Abstract
NanoRNase (Nrn) specifically degrades nucleoside 3′,5′-bisphosphate and the very short RNA, nanoRNA, during the final step of mRNA degradation. The crystal structure of Nrn in complex with a reaction product GMP was determined. The overall structure consists of two domains that are interconnected by a flexible loop and form a cleft. Two Mn2+ ions are coordinated by conserved residues in the DHH motif of the N-terminal domain. GMP binds near the DHHA1 motif region in the C-terminal domain. Our structure enables us to predict the substrate-bound form of Nrn as well as other DHH/DHHA1 phosphoesterase family proteins.
Keywords: nanoRNA, nanoRNase, DHH/DHHA1 family, crystal structure, mRNA degradation
1. Introduction
RNA metabolism is important for the regulation of gene expression. The mRNA levels are controlled by both the rate of their synthesis and degradation. RNA degradation is mediated by many different ribonucleases (RNases) [1]. In Escherichia coli, the degradation of transcripts is initiated by the endonucleolytic cleavage by RNase E [2]. The resulting fragments are then degraded by 3′–5′ exoribonucleases, RNase II, RNase R and polynucleotide phosphorylase. However, these RNases cannot completely degrade RNA to monoribonucleotides [3–5], but instead generate “nanoRNAs” that comprise very short RNA species (2 to 5-mers) [6]. Complete degradation of nanoRNAs is mediated by oligoribonuclease (Orn), one of the two essential RNases in E. coli [7,8]. Recently, it was shown that nanoRNA can act as a primer for transcription initiation and thereby alter global gene expression [9,10]. Although the degradation of nanoRNA is important, many bacteria lack a homologue of Orn. NanoRNase (Nrn), which was first identified in Bacillus subtilis, is the functional homologue of Orn [11]. Nrn homologues are found in Firmicutes, Bacteroides, Chlorobi, the σ and ε subdivisions of Proteobacteria, Actinobacteria, Crenarchaeota and Euryarchaeota, whereas Orn homologues are absent in these species [11].
Nrn is a member of DHH phosphoesterase superfamily, one of the phosphodiesterase superfamilies [12]. The DHH superfamily is divided into two subfamilies, DHHA1 and DHHA2, which share conserved N-terminal motifs but possess different C-terminal motifs. The DHH/DHHA1 subfamily includes Nrn as well as RecJ, a 5′–3′ single-strand DNA (ssDNA)-specific exonuclease, which acts in DNA repair and homologous recombination. We previously reported that TTHA0118 (ttNrn), an Nrn homologue from Thermus thermophilus HB8, degrades nanoRNAs and very short ssDNA in a 5′–3′ direction and displays a preference for shorter nucleotides [13]. ttNrn also hydrolyzes adenosine 3′,5′-bisphosphate (pAp) to give 5′-AMP [13]. The biochemical characteristics of some Nrn proteins have also been investigated [14–16]. However, the molecular mechanism of the activity of Nrn, including its preference for short nucleotides, remains unclear. In addition, another DHH/DHHA1 family protein YybT hydrolyzes bacterial signaling molecules, bis-(3′,5′)-cyclic diadenylic acid (c-di-AMP) and bis-(3′,5′)-cyclic diguanylic acid (c-di-GMP) yielding 5′-pApA and 5′-pGpG, respectively, but does not act on pAp, nanoRNA or ssDNA [17,18]. The molecular basis for this difference in substrate specificity between DHH/DHHA1 family proteins is not understood.
To date, two crystal structures of Nrn homologues from Bacteroides fragilis (PDB ID: 3DMA) and Staphylococcus haemolyticus (PDB ID: 3DEV) have been determined, although these findings are not reported in the literature. We previously described the crystal structure of full-length RecJ from T. thermophilus HB8 (ttRecJ) [19]. These studies revealed the active site includes two metal cations, but the requirements for substrate recognition remain unclear. The C-Ala domain of alanyl-tRNA synthetase is also annotated as a DHHA1 domain in the Pfam database. Moreover, this C-Ala domain alone has been shown to bind tRNA with high affinity [20]. Although the DHHA1 domain is thought to have RNA-binding function, the structural basis for this interaction is unknown.
