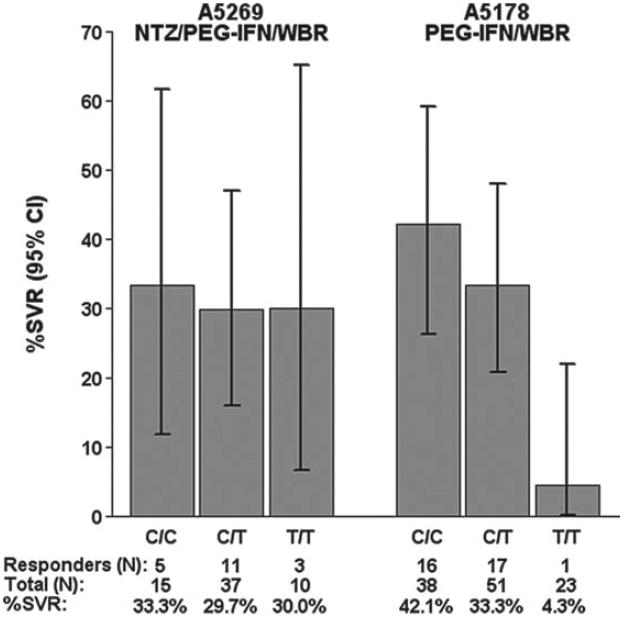
Number and percent of subjects achieving sustained viro-logic response (SVR) by IL28B rs12979860 genotype in A5269 and A5178. NTZ/PEG-IFN/WBR = nitazoxanide/pegylated interferon alfa-2a/weight-based ribavirin; PEG-IFN/WBR = pegylated interferon alfa-2a/weight-based ribavirin.
Metabolic Results
We examined whether pretreatment insulin resistance would predict virologic response. Six subjects had self-reported diabetes or were on antidiabetic medications (1 on metformin for lipoatrophy). Insulin resistance (HOMA-IR >2.5) was observed in 56.5% (35 of 62) of subjects with metabolic data at baseline. Subjects with higher HOMA-IR were less likely to achieve SVR (P = .05).
Achieving SVR was not associated with improved HOMA-IR in the 15 subjects for whom week 76 data were available. In fact, among those who went on to achieve SVR, both insulin and HOMA-IR levels increased while they were on therapy; there was a median insulin increase of 43% and HOMA-IR increase of 50% by the end of treatment. At 24 weeks following therapy, insulin levels were 12% higher and HOMA-IR levels were 14% higher than baseline. However, none of the results were statistically significant.
Weight loss is common on PEG-IFN–based therapy.13 Because body mass index (BMI) can affect insulin sensitivity, we examined changes in BMI during and after therapy to explore whether this could account for the insulin resistance findings. BMI decreased more in subjects who did not achieve SVR than in those who did (median percent decrease in BMI of 10.0% vs 8.6% by the end of treatment, P = .09). This trend remained following therapy.
Discussion
NTZ was safe and was well tolerated overall in this HIV-1/HCV GT-1 coinfected A5269 study population. Although nearly half of subjects experienced GI toxicity on therapy, the overall rate was not greater than that seen with PEG-IFN and RBV alone among historical controls from A5178. Although EVR improved as hypothesized, we did not observe a sustained HCV virologic benefit (ie, SVR) from adding NTZ to PEG-IFN and WBR in the treatment of HIV-1/HCV GT-1 coinfected subjects naïve to HCV therapy in comparison to historical control data.
