Abstract
The biosynthesis and export of bacterial cell-surface polysaccharides is known to occur through several distinct mechanisms. Recent advances in the biochemistry and structural biology of several proteins in synthase-dependent polysaccharide secretion systems have identified key conserved components of this pathway in Gram-negative bacteria. These components include an inner-membrane-embedded polysaccharide synthase, a periplasmic tetratricopeptide repeat (TPR)-containing scaffold protein, and an outer-membrane β-barrel porin. There is also increasing evidence that many synthase-dependent systems are post-translationally regulated by the bacterial second messenger bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP). Here, we compare these core proteins in the context of the alginate, cellulose, and poly-β-D-N-acetylglucosamine (PNAG) secretion systems.
Keywords: synthase, exopolysaccharide, alginate, cellulose, poly-β-D-N-acetylglucosamine
Role of exopolysaccharide production
The secretion of polysaccharides by bacteria is a physiological process that occurs under a multitude of different environmental circumstances. The role of these polysaccharides includes basic functions such as maintaining the structural integrity of the cell envelope and preventing cellular desiccation, as well as more complex functions such as facilitating interactions within bacterial communities, and between bacteria and eukaryotes. Bacterial polysaccharide production plays a direct role in human health, in part because of the ability of many pathogenic bacteria to form multicellular conglomerates called bio-films. The structural integrity of bacterial biofilms is highly dependent on a self-produced extracellular matrix that may comprise exopolysaccharides, nucleic acids, and proteins [1–3]. The major components of the biofilm matrix differ, depending on the bacterial species and strain, the stage of biofilm development, and the environmental conditions. In Pseudomonas aeruginosa, a model organism that has been extensively used to study biofilm development, the exopolysaccharide component of the biofilm matrix predominates and protects the bacteria from host defense mechanisms and administered antibiotics [4–7]. Exopolysaccharides produced by bacteria also have industrial applications in the paper, food, and health industries, and thus an understanding of how these long polymers are secreted may lead to further efficiencies in commercial polysaccharide production.
Secretion of cell-surface polysaccharides in Gram-negative bacteria
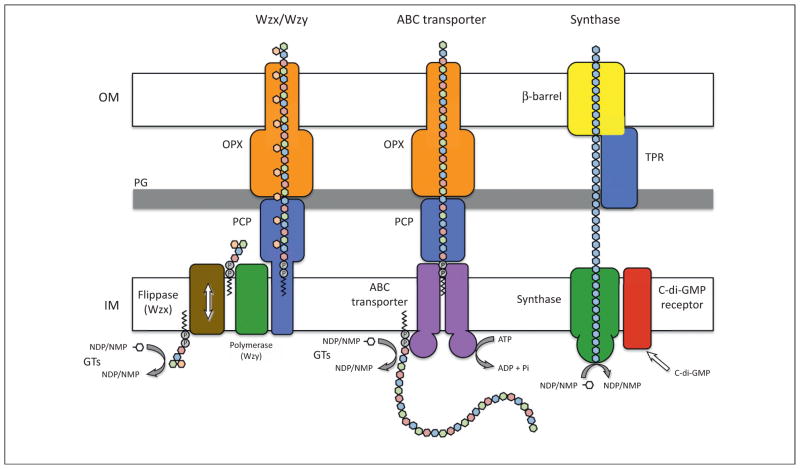
Despite the enormous chemical diversity among the carbohydrate building blocks of bacterial exopolysaccharides, the molecular mechanism by which these biopolymers are assembled and exported from the cell can currently be categorized into three distinct mechanisms (Figure 1). These include the Wzx/Wzy- and ATP-binding cassette (ABC) transporter-dependent pathways, both of which use a lipid acceptor to initiate polysaccharide synthesis, and a synthase-dependent pathway, for which the requirement for a lipid acceptor molecule depends on the polysaccharide in question. In Wzx/Wzy-dependent secretion systems, such as Escherichia coli group 1 capsular polysaccharides (CPS) and lipopolysaccharide (LPS) O-antigen, the polysaccharide repeat unit is assembled on an undecaprenyl phosphate acceptor moiety by various inner-membrane-embedded or -associated glycosyl transferases [8,9]. This synthesized precursor is then transported across the inner membrane by a flippase, Wzx [10–12], before being polymerized into a high-molecular-weight polysaccharide by the periplasmic polymerase Wzy [13]. In comparison, the ABC transporter-dependent systems, such as E. coli group 2 CPS and LPS common antigen, assemble the entire polysaccharide chain on a lipid acceptor, the identity of which varies depending on the polysaccharide being synthesized, before transporting the polymer across the inner membrane via an ABC transporter [14–16]. Despite these mechanistic differences in their modes of polysaccharide assembly, both the Wzx/ Wzy- and ABC transporter-dependent secretion systems use similar protein families to facilitate exopolysaccharide export across the periplasm and through the outer membrane. This process involves proteins from the outer-membrane polysaccharide export (OPX) and polysaccharide copolymerase (PCP) protein families [17,18]. The X-ray crystal structures of E. coli Wza [19], an OPX protein, and several PCPs [20,21] have demonstrated that both of these proteins arrange into homo-oligomeric assemblies that interact with one another to form a protein channel through which the nascent polysaccharide chain is exported from the cell [22].