Here, we report the crystal structure of Nrn from B. fragilis (bfNrn) as a Mn2+-GMP complex. This is the first study to determine the ligand-bound form of a DHH/DHHA1 phosphoesterase family protein and demonstrate that the DHHA1 motif is required for ligand binding.
2. Materials and Methods
2.1. Materials and assays
Polypeptone and yeast extract for cultivating E. coli were obtained from Difco (Detroit, MI). The oligoribonucleotides were synthesized by BEX Co. Ltd. (Nagoya, Japan). All other reagents used were of the highest commercially available grade. Purification and enzymatic assay of bfNrn were performed as described in Supplementary methods.
2.2. Crystallization, data collection, and structure determination
Crystallization of bfNrn-Mn2+-GMP complex was performed by the hanging drop vapor diffusion method. Drops (1 μl) of 3.5 mg/ml protein solution were mixed with 1 μl of 0.1 M sodium acetate trihydrate (pH 5.3), 1.6 M NaCl, 20 mM MnCl2 and 10 mM pGp–and equilibrated against 0.1 ml of the reservoir solution at 20°C. For cryoprotection, the crystal was soaked in a drop of the crystallization solution containing 20 mM MnCl2 and 25% glycerol. The crystals were then flash-frozen in a stream of liquid nitrogen (−180°C). Single-wavelength diffraction data at 1.000 and 1.892 Å were collected at beamline BL26B2 at SPring-8 (Hyogo, Japan). The diffraction images were processed using the program HKL2000 [21]. An examination of the data using phenix.xtriage [22] indicated that the crystal was not twinned. The structure was determined by molecular replacement with MOLREP [23] using the free form structure of bfNrn (PDB ID: 3DMA) as the model. The refinement was carried out using the following programs; CCP4 suite [24], Refmac5 [25], CNS [26] and Coot [27]. In the refinement process, we first built the structure with the 2Fo-Fc and Fo-Fc maps calculated by Refmac5. We then carried out iterative rounds of energy minimization, individual B-factor refinement using CNS, and manual model correction using Coot with omit map calculated by CNS to reduce the effects of model bias. The model structure of bfNrn-Mn2+-4 mer RNA (5′-UUUU-3′) was built using Pymol [28].
3. Results and discussion
3.1. Activity of bfNrn
We first measured exonuclease activity of bfNrn against ssDNA of different length. Prior to this study, there was only one report describing the biochemical activity of bfNrn, which stated the enzyme ‘hydrolyzes 2′,3′-cAMP to 3′-AMP’ [29]. Here, we found bfNrn degraded 11-mer ssDNA to mononucleotide in a Mn2+-dependent manner (Supplementary Fig. S1A). The 5′ end of the substrate was labeled with 32P, and mononucleotide as the product was found predominantly. This result suggested that bfNrn has exonuclease activity with 5′ to 3′ polarity. It should be noted, however, that a faint band seemed to be found just below the band of the substrate 11-mer ssDNA. Mass spectrometric analysis of the reaction product also suggested that the substrate was degraded both with 5′–3′ and 3′–5′ exonuclease activity (Supplementary Fig. S2) although it was difficult to determine quantitatively which activity was primary. It was previously suggested that an Nrn homologue (Mpn140) of Mycoplasma pneumoniae changes its polarity of exonuclease activity with the nature of the substrate [16]. In addition, bfNrn hydrolyzed 3-mer more efficiently than 6- and 11-mer (Supplementary Fig. S1B). Thus, bfNrn preferentially hydrolyzes short ssDNA, which is similar to other Nrn homologues.