There are potentially intriguing findings in this study. The use of NTZ may attenuate the negative effect of an unfavorable IL28B T/T genotype on achieving SVR. The pathway leading to differences in HCV clearance among those with IL28B polymorphisms is thought to be mediated by the IL28B gene product IFN-λ3's downstream immune effects.14 Because NTZ's mechanism of action is via upregulation of intracellular IFN-responsive elements and NTZ has been demonstrated to have synergy with PEG-IFN against HCV in vitro,1 the addition of NTZ to PEG-IFN/WBR could lead to an immunologic compensation for the lack of PEG-IFN–initiated intrinsic immune upregulation among individuals with an unfavorable IL28B genotype. However, because of the limited sample size of this study, we cannot make firm conclusions that NTZ has any benefit in regard to IL28B genotype. Moreover, initial HCV viral load declines were still significantly greater in subjects with the favorable IL28B polymorphism, so that one would have to posit an effect on interferon responsiveness that emerges over the course of therapy. In a small study among HCV GT-1 monoinfected, prior nonresponders (presumably a group enriched with unfavorable IL28B T/T genotype), the addition of NTZ did not improve SVR over PEG-IFN/WBR alone.15 We are studying NTZ's effect on in vitro immune correlates of virologic response in relation to interferon-mediated immune upregulation and in the context of IL28B polymorphisms and downstream effects.
Consistent with prior observations,16,17 subjects with baseline insulin resistance in our study were less likely to respond to HCV therapy. The successful treatment of HCV was not associated with improvement in insulin resistance, but the analysis was limited by small numbers. A more prolonged follow-up among individuals who achieve SVR may be necessary to determine whether long-term improvement in insulin resistance could be obtained by HCV cure.18
Limitations of our study are the small sample size and the use of a historical comparison, given the nature of pilot studies. Our historical comparator group within the ACTG network was well matched for several factors associated with SVR, so our conclusions are likely to be robust.
Conclusion
Despite improved EVR with the addition of NTZ to PEG-IFN/WBR-based therapy, we did not observe a SVR benefit from adding NTZ to PEG-IFN and WBR in the treatment of HIV-1/HCV GT-1 coinfected subjects naïve to HCV therapy. NTZ's overall safety profile was favorable. Our findings do not suggest that NTZ should be considered a promising agent in the treatment of HCV in HIV/HCV coinfected treatment-naïve subjects, especially in light of promising HCV direct-acting agents that were not available at the time of this study. The study did not address other genotypes such as GT-4 in which NTZ has shown favorable results in HCV monoinfected persons. The suggestion that NTZ may attenuate the effects of the unfavorable IL28B T/T genotype is undergoing further exploration to elucidate potential underlying immunologic mechanisms of these findings.
Acknowledgments
ACTG A5269 members and funding: ACTG Network Leadership Grant 1U01AI068636. ACTG Statistical and Data Management Center Grant 1U01AI068634. For HCV RNA and HCV genotyping efforts performed at Dr. Victoria A. Johnson's UAB Virology Laboratory 54: James Darren Hazelwood. UAB VSL grant (NIH/NIAID 7UM1AI068636). UAB Center for AIDS Research laboratory facilities (UAB CFAR grant P30AI27767-24) and Birmingham Veterans Affairs Medical Center laboratory facilities. Hospital of the University of Pennsylvania (A6201): Deb Kim, RPh, and Kathleen Maffei, BSN, RN. ACTG grant: U01 – AI – 096497 – 07.
Anna Smith, RN: UCSF AIDS CRS (Site 801), ACTG CTU Grant 5UO1 AI069502-06;