Figure 1.
Mechanisms of polysaccharide secretion. Cartoon schematic of the Wzx/Wzy-, ABC-transporter- and synthase-dependent pathways for exopolysaccharide biosynthesis and export. The key protein components for each pathway are indicated on the diagram. Glycosyl transferases (GTs) synthesize the lipid-linked polysaccharide repeat units from nucleotide diphosphates (NDPs) or nucleotide monophosphates (NMPs) in the Wzx/Wzy- and ABC-transporter-dependent systems. For Wzx/Wzy-dependent secretion, the polysaccharide repeat unit is assembled on an undecaprenyl phosphate carrier located in the inner leaflet of the inner membrane before being transported across the inner membrane by the flippase Wzx. In the periplasm, the repeat units are assembled into the mature polysaccharide by the polymerase Wzy before being exported through the periplasm and across the outer membrane by a translocation pathway formed by members of the PCP and OPX families of proteins. For ABC transporter-dependent secretion, the entire polysaccharide is assembled on a lipid carrier located in the inner leaflet of the inner membrane before being transported across the inner membrane by an ABC transporter. The polysaccharide is then exported through the periplasm and across the outer membrane by a translocation pathway formed by members of the PCP and OPX families of proteins. For synthase-dependent secretion, the polysaccharide is polymerized and exported across the inner membrane by an inner-membrane synthase protein. In some instances, the activity of the polysaccharide synthase is post-translationally regulated by an inner-membrane c-di-GMP receptor. The polysaccharide is then exported across the outer membrane by a periplasmic TPR-containing protein and an integral outer-membrane β-barrel. Abbreviations: OPX, outer membrane polysaccharide export; PCP, polysaccharide copolymerase; TPR, tetratricopeptide repeat proteins; IM, inner membrane; PG, peptidoglycan sacculus; OM, outer membrane; c-di-GMP, bis-(3′-5′)-cyclic dimeric guanosine monophosphate; ABC transporter, ATP-binding cassette transporter.
Synthase-dependent exopolysaccharide secretion can occur in the presence or absence of a lipid acceptor molecule, depending on the polysaccharide [23–25]. In these systems, it appears that a membrane-embedded glycosyl transferase is able to facilitate simultaneous polymer formation and translocation across the inner membrane [25]. In some of the better-characterized Gram-negative synthase-dependent secretion systems such as P. aeruginosa alginate and Gluconacetobacter xylinus cellulose, polymerization is also regulated by an inner-membrane receptor, sometimes referred to as a co-polymerase, that binds the bacterial second messenger bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) to activate polysaccharide production [26,27]. Once the polymer reaches the periplasm, a tetratricopeptide repeat (TPR)-containing scaffold protein is thought to protect it from degradation before it is exported across the outer membrane through a β-barrel porin [28–30]. This export apparatus requires protein families that are clearly distinct from the OPX and PCP proteins used by the Wzx/Wzy- and ABC transporter-dependent pathways, and given the predictability of these protein domain families by sequence analysis programs such as SMART [31,32] and Phyre2 [33], they serve as identifiable hallmarks of synthase-dependent exopolysaccharide secretion systems in Gram-negative bacteria. Here, we summarize the current understanding of synthase-dependent polysaccharide production in the context of the secreted exopolysaccharides alginate, cellulose, and poly-β-D-N-acetylglucosamine (PNAG).
Synthase-dependent production of alginate
Alginate is a random linear polymer of 1,4-linked β-D-mannuronic acid and its C5 epimer α-L-guluronic acid [(β-D-ManUA-(1→4)-β-L-GulUA)n], which was originally identified in brown seaweeds and subsequently in several species of Gram-negative bacteria [34–36]. Interest in alginate biosynthesis has been driven by the observation that mucoid isolates of the opportunistic pathogen P. aeruginosa found in the lungs of cystic fibrosis (CF) patients secrete copious amounts of the polysaccharide [37]. Moreover, conversion of P. aeruginosa from a non-mucoid to a mucoid phenotype correlates with poor prognosis among these patients [38,39]. Current understanding of the mechanism of alginate biosynthesis and secretion has been gained from studies of the alginate-producing bacteria P. aeruginosa and Azotobacter vinelandii. In these organisms, alginate is first synthesized as a homopolymer of 1,4-linked β-D-mannuronic acid before being epimerized at the polymer level to form the mature polysaccharide [40,41]. In addition, it has been found that the O2 and/or O3 positions of β-D-mannuronic acid residues in bacterial alginates can be acetylated [42,43], which increases the water-binding capacity of the polymer and, during infection, protects P. aeruginosa from opsonic phagocytosis by the immune system of the host [44,45]. Moreover, O-acetylated alginate has been identified as an important structural component of biofilms produced by mucoid strains of P. aeruginosa [46].