Next, we examined phosphatase activity. bfNrn degraded pAp (3′,5′-pAp) to 5′-AMP, but not adenosine 2′,5′-bisphosphate (2′,5′-pAp) in the presence of Mn2+ ions (Supplementary Fig. S3A, B), indicating 3′ phosphatase activity. Moreover, bfNrn showed phosphatase activity against 3′-AMP and yielded adenosine (Supplementary Fig. S3C), but the amount of the product was less for 3′-AMP than pAp after the same reaction time (Supplementary Fig. S3F). These results indicate that the 5′-phosphate group contributes to the phosphatase activity of bfNrn, but is not essential. bfNrn also degraded pGp to 5′-GMP (Supplementary Fig. S3D), suggesting no base specificity of the phosphatase activity of bfNrn. Interestingly, bfNrn degraded c-di-GMP to generate 5′-GMP (Supplementary Fig. S3E). We conclude that 3′-phosphatase activity produces two 5′-GMP molecules from one c-di-GMP molecule. This degradation pattern is different from that of YybT, which generates 5′-pGpG from c-di-GMP [18]. Except for pAp, the activity of Nrn against these substrates has not been reported. c-di-GMP is a signaling molecule involved in bacterial virulence, infectiousness, pathogenesis and biofilm formation [30]. pGp is the product of hydrolysis of bacterial alarmone ppGpp by Ndx8 in T. thermophilus HB8 [30,31]. Thus, it is possible that Nrn acts not only in the mRNA degradation pathway but also in signaling pathway.
3.2. Overall structure of bfNrn
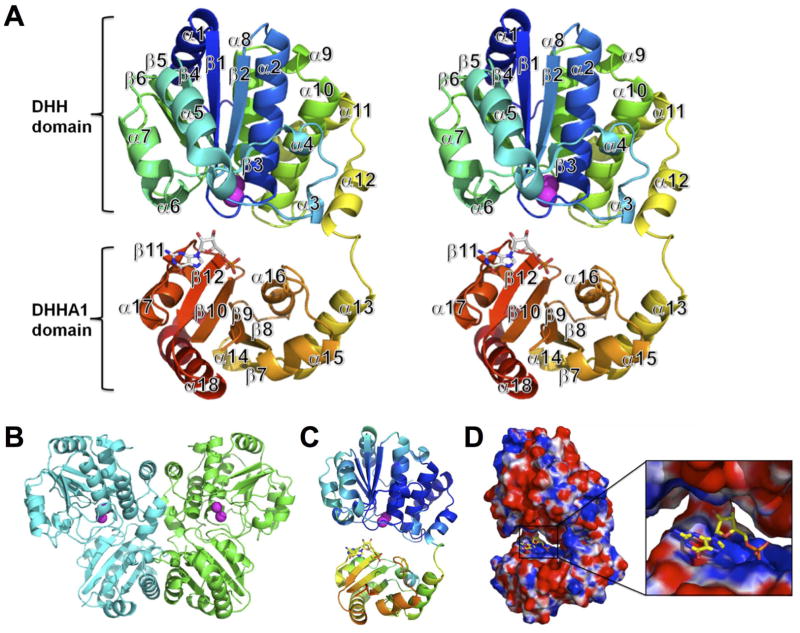
We determined the crystal structure of bfNrn in complex with Mn2+ and GMP at 2.95 Å resolution (Fig. 1). The crystal was obtained by co-crystallization with pGp and MnCl2 i.e., a reaction product GMP was generated during crystallization. Data collection and refinement statistics are shown in Supplementary Table S1. The overall structures of bfNrn in a ligand-free form (PDB ID: 3DMA) and as a Mn2+-GMP complex are similar to each other (the R.M.S.D. of 340 Cα atoms is 0.461). By size exclusion chromatography, the apparent molecular mass of bfNrn was estimated to be approximately 80,000, suggesting that bfNrn exists in a dimeric form in solution (data not shown). Although the asymmetric unit contains only one protein chain, the results of the size exclusion chromatography together with the analysis of the interface properties suggest that the two monomers related by the crystallographic 2-fold might form the biologically active entity (Fig. 1B).
Fig. 1.
Crystal structure of the bfNrn-Mn2+-GMP complex. (A) Stereo-view of the cartoon representation. The chain is color ramped from blue (N terminus) to red (C terminus). Mn2+ ions are shown as spheres. GMP is represented by a stick model. (B) A homodimer in the crystal by crystallographic 2-fold axis. (C) Distribution of the B-factor of main chain atoms. The B-factor is colored from high (red) to low (blue). (D) Electrostatic potential surface (positive potential, blue; negative potential, red). The GMP-binding site is expanded (inset).