Susan Blevins, RN, MS, ANP-C, and Carolyn Coffay, BS, MA: UNC AIDS Clinical Trials Unit (Site 3201), ACTG CTU Grant AI069423, UNC Clinical Trials Research Center of the Clinical and Translational Science Award RR 025747, UNC Center for AIDS Research Grant AI050410; Jan Fuller, MSN, RN (study nurse) and Tamara James, BS (data manager): Alabama Therapeutics CRS (Site 5801), ACTG CTU Grant U01 AI069452; Richard Sterling, MD, MSc, and Kathleen Genther, BSN, RN: VCU CRS (Site 31475), ACTG CTU Grant 1-UO1-AI069503 to Fred Gordin (PI) at GWU; VCU Clinical Translational Research Center is supported by UL1-TR000058; Karen T. Tashima, MD, and Deborah Perez, RN: The Miriam Hospital (Site 2951), ACTG CTU Grant U01 A1069472; Jorge L. Santana Bagur, MD, and Olga I. Méndez Flores, MD: PR-ACTU (Puerto Rico-AIDS Clinical Trials Unit) (Site 5401), ACTG CTU Grant 5UM1AI069415-06; Nancy Reilly, RN, and Shobha Swaminathan, MD: NJMS (Site 31477), ACTG CTU Grant 5 UM1 AI069053-07; Valery Hughes, NP, and Christina Megill, PA-C:Cornell CTU (Site 7804), CTU Grant AI069419, CTSC Grant RR024996;Peter Gordon, MD, and Madeline Torres, RN: Columbia University HPTR (Site 30329), ACTG CTU Grant 5U01 AI069470-01, GCRC Grant 5 UL1 RR021456-07; Debbie Slamowitz, BSN, ACRN, and Sandra Valle, PA-C: Stanford University AIDS Clinical Trials Unit (Site 501), ACTG CTU Grant AI069556; Julie Hoffman, RN, and Susan Cahill, RN: UCSD Antiviral Research Center (Site 701), ACTG CTU Grant AI69432; Christine Hurley, RN, and Roberto Corales, DO: AIDS Care (Site 1108), ACTG CTU Grant U01AI069511-02 (as of 2/12/08), CTSI Grant UL1 RR 024160;
Amy Sbrolla and Gilbert Roy: Massachusetts General Hospital (Site 101), ACTG CTU Grant 1U01AI069472; Debika Bhattacharya, MD, and Maria Palmer: UCLA CARE Center (Site 601), ACTG CTU Grant AI 069424; Mary Adams RN, MPh, and Amneris Luque MD: University of Rochester CRS (Site 1101), ACTG CTU Grant U01AI069511-02 (as of 2/12/08), CRC Grant UL1 RR 024160; Kenneth Sherman, MD, and Michelle Saemann, RN: University of Cincinnati (Site 2401), ACTG CTU Grant AI-069513; Julie Ziegler and Kim Whitely: MetroHealth (Site 2503), ACTG CTU Grant AI069501.
Financial support/disclosures: This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland grant UL1RR024989 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Victoria Johnson's laboratory receives research support from Siemens Medical Solutions Diagnostics, Roche Molecular Diagnostics, Abbott Molecular Inc. David Haas has received research grants from Bavarian Nordic, Boehringer-Ingel-heim, Bristol-Myers Squibb, Gilead Sciences, Merck, Tanox, and Tibotec. He has been on scientific advisory boards for GlaxoSmithKline and Tibotec. Matthew Bardin is employed by Romark, Inc.
Raymond Chung has had grant support from Romark, Merck, Vertex, Gilead, and Mass Biologic, and has consulted for Enanta. Pablo Tebas is a consultant for Merck and has served on data safety monitoring boards for GlaxoSmithKline and Cytheris. Marion Peters is on the advisory board of Biotron. She has received honoraria from Janssen. Her spouse is an employee of Genentech Research and Development (subsidiary of Roche).
Additional contributions: We would also like to acknowledge the late Emmet B. Keeffe, MD, for his insights and work on the development of the protocol.
Footnotes
Conflicts of interests: Valerianna Amorosa, Anne Luetkemeyer, Minhee Kang, Triin Umbleja, Suria Yesmin, and Beverly Alston-Smith: No conflicts to declare.
Previous publication: This study was presented in part at the 2011 International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention in Rome (Nitazoxanide improves the early virological response to peginterferon alfa-2a and ribavirin in HIV/HCV treatment-naïve genotype 1 infected subjects: Results of ACTG 5269 [abstract TUPE176] presented at the 6th IAS Conference on HIV Pathogenesis and Treatment) and the 2012 International AIDS Conference in Washington, DCThe addition of nitazoxanide to peginterferon alfa-2a and ribavirin does not significantly improve sustained virologic response in HCV treatment-naïve genotype 1 HIV-1/HCV co-infected subjects: Results of ACTG 5269 [abstract WEAB0103] presented at the XIX International AIDS Conference).