The proteins responsible for the polymerization and export of alginate are encoded on the algD operon (Figure 2). Synthesis of poly-β-D-mannuronic acid from GDP-mannuronic acid occurs at the inner membrane and requires the integral inner membrane proteins Alg8 and Alg44 (Figures 3 and 4; Table 1). Alg8 is a putative alginate synthase because it contains five predicted trans-membrane domains and a large cytoplasmic synthase domain that shares homology with family 2 glycosyl transferases [47,48]. The polymerization reaction also requires the c-di-GMP-binding activity of the inner-membrane protein Alg44 [26]. Alg44 is a single-pass transmembrane protein that is predicted to have an N-terminal cytoplasmic c-di-GMP-binding PilZ domain and C-terminal periplasmic region that may resemble a membrane fusion protein (MFP) domain [47]. It is thought that these two proteins act in concert to facilitate the polymerization and export of poly-β-D-mannuronic acid across the inner membrane by a mechanism that is regulated by c-di-GMP binding to Alg44. This hypothesis was derived, in part, from the observation that site-specific mutations that compromise the c-di-GMP-binding ability of Alg44 abrogate alginate production in vivo [26]. The role of MFP domains as periplasmic adaptors that join inner-membrane transport proteins to outer-membrane export proteins in bacterial tripartite drug-efflux pumps suggests that the MFP domain of Alg44 may take part in periplasmic protein–protein interactions (Figure 3) [49]. At present, there are no experimental data confirming that Alg8 and Alg44 are directly responsible for export of the newly synthesized polymer across the inner membrane or if the MFP domain of Alg44 couples alginate polymerization to its export.
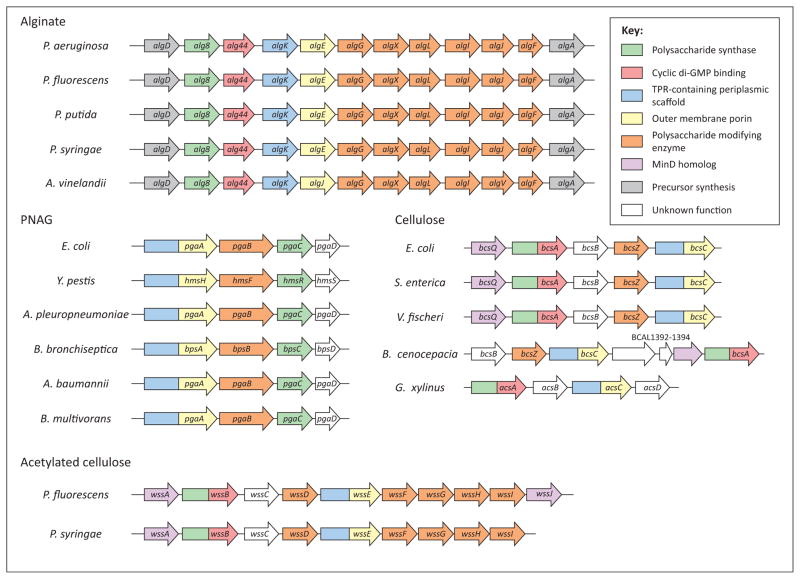
Figure 2.
Synthase-dependent exopolysaccharide loci in Gram-negative bacteria. Operons for which there is experimental confirmation of alginate, cellulose, and poly-β-D-N-acetylglucosamine (PNAG) production in Gram-negative bacteria. Each open reading frame is shown as an arrow (not drawn to scale) and named as labeled. The predicted or confirmed function of each open reading frame is indicated by its color as described by the legend in the figure. Open reading frames that have two colors represent two attributed functions. Abbreviations: P. aeruginosa, Pseudomonas aeruginosa; P. fluorescens. Pseudomonas fluorescens; P. putida, Pseudomonas putida; P. syringae, Pseudomonas syringae; A. vinelandii, Azotobacter vinelandii; E. coli, Escherichia coli; Y. pestis, Yersinia pestis; A. pleuropneumoniae, Actinobacillus pleuropneumoniae; B. bronchiseptica, Bordetella bronchiseptica; A. baumannii, Acinetobacter baumannii; B. multivorans, Burkholderia multivorans; S. enterica, Salmonella enterica; V. fischeri, Vibrio fischeri; B. cenocepacia, Burkholderia cenocepacia; G. xylinus, Gluconacetobacter xylinus.
Figure 3.