The structure of the subunit can be divided into two structural domains that are interconnected by a flexible loop to form a cleft. The N-terminal domain (residues 2–212) comprises parallel β-sheet (β1–β6) surrounded by twelve α-helices. The C-terminal domain (residue 213–343) comprises anti-parallel β-sheet (β7–β12) surrounded by six α-helices. The first methionine, residues (283 and 284) in the loop regions and C-terminal His6-tag and a linker (344–351) were found to be disordered. The average B-factor of the C-terminal domain was higher than that of the N-terminal domain (Fig. 1C). The B-factor also tends to increase with distance from the N-terminal domain, implying rigid body vibration of an otherwise well-ordered C-terminal domain. The N-terminal domain contains DHH motifs I–IV forming the active site and coordinating two Mn2+ ions, whereas the C-terminal domain contains the DHHA1 motif. GMP is located in the cleft between the two domains and bound to the positive region formed by conserved basic residues near the DHHA1 motif (Supplementary Fig. S4) in the C-terminal domain (Fig. 1D). In the following discussion, the N- and C-terminal domains are referred to as the DHH and DHHA1 domain, respectively.
In the active site there are two Mn2+ ions (Mn1 and Mn2) in the concave patch composed of residues from motifs I–IV. An anomalous difference Fourier map at 1.892 Å (the Mn absorption edge) showed two obvious peaks in one subunit above 5 σ (Fig. 2A). Both Mn1 and Mn2 are coordinated by one nitrogen atom and three oxygen atoms. Mn1 is coordinated by Asp33 (2.2 Å, motif I), Asp99 (2.3 Å, motif II), His125 (2.6 Å, motif III) and Asp177 (2.3 Å, motif IV). Mn2 is coordinated by the side chains of Asp33 (2.2 Å and 2.6 Å, motif I), Asp99 (2.3 Å, motif II) and His29 (2.7 Å). The coordination of metal ions in bfNrn is similar to that of ttRecJ (Fig. 2C), except the coordinating residue His29 in bfNrn is replaced by Asp80 in ttRecJ. Also in the crystal structure of S. haemolyticus Nrn (shNrn), this position is occupied by His23 (although the structure only has one metal ion) (see Fig. 2B). The His residue at this position is highly conserved among Nrn homologues, whereas the Asp residue at the corresponding position is conserved among RecJ residues (Supplementary Fig. S4). Therefore, we conclude that this position in motif I of Nrn homologues is occupied by His, but not Asp. Nevertheless, overall coordination geometry is common to both Nrn and RecJ. Our findings suggest that the hydrolysis reaction catalyzed by Nrn proceeds via a two-metal ion mechanism, as is the case for RecJ [19]. After we positioned two Mn2+ ions, there still remained the third peak in the 2Fo-Fc map. Because there was no anomalous signal for this peak, we put a water molecule in this density. It should be mentioned that the position of the water molecule in the active site of bfNrn is different from that in ttRecJ (Fig. 2A and C). Interestingly, the corresponding position to His29 in bfNrn is occupied by His and Asn in type II inorganic pyrophosphatase and exopolyphosphatase, respectively, which belong to the DHH family [32]. The substrate of the former, pyrophosphate, is smaller than that of the latter, polyphosphate. The relationship between His/Asn residue and short/long polyphosphate in these enzymes seems similar to that between His/Asp and short/long nucleic acid in Nrn/RecJ. Hence, the residue at this position could be important in determining substrate preference.
Fig. 2.