References
- 1.Korba BE, Montero A, Farrar K, et al. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B and hepatitis C replication. Antivir Res. 2008;77:56–63. doi: 10.1016/j.antiviral.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Rossignol JF, Elfert A, Keeffe EB. Treatment of chronic hepatitis C using a 4-week lead-in with nitazoxanide before peginterferon plus nitazoxanide. J Clin Gastroenterol. 2010;44(7):504–509. doi: 10.1097/MCG.0b013e3181bf9b15. [DOI] [PubMed] [Google Scholar]
- 3.Rossignol JF, Elfert A, El-Gohary Y, Keeffe EB. Improved virologic response in chronic hepatitis C genotype 4 treated with nitazoxanide, peginterferon, and ribavirin. Gastroenterology. 2009;136(3):856–862. doi: 10.1053/j.gastro.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Rossignol JF. Nitazoxanide in the treatment of acquired immune deficiency syndrome-related cryptosporidiosis: Results of the United States compassionate use program in 365 patients. Aliment Pharmacol Ther. 2006;24(5):887–894. doi: 10.1111/j.1365-2036.2006.03033.x. [DOI] [PubMed] [Google Scholar]
- 5.Elazar M, Liu M, McKenna SA, et al. The anti-hepatitis C agent nitazoxanide induces phosphorylation of eukaryotic initiation factor 2alpha via protein kinase activated by double-stranded RNA activation. Gastroenterology. 2009;137(5):1827–1835. doi: 10.1053/j.gastro.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 6.Chung RT, Umbleja T, Chen JY, Andersen JW, Butt AA, Sherman KE. ACTG A5178 Study Team. Extended therapy with pegylated interferon and weight-based ribavirin for HCV-HIV coinfected patients. HIV Clin Trials. 2012;13(2):70–82. doi: 10.1310/hct1302-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Division of AIDS table for grading the severity of adult and pediatric adverse events. Version 1.0. [Accessed October 10, 2013];2004 Dec; clarifi cation August 2009. http://rsc.tech-res.com/Document/safetyandpharmacovigilance/Table_for_Grad-ing_Severity_of_Adult_Pediatric_Adverse_Events.pdf.
- 8.Sizmann D, et al. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol. 2007;38(4):326–333. doi: 10.1016/j.jcv.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Wai CT, Greenson J, Fontana RJ, Kalbfleisch JD, Marrero JA, Conjeevaram HS, Lok AS. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–526. doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- 10.Sterling RK, Lissen E, Clumeck N, et al. APRICOT Clinical Investigators. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 11.Levy JC, Matthews D, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998;21(12):2191–2192. doi: 10.2337/diacare.21.12.2191. [DOI] [PubMed] [Google Scholar]
- 12.Matthews D, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 13.Lo Re V, 3rd, Kostman J, Gross R, et al. Incidence and risk factors for weight loss during dual HIV/hepatitis C virus therapy. J Acquir Immune Defic Syndr. 2007;44(3):344–350. doi: 10.1097/QAI.0b013e31802f12d3. [DOI] [PubMed] [Google Scholar]
- 14.Balagopal A, Thomas D, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139(6):1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiffman ML, Ahmed A, Jacobson IM, et al. Phase 2 randomized, double-blind, placebo-controlled study of nitazoxanide with peginterferon alfa-2a and ribavirin in nonresponders (NR) with chronic hepatitis C genotype 1: Final report. 45th Annual Meeting of the European Association for the Study of the Liver (EASL 2010); 2010; Vienna. [Google Scholar]
- 16.Nasta P, Gatti F, Puoti M, et al. Insulin resistance impairs rapid virologic response in HIV/hepatitis C virus coinfected patients on peginterferon-alfa-2a. AIDS. 2008;22(7):857–861. doi: 10.1097/QAD.0b013e3282fbd1c4. [DOI] [PubMed] [Google Scholar]
- 17.Caronia S, Taylor K, Pagliaro L, et al. Further evidence for an association between non-insulin-dependent diabetes mellitus and chronic hepatitis C virus infection. Hepatology. 1999;30(4):1059–1063. doi: 10.1002/hep.510300416. [DOI] [PubMed] [Google Scholar]
- 18.Arase Y, Suzuki F, Suzuki Y, et al. Sustained virological response reduces incidence of onset of type 2 diabetes in chronic hepatitis C. Hepatology. 2009;49(3):739–744. doi: 10.1002/hep.22703. [DOI] [PubMed] [Google Scholar]