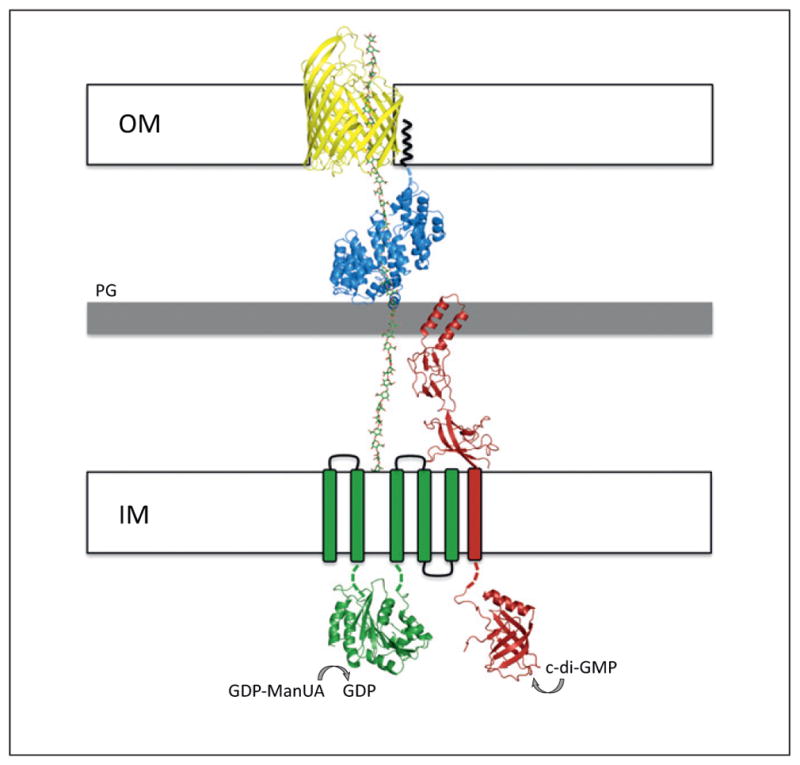
Model of the polymerization and export of alginate. Structural representation of the inner- and outer-membrane components required for alginate secretion. AlgE, AlgK, Alg8, and Alg44 are displayed in cartoon representation in yellow, blue, green, and red, respectively. The AlgE (Pseudomonas aeruginosa) and AlgK (Pseudomonas fluorescens) models are derived from the recent crystal structures determined by Whitney et al. (PDB 3RBH) [30] and Keiski et al. (PDB 3E4B) [29]. The synthase domain of Alg8 was modeled using the first GT-A domain of the unpublished crystal structure of Escherichia coli chondroitin polymerase (PDB 2Z86). The cytoplasmic PilZ domain of Alg44 was modeled using the apo structure of P. aeruginosa PA4608 (PDB 1YWU) solved by the Northeast Structural Genomics Consortium [92] and the periplasmic MFP domain was modeled using E. coli MacA (PDB 3FPP), the periplasmic component of a tripartite macrolide-specific efflux pump [93]. The Alg8 and Alg44 models were generated using the Protein Homology/analogY Recognition Engine (Phyre2) server [33]. 1,4-Linked β-D-mannuronic acid was modeled in the diagram (shown as green sticks) to demonstrate the role of AlgK as a protective scaffold protein and of AlgE as the outer-membrane alginate export protein. GDP mannuronic acid (GDP-ManUA) is the activated sugar nucleotide precursor. The black squiggly line indicates the N-terminal lipid anchor of AlgK. IM, PG, and OM refer to the inner membrane, the peptidoglycan sacculus, and the outer membrane, respectively.
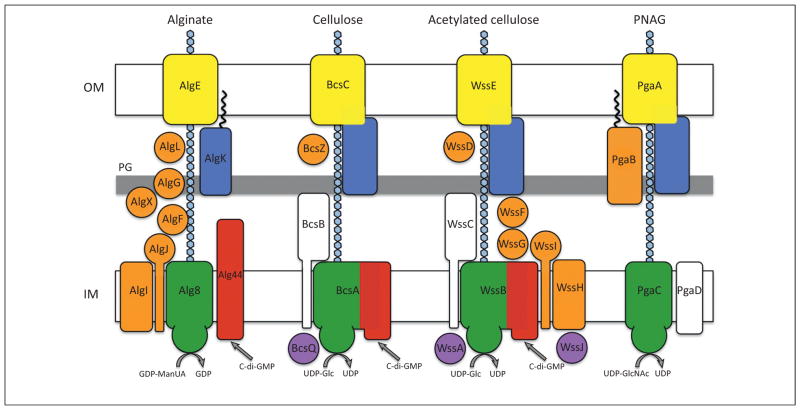
Figure 4.
Schematic representation of the alginate, cellulose, acetylated cellulose, and poly-β-D-N-acetylglucosamine (PNAG) exopolysaccharide secretion systems. The components for each pathway are indicated on the diagram and color-coded according to similar predicted function as follows: green, synthase; red, c-di-GMP, bis-(3′-5′)-cyclic dimeric guanosine monophosphate; blue, tetratricopeptide repeat; yellow, β-barrel porin; orange, exopolysaccharide-modifying enzyme; purple, MinD homolog; and white, unknown function. The black squiggly line indicates the N-terminal lipid anchors of AlgK and PgaB. In each system, the polysaccharide indicated is polymerized and transported across the inner membrane by its respective synthase. For alginate, cellulose, and acetylated cellulose, this process also requires the c-di-GMP receptor indicated. Once in the periplasm, various polysaccharide-modifying enzymes act on each of the polysaccharides before they are exported across the outer membrane by a tetratricopeptide repeat (TPR)-containing protein and an integral outer-membrane β-barrel. Abbreviations: IM, inner membrane; PG, peptidoglycan sacculus; OM, outer membrane; GDP-ManUA, GDP mannuronic acid; UDP-Glc, UDP glucose; UDP-GlcNAc, UDP N-acetylglucosamine.
Table 1.