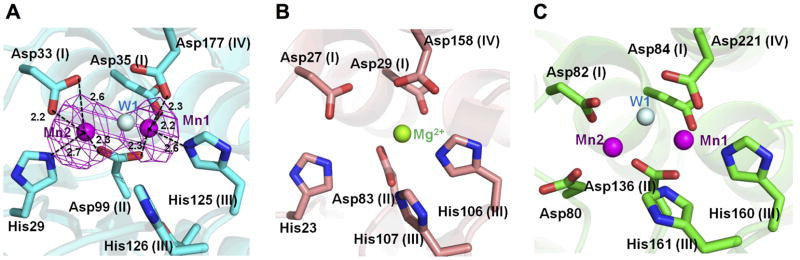
Metal ion-binding site. (A) Mn2+-binding site of bfNrn. Electron density (magenta) of the Mn2+ ions derived from an anomalous difference Fourier map (λ = 1.892000 Å) is superimposed on the model. The positions of the Mn2+ ions are shown as a magenta sphere. Water molecules are shown as cyan spheres. The dashed lines show the coordination with the metal ion (distances given in Å). (B) Mg2+-binding site of shNrn (PDB ID: 3DEV). The position of the Mg2+ ion is shown by a green sphere. (C) Mn2+-binding site of ttRecJ ttRecJ (PDB ID: 2ZXP). The motif names are indicated in parentheses.
3.4. Ligand-binding site
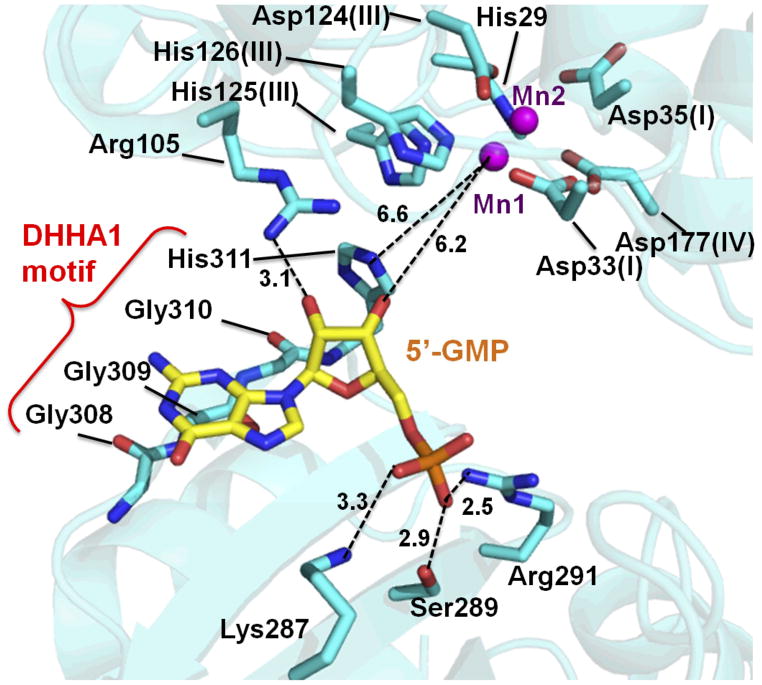
GMP (guanosine 5′-phosphate) is located near the GGGH residues (Supplementary Fig. S5A), which comprise the conserved motif in the DHHA1 family (Supplementary Fig. S4). GMP interacts with Arg105 in the DHH domain and Lys287, Ser289 and Arg291 in the DHHA1 domain (Fig. 3). To our knowledge, this is one of the first structural studies to show that the DHHA1 domain is directly involved in ligand binding, which was suggested previously [20]. As mentioned about C-Ala domain of alanyl-tRNA synthetase [20], the short β-strand (β11) formed by the DHHA1 motif (GGGH residues) in bfNrn is bent downwards (Supplementary Fig. S5B). Moreover, the conformation of β11 facilitates the formation of the concave patch on the molecular surface.
Fig. 3.
GMP-binding site. The position of the Mn2+ ion is shown by magenta spheres. The dashed lines indicate the distances (Å).
The oxygen atom of the 5′-phosphate group of GMP interacts with Lys287 (3.3 Å), Ser289 (2.9 Å) and Arg291 (2.5 Å) (Fig. 3). Arg291 is completely conserved among Nrn proteins. The side chain of Arg291 is closer to GMP in the Mn2+-GMP complex compared to the ligand-free form (Supplementary Fig. S5B), which is the only movement observed upon GMP binding. The 2′-hydroxyl group of the ribose moiety makes a hydrogen bond (3.1 Å) with the side chain of Arg105, which is highly conserved in Nrn. The 3′-hydroxyl group, which is generated by hydrolysis of the 3′-phosphate of pGp have no contact with the protein. In the guanine moiety, the main-chain carbonyl group of Gly309 is at a distance of 3.1 Å from N-3 atom. This is consistent with our finding that the phosphatase activity of bfNrn displays no base specificity.
The 3′-hydroxyl group of the ribose moiety and Mn2+ ions in the DHH domain are too far apart to interact (>6 Å) (Fig. 3). Given that GMP is a product of pGp hydrolysis, the substrate-bound form of the enzyme could be different from the determined structure. The 3′ scissile phosphate is predicted to be close to the metal-binding site for catalysis. Thus, we hypothesize that the two domains move close to each other to form the catalytically active conformation. Indeed, the average B-factor of the DHHA1 domain is higher than that of the DHH domain (Fig. 1C). Inorganic pyrophosphatases belonging to the DHH superfamily also fold into two domains and the C-terminal domain move closer to the N-terminal domain by rigid-body rotation around the hinge axis upon substrate binding [33]. Thus, the DHHA1 domain could move close to the DHH domain. If such domain movement occurs, the 3′-hydroxl group would be located closer to Mn1 than the 5′-hydroxyl group. This domain movement is in accordance with the observed 3′ phosphatase activity against pGp as well as pAp and 3′-AMP. Assuming rigid body movement of the DHHA1 domain relative to the DHH domain, molecular recognition of the product GMP in the determined structure would reflect that of the substrate. The following discussion concerning structure-function relationship is based on these assumptions.
Recognition of the 5′-phosphate group by Lys287, Ser289 and Arg291 is consistent with the result of the phosphatase assay, which indicated some contribution of the 5′-phosphate to the enzyme activity (Fig. S2). However, the 5′-phosphorylation does not affect the exonuclease activity of ttNrn against 11-mer ssDNA [13]. This observation suggests a difference in the detailed mechanism of substrate recognition between mononucleotide and oligoribonucleotide. Lys287 and Arg291 of bfNrn correspond to Lys366 and Arg370 of ttRecJ, respectively (Fig. S5C). Although these residues are also conserved in RecJ (Fig. S4), the 5′-phosphorylation status does not affect its exonuclease activity [34]. This observation suggests other residues contribute to substrate recognition in RecJ [19].
The side chain of Arg105 makes a hydrogen bond (3.3 Å) with the 2′-hydroxyl group of the ribose moiety. This seems inconsistent with the observation that Nrn degrades both ssDNA and RNA to a similar degree [13]. The distance between Arg105 and the 3′-hydroxyl group is 4.1 Å. If the DHHA1 domain moves closer to the DHH domain, it is possible that Arg105 could make contact with the 3′-phosphate group of the substrate. Arg105 is highly conserved among Nrn enzymes, but not among other DHH/DHHA1 family proteins such as YybT and RecJ (Supplementary Fig. S4). YybT degrades c-di-GMP and c-di-AMP but not nanoRNA and pAp [18], whereas RecJ degrades only ssDNA [19]. Therefore, Arg105 might play an important role in substrate recognition of Nrn.
The lack of interaction with the guanine moiety of GMP is consistent with our result of the phosphatase assay (Supplementary Fig. S4).
The β strand (β11), which is made up of residues from the DHHA1 motif, appears to be responsible for ligand binding (Fig. 3). Among these residues, the side chain of His311 is located at a distance of 4.1 Å from the 3′-hydroxyl group of GMP (Fig. 3). If the phosphate group is attached to the 3′ position, His311 would make contact with the substrate. Therefore, it is possible that His311 is involved in substrate recognition even in the absence of a conformational change. However, mutation of the corresponding His residue in E. coli RecJ decreased the exonuclease activity [35], suggesting that His311 plays a critical role in the activity. This raises the possibility that His311 contributes to the catalytic step. The distance between His311 and Mn1 is 6.6 Å (Fig. 3). Moreover, the conserved His in the DHHA1 motif of RecJ is located far away (10 Å) from the metal ions in the DHH domain [19]. However, based on the “domain closure” hypothesis outlined earlier, this distance would decrease upon substrate binding (Fig. S6). Indeed, the C-Ala domain of alanyl-tRNA synthetase does not have this conserved His residue but can still bind tRNA with high affinity [20].