Proteins involved in alginate, cellulose, and PNAG polymer biosynthesis and export
| Protein | Predicted or demonstrated function | Subcellular localization | PDB IDa | Refs |
|---|---|---|---|---|
| Alginate | ||||
| Alg8 | Synthase | Inner membrane | N/A | [41,42] |
| Alg44 | c-Di-GMP receptor | Inner membrane | N/A | [22,41] |
| AlgK | TPR scaffold | Outer membrane | 3E4B | [24,25] |
| AlgE | β-Barrel porin | Outer membrane | 3RBH | [26] |
| AlgG | C5 mannuronan epimerase | Periplasm | N/A | [35,36,43] |
| AlgX | O-Acetylation? | Periplasm | Forthcoming | [48,49] |
| AlgL | Alginate lyase | Periplasm | Forthcoming | [50,51] |
| AlgI | O-Acetylation | Inner membrane | N/A | [45–47] |
| AlgJ | O-Acetylation | Inner membrane | N/A | [45–47] |
| AlgF | O-Acetylation | Periplasm | N/A | [44,46] |
| Cellulose | ||||
| BcsQ/WssA | Polar localization of apparatus? | Cytoplasm | N/A | [61] |
| BcsA/WssB | Synthase/c-di-GMP receptor | Inner membrane | N/A | [23,53] |
| BcsB/WssC | Unknown | Inner membrane | N/A | [59] |
| BcsZ/WssD | Glycosyl hydrolase | Periplasm | 3QXF, 3QXQ | [60] |
| BcsC/WssE | TPR scaffold/β-barrel porin | Outer membrane | N/A | [25,26,59] |
| WssF | O-Acetylation? | Periplasm | N/A | [56] |
| WssG | O-Acetylation? | Periplasm | N/A | [47,56] |
| WssH | O-Acetylation? | Inner membrane | N/A | [47,56] |
| WssI | O-Acetylation? | Inner membrane | N/A | [47,56] |
| WssJ | Unknown | Cytoplasm | N/A | [56] |
| AcsD | Cellulose fiber formation | Extracellular | 3AJ1, 3A8E | [62,63] |
| PNAG | ||||
| PgaA | TPR scaffold/β-barrel porin | Outer membrane | N/A | [65,69] |
| PgaB | De-N-acetylase | Outer membrane | 4F9D, 4F9J | [65,69,71] |
| PgaC | Synthase | Inner membrane | N/A | [65,69] |
| PgaD | c-Di-GMP binding? | Inner membrane | N/A | [65,69] |
N/A, structure not available in PDB.
Once in the periplasm, poly-β-D-mannuronic acid is epimerized by the polymer-level mannuronan C5 epimerase AlgG [40]. AlgG is thought to play dual roles in both C5 epimerization and secretion of the nascent alginate polymer across the periplasm [50]. An algG deletion mutant produces degraded alginate fragments that are characteristic of digestion products of the periplasmic alginate lyase AlgL, suggesting that AlgG also plays a role in protecting the alginate polymer as it traverses the periplasm [50].
The algI, algJ, and algF gene products have all been implicated in the O-acetylation of mannuronic acid residues [51,52]. AlgI and AlgJ are integral inner-membrane proteins, whereas AlgF localizes to the periplasm. Topology modeling of AlgI suggests that it contains at least seven transmembrane helices, whereas except for a single trans-membrane helix at its N-terminus, the majority of AlgJ resides in the periplasm [53]. The current model of alginate O-acetylation involves export of an unknown cytoplasmic acetyl donor by AlgI across the inner membrane to the periplasm, where the acetate group is then transferred to alginate by the activities of AlgJ and AlgF [54]. AlgX is a periplasmic protein of unknown function; however, its high sequence similarity to AlgJ suggests that it may also play a role in alginate O-acetylation [55,56]. As with the algG deletion mutant, an algX deletion mutant produces AlgL-degraded alginate, suggesting that AlgX may also play a role in guiding the mature alginate polymer across the periplasm [55].
The TPR-containing outer-membrane lipoprotein AlgK is believed to guide the mature polymer towards the integral outer-membrane protein AlgE, which facilitates translocation of alginate across the outer membrane [29,30]. The proposed function of AlgK was derived, in part, from phenotypic studies demonstrating that in the absence of AlgK, alginate is degraded by AlgL [28,57,58]. The presence of TPRs in the structure of AlgK suggests that it may serve as a scaffold to which the other periplasmic Alg proteins interact to form a multiprotein complex (Figure 3) [29]. TPR-containing proteins often function as protein–protein interaction modules and are involved in a variety of different cellular processes in all domains of life [59]. In Gram-negative bacteria, TPR domains facilitate protein–protein interactions within the outer-membrane β-barrel assembly machinery (BAM) complex, and between the chaperone and substrate for several translocator proteins exported by the type III secretion system (T3SS) apparatus [60,61].
The recent X-ray crystal structure of AlgE shows that it adopts an 18-stranded β-barrel with a highly electropositive interior (Figure 3) [30]. Given the strict conservation of many of the pore-forming residues, it has been suggested that AlgE forms a highly specific translocation pathway for the negatively charged alginate polymer. On the basis of subcellular fractionation experiments using an algK deletion mutant, it has also been proposed that AlgE interacts directly with AlgK because a significant proportion of AlgE mislocalizes to the inner membrane in this mutant [29]. Lending further support to this hypothesis is the prediction that the outer-membrane proteins involved in cellulose and PNAG secretion (discussed in the following sections) each form large two-domain proteins with a periplasmic TPR-containing domain and a transmembrane β-barrel (Figure 4) [30].