3.5. Model for the bfNrn-nanoRNA complex
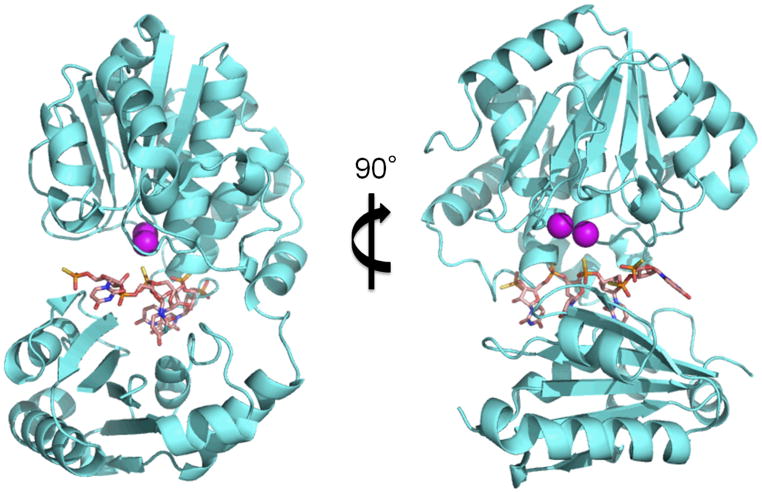
Based on this study and previous studies, we construct a model structure of the bfNrn-nanoRNA (4-mer RNA) complex (Fig. 4). First we used a model structure of ttRecJ-ssDNA complex as a reference [19]. In addition, we utilized structural information of shNrn for positioning the substrate. In the structure of shNrn (PDB ID: 3DEV), there are three sulfate ions per two subunits (Supplementary Fig. S7). Two of the sulfate ions are located at almost the same position as the 5′-phosphate group of GMP in the bfNrn-Mn2+-GMP complex, while the other sulfate ion is located on the dimer axis. We assume that the positions of the sulfate ions correspond to those of the phosphate groups of the substrate RNA. Based on this assumption, the 5′-end of the substrate RNA is positioned at that of GMP in the bfNrn-Mn2+-GMP complex i.e., GMP corresponds to the nucleotide at the 5′-end of the substrate. Moreover, the position of an internal phosphate group of the substrate was aligned to that of the sulfate ion on the dimer axis. In the constructed model structure, four nucleotide units are fitted to the length of the cleft between the two domains (Fig. 4).
Fig. 4.
A model structure of the bfNrn-nanoRNA complex. 4-mer RNA is shown as pink sticks. The position of each Mn2+ ion is shown as a magenta sphere.
Based on these findings, we can speculate on the structure-function relationship of Nrn, especially in terms of preference for substrate length. Our previous study on the exonuclease activity of ttNrn revealed that the Km values for the substrates were all within a relatively narrow range, whereas kcat values increased by ~5 orders of magnitude as ssDNA length decreased from 21-mer to 3-mer [13]. Analysis of the model structure shows the substrate-binding site (i.e., cleft between DHH and DHHA1 domains) has sufficient length to accommodate a 4-mer oligonucleotide. This is consistent with similar Km values for ssDNA of various lengths. However, our reaction model necessitates domain closure before the catalytic reaction can proceed. For substrates shorter than a 4-mer, domain closure can occur. However, the 3′-tail portion of a substrate longer than a 4-mer might protrude from the cleft near the linker (hinge) region. This mode of binding would hamper domain closure and inhibit the catalytic reaction. Further study is needed to verify our structural model and test our hypothesis concerning the reaction mechanism.
4. PDB accession numbers
Coordinates and structure factors for the crystal structure of bfNrn-Mn2+-GMP have been deposited in the PDB with PDB ID: 3W5W.
Supplementary Material
Highlights.
nanoRNase (Nrn) specifically degrades very short RNA in the mRNA degradation pathway.
The crystal structure of Nrn complexed with a reaction product GMP was determined.
Two Mn2+ ions are coordinated by the DHH motif residues in the N-terminal domain.
GMP binds to the DHHA1 motif region in the C-terminal domain.
This structure enables us to predict the substrate-bound form of Nrn.
Abbreviations
- Nrn
nanoRNase
- bfNrn
nanoRNase of Bacteroides fragilis
- shNrn
nanoRNase of Staphylococcus haemolyticus
- ttNrn
nanoRNase of Thermus thermophilus HB8
- ttRecJ
RecJ exonuclease of T. thermophilus HB8
- pAp
adenosine 3′,5′-bisphosphate
- pGp
guanosine
- 3′,5′
-bisphosphate
References
- 1.Arraiano C, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amblar M, Barbas A, Fialho A, Arraiano C. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 5.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 6.Yu D, Deutscher M. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J Bacteriol. 1995;177:4134–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′–5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime transcription initiation in vivo. Mol Micribiol. 2011;42:817–825. doi: 10.1016/j.molcel.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vvedenskaya IO, Sharp JS, Goldman SR, Kanabar PN, Livny J, Dove SL, Nickels BE. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012;26:1498–1507. doi: 10.1101/gad.192732.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 13.Wakamatsu T, Kim K, Uemura Y, Nakagawa N, Kuramitsu S, Masui R. Role of RecJ-like protein with 5′–3′ exonuclease activity in oligo(deoxy)nucleotide degradation. J Biol Chem. 2011;286:2807–2816. doi: 10.1074/jbc.M110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37:5114–5125. doi: 10.1093/nar/gkp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Cescau S, Mechold U, Wang J, Cohen D, Danchin A, Boulouis H, Biville F. Identification of a novel nanoRNase in Bartonella. Microbiology. 2012;158:886–895. doi: 10.1099/mic.0.054619-0. [DOI] [PubMed] [Google Scholar]
- 16.Postic G, Danchin A, Mechold U. Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae. RNA. 2012;18:155–165. doi: 10.1261/rna.029132.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao F, Ji Q, Soehano I, Liang Z. Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol. 2011;193:1543–1551. doi: 10.1128/JB.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakamatsu T, Kitamura Y, Kotera Y, Nakagawa N, Kuramitsu S, Masui R. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J Biol Chem. 2010;285:9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo M, Chong YE, Beebe K, Shapiro R, Yang XL, Schimmel P. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–747. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zbyszek O, Wladek M. Processing of X-ray diffraction data collected in oscillation code. Methods Enzymol. 1991;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 24.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Cryst. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudova GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 29.Rao F, Qi Y, Murugan E, Pasunooti S, Ji Q. 2′,3′-cAMP hydrolysis by metal-dependent phosphodiesterases containing DHH, EAL, and HD domains is non-specific: Implications for PDE screening. Biochem Biophys Res Commun. 2010;398:500–5. doi: 10.1016/j.bbrc.2010.06.107. [DOI] [PubMed] [Google Scholar]
- 30.Kalia D, et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 31.Ooga T, Ohashi Y, Kuramitsu S, Koyama Y, Tomita M, Soga T, Masui R. Degradation of ppGpp by Nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J Biol Chem. 2009;284:15549–15556. doi: 10.1074/jbc.M900582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tammenkoski M, Moiseev VM, Lahti M, Ugochukwu E, Brondijk THC, White SA, Lahti R, Baykov AA. Kinetic and mutational analyses of the major cytosolic exopolyphosphatase from Saccharomyces cerevisiae. J Biol Chem. 2007;282:9302–9311. doi: 10.1074/jbc.M609423200. [DOI] [PubMed] [Google Scholar]
- 33.Ahn S, Milner AJ, tterer KF, Konopka M, Ilias M, Young TW, White SA. The “open” and “closed” structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii. J Mol Biol. 2001;313:797–811. doi: 10.1006/jmbi.2001.5070. [DOI] [PubMed] [Google Scholar]
- 34.Han ES, Cooper DL, Persky NS, Vincent A, Sutera J, Whitaker RD, Montello ML, Lovett ST. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutera VA, Jr, Han ES, Rajman LA, Lovett ST. Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J Bacteriol. 1999;181:6098–6102. doi: 10.1128/jb.181.19.6098-6102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.