At present, there is a lack of definitive evidence that the alginate biosynthesis and export proteins interact to form a trans-envelope complex, although initial studies showing that in vitro alginate polymerization requires both inner-and outer-membrane fractions suggest that this may be the case [48]. Preliminary evidence demonstrating direct interactions between AlgX, AlgK, and the regulatory protein MucD has also been reported recently [62]. Although an AlgK–AlgX interaction is logical given the involvement of the two proteins in the alginate secretion process, MucD is a homolog of E. coli HtrA/DegP and a negative regulator of alginate biosynthesis [63]. The proposed function of MucD is to degrade cell-wall stress signals that activate transcription of the algD operon [64]. The proposed role of MucD is to function upstream of algX and algK translation, so the biological significance of the MucD–AlgX–AlgK complex observed in vitro is unclear at this point. Although the individual functions of the proteins involved in the polymerization, modification, and export of alginate have been elucidated, future research needs to address which of these proteins interact with one another to facilitate the overall synthesis and secretion process.
Synthase-dependent production of cellulose
Produced by higher plants, fungi, algae, and bacteria, the β-1,4-linked D-glucose homopolymer cellulose [(β-D-Glc-(1→4)-β-D-Glc)n] is one of the most abundant polysaccharides found in nature. Despite this, little is known about the molecular mechanism of its biosynthesis and export. In bacteria, cellulose biosynthesis was first described in G. xylinus (formerly Acetobacter xylinum) and has since been described in a variety of Gram-negative bacteria including E. coli, Salmonella enterica, and Vibrio fischeri among others (Figure 2) [65–68].
Although the order of genes required for cellulose production exhibits more variability among bacteria compared to those involved in alginate production, the same core protein components involved in its biosynthesis and export appear to be conserved between the two polysaccharide secretion systems (Figures 2 and 4; Table 1). For example, the E. coli bcsA gene (acsA in G. xylinus) encodes the cellulose synthase protein. Similarly to the alginate synthase Alg8, BcsA is an inner-membrane protein with multiple transmembrane domains and a cytoplasmic family 2 glycosyl transferase domain [67,69]. BcsA is thought to both catalyze cellulose polymerization from UDP-glucose and facilitate translocation of the newly formed polymer across the inner membrane. In addition, BcsA contains a PilZ domain at its C terminus, whose c-di-GMP binding activity activates cellulose production [27]. Thus, unlike alginate secretion, in which Alg8 and Alg44 are responsible for the polymerization and c-di-GMP binding activities, respectively, in cellulose biosynthesis both of these functions are carried out by a single protein (Figures 2 and 4). The bcsB gene product localizes to the inner membrane and is required for both in vitro and in vivo cellulose production; however, its specific role in cellulose production is unclear at this point [70]. It has been proposed that the bcsC gene encodes a large outer-membrane protein that contains an N-terminal TPR-containing domain that resides in the periplasm and a C-terminal porin domain that facilitates cellulose export across the outer membrane [29]. Thus, it is thought that BcsC contains a domain architecture that resembles AlgK and AlgE from the alginate secretion system [30]. This putative function would also explain why BcsC is necessary for cellulose production in vivo but is dispensable for its production in vitro [70]. The bcsZ gene is located within the cellulose biosynthetic operon in some bacteria or elsewhere in the genome in others. bcsZ encodes a periplasmic enzyme with endo-β-1,4-glucanase activity that may be required for degradation of accumulated cellulose in the periplasm and/or cleavage of nascent cellulose chains to allow micro-fibril formation to occur outside the cell [71]. BscQ, which is homologous to the E. coli cell division protein MinD, localizes to the cell pole and thus may be required for polar localization of the cellulose biosynthesis apparatus in E. coli, S. enterica, and Burkholderia cenocepacia [72]. However, it should be noted that the authors of this study were not able to detect polar localization of the other Bcs proteins even though cellulose itself was produced at the cell pole. The AcsD protein, which is unique to G. xylinus, arranges into a homo-octomeric assembly that is capable of binding cellulose [73]. It is thought that the AcsD multimer exists extracellularly and that its function is to twist the newly synthesized glucan polymers into higher-order cellulose fibrils [73,74]. This hypothesis would also help to explain why G. xylinus produces fibrillar cellulose whereas other bacteria that lack an AcsD homolog, such as E. coli, produce amorphous cellulose.
P. fluorescens SBW25 and Pseudomonas syringae patho-var tomato DC3000 produce an acetylated form of cellulose whose production requires the wss operon (Figure 2) [75,76]. This operon is predicted to encode not only proteins involved in the secretion of non-acetylated cellulose (WssA/ BcsQ, WssB/BcsA, WssC/BcsB, WssD/BcsZ, and WssE/ BcsC) but also proteins that resemble those involved in the O-acetylation of alginate (WssG/AlgF, WssH/AlgI, and WssI/AlgJ) (Figure 4 and Table 1). WssF is also involved in the O-acetylation of cellulose; however, there does not appear to be a functionally similar protein involved in alginate O-acetylation [75]. In addition, the wss operon of P. fluorescens SBW25 contains a second MinD homolog, WssJ; it is speculated that WssJ plays a role in cellular localization of the cellulose O-acetylation proteins, as opposed to the cellulose synthase proteins, whose localization is more likely to be regulated by WssA given its similarity to BcsQ [75]. However, characterization of the acetylated cellulose produced by these bacteria is still in its infancy and requires experimental confirmation of the proposed functions for each of the proteins.
Synthase-dependent production of PNAG
The genes responsible for synthesis of PNAG have recently been identified in a number of Gram-negative bacteria including E. coli, Yersinia pestis, Actinobacillus pleuropneumoniae, and Bordetella bronchiseptica, among others (Figure 2) [77–80]. PNAG is a β-1,6-linked N-acetyl-D-glucosamine homopolymer [(β-D-GlcNAc-(1→6)-β-D-GlcNAc)n] that functions as an important component of the biofilm matrix produced by these organisms and thus contributes to their overall persistence during infection. Unlike alginate and cellulose, which are synthesized as non-acetylated polymers by their respective synthases and subsequently O-acetylated in the periplasm, PNAG is assembled as a fully N-acetylated precursor starting from a UDP-N-acetylglucosamine precursor. The PgaC protein (Y. pestis HmsR and B. bronchiseptica BpsC), similar to Alg8, BcsA, and WssB, is predicted to contain multiple transmembrane domains and a large cytoplasmic domain that shares homology with family 2 glycosyl transferases (Figure 4). Therefore, PgaC is a putative PNAG synthase that is thought to catalyze the polymerization of β-1,6-linked N-acetyl-D-glucosamine and facilitate its export across the inner membrane. The precise function of the predicted inner-membrane protein PgaD/ HmsS/BpsD is unknown; however, it is thought to play a role in the polymerization process because, similar to PgaC, its deletion abrogates the production of PNAG [81]. Although it is not predicted to contain a PilZ domain, the possibility that PgaD binds c-di-GMP to post-translationally regulate PNAG production cannot be ruled out given the diversity of protein folds that are capable of binding to the dinucleotide [82]. Once in the periplasm, the PNAG polymer is partially de-N-acetylated by the carbohydrate esterase PgaB/HmsF/BpsB. A recent structure–function characterization of E. coli PgaB has shown that it harbors a de-N-acetylase domain with low catalytic efficiency [83]. These observations corroborate the relatively low amount (~22%) of de-N-acetylation observed in vivo and has led to speculation that the differing amounts of de-N-acetylation observed among PNAG-producing bacteria are probably related to the enzyme activity of their PgaB homolog. PgaA is a predicted outer-membrane protein that is proposed to have a similar domain arrangement as BcsC/AlgK and AlgE. In the absence of PgaB, PNAG accumulates in the periplasm of E. coli, leading to speculation that the putative export function of PgaA is specific for partially de-N-acetylated (mature) PNAG [81]. Aside from PgaB, the proteins involved inPNAG polymerization and export have not been functionally characterizedtodate. Structural and functional characterization of the PgaA, PgaC, and PgaD proteins will be required to fully understand the mechanism of PNAG secretion across the cell envelope.
Additional synthase-dependent polysaccharide secretion systems
Alginate, cellulose, and PNAG are the most studied synthase-dependent secretion systems in Gram-negative bacteria, but additional polysaccharides appear to use a similar mechanism of biosynthesis and export. Pasteurella multocida produces hyaluronan (HA), which comprises repeating units of D-glucuronic acid and N-acetyl-D-glucosamine [(1→4)-β-D-GlcUA-(1→3)-β-D-GlcNAc-(1→4)] and requires hyaluronan synthase (HAS) for its production [84,85]. This synthase is the fusion of two glycosyl transferases, one with specificity towards β-D-N-acetylglucosamine and the other towards β-D-glucuronic acid. The details of HA export through the periplasm and across the outer membrane are unclear at this point so it is difficult to ascertain if the same protein components as for alginate, cellulose, and PNAG are utilized. There is also controversy as to whether HA assembly requires a lipid precursor. However, a recent study on a HAS from the Gram-positive bacterium Streptococcus equisimilis indicates that the polymerization and membrane translocation processes do not require a lipid-linked precursor in this bacterium [25].
The PEL polysaccharide, whose chemical structure has yet to be identified, is produced by some strains of P. aeruginosa and Ralstonia solanacearum [86]. Similar to alginate and cellulose, PEL is regulated post-translationally by c-di-GMP [87]. However, this regulation occurs via the degenerate GGDEF domain of PelD and not a PilZ domain as in alginate and cellulose production [88]. Therefore, the mechanism by which PelD exerts its regulation may differ from the PilZ domains of Alg44 and BcsA. In addition, the putative PEL polymerase PelF is predicted to localize to the cytoplasm and is thus unlikely to facilitate polysaccharide translocation across the inner membrane as has been proposed for Alg8 and BcsA [89]. At present, characterization of the Pel proteins is still in its early stages and it remains to be determined if PEL polysaccharide assembly occurs on a lipid carrier.
Curdlan [β-D-Glc-(1→3)-β-D-Glc)n] production by Agro-bacterium tumefaciens requires the curdlan synthase protein CrdS, which probably uses a synthase-dependent pathway given its sequence similarity to cellulose synthases [90]. However, lack of experimental characterization of this system makes it difficult to determine its mechanism of biosynthesis and secretion.
Concluding remarks
Although this review highlights some interesting commonalities between the better-studied synthase-dependent exopolysaccharide secretion systems, it is clear that more research is required to tease out the molecular details of this process. One central question that has yet to be addressed is whether or not the synthase-dependent secretion systems highlighted here form a trans-envelope multiprotein complex as observed in the Wzx/Wzy- and ABC-transporter-dependent systems [22]. Although this hypothesis seems feasible for alginate and cellulose secretion, for which there are a significant number of proteins in the periplasm that could facilitate the formation of such a complex, the apparent simplicity of the PNAG secretion system suggests that it may not have enough protein components to span the ~200-Å periplasmic space unless a constriction occurs [91].
Post-translational regulation by c-di-GMP is another process that appears widespread among synthase-dependent polysaccharide systems. However, the mechanism by which c-di-GMP exerts its effect has not been addressed for any of these systems. Is binding of c-di-GMP to its receptor a switch that stimulates the catalytic activity of its associated synthase or does it also serve to couple exo-polysaccharide polymerization to export through alteration of protein–protein interactions by an as yet unidentified allosteric mechanism? Moreover, how does the synthase itself facilitate extrusion of the nascent polymer across the inner membrane? To date, the only synthase whose polysaccharide export activity has been definitively shown is the aforementioned bacterial HAS from S. equisimilis. To address these questions and many others (Box 1), in vitro reconstitution of these systems using purified components will be required. High-resolution crystal structures of these components will also be critical, because they will help in generating mechanistic hypotheses that can then be tested experimentally.
Box 1. Outstanding questions.
How is synthase-dependent exopolysaccharide polymerization initiated? If, as proposed, some synthase-dependent secretion systems do not require a lipid acceptor molecule, then what molecular entity serves as the nucleophile for the first sugar–nucleotide precursor molecule? As has been shown for some UDP-glucosyl transferases [94], is water capable of functioning as a sugar–nucleotide acceptor?
By what mechanism does c-di-GMP post-translationally regulate exopolysaccharide polymerization? Does c-di-GMP binding to its target receptor alleviate inhibition of its associated synthase or does it allosterically activate synthase activity through conformational changes? Does c-di-GMP binding also serve to couple exopolysaccharide polymerization to export through protein–protein interactions?
How does the exopolysaccharide cross the cytoplasmic inner membrane? Are the transmembrane domains of the synthase sufficient for inner membrane transport of the polymer [25] or are additional protein components required? As described above, what is the role of c-di-GMP binding in this process?
In the alginate and acetylated cellulose systems, what is the chemical identity of the acetyl donor? Mechanistically, how is the acetate group transported across the inner membrane and transferred to the nascent polymers? Identification of this acetyl donor may also shed light on the mechanism of O-acetylation in other polysaccharides such as bacterial (peptidoglycan) and plant (xyloglucan) cell-wall polymers [95,96].
Do the inner and outer membrane components of synthase-dependent polysaccharide secretion systems interact with one another to form a trans-envelope complex as observed for OPX and PCP proteins of the Wzx/Wzy-dependent polysaccharide secretion systems?
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR #13337 and #43998) to P.L.H. J.C.W. has been supported by graduate scholarships from the Natural Science and Engineering Research Council of Canada (NSERC), Cystic Fibrosis Canada, the Ontario Graduate Scholarship Program, the Ontario Student Opportunities Trust Fund, and The Hospital for Sick Children Foundation Student Scholarship Program. P.L.H. is the recipient of a Canada Research Chair.
Footnotes
Note added in proof
Since this paper first appeared online (30 October 2012), the structure of the BcsA/BcsB inner membrane cellulose synthase complex has been determined (PDB 4HG6 [97]), and evidence that PgaC and PgaD interact with each other, and c-di-GMP to form a functional PNAG synthase complex reported [98]. The conclusions of these two studies provide strong support for the mechanisms of cellulose and PNAG secretion proposed herein.
References
- 1.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 2.Whitchurch CB, et al. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 3.Borlee BR, et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colvin KM, et al. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 6.Mah TF, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 7.Costerton JW, et al. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Mulford CA, Osborn MJ. An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc Natl Acad Sci USA. 1983;80:1159–1163. doi: 10.1073/pnas.80.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGrath BC, Osborn MJ. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:649–654. doi: 10.1128/jb.173.2.649-654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, et al. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol. 1996;178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman MF, et al. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 12.Alaimo C, et al. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 2006;25:967–976. doi: 10.1038/sj.emboj.7601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodward R, et al. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat Chem Biol. 2010;6:418–423. doi: 10.1038/nchembio.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 15.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuthbertson L, et al. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 2010;74:341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuthbertson L, et al. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morona R, et al. Sequence–structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem Sci. 2009;34:78–84. doi: 10.1016/j.tibs.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Dong C, et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tocilj A, et al. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol. 2008;15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 21.Kalynych S, et al. Structural characterization of closely related O-antigen LPS-chain length regulators. J Biol Chem. 2012;287:15696–15705. doi: 10.1074/jbc.M112.354837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins RF, et al. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cartee RT, et al. Expression of the Streptococcus pneumoniae type 3 synthase in Escherichia coli. Assembly of type 3 polysaccharide on a lipid primer. J Biol Chem. 2001;276:48831–48839. doi: 10.1074/jbc.M106481200. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto A, et al. Extraction of cellulose-synthesizing activity of Gluconacetobacter xylinus by alkylmaltoside. Carbohydr Res. 2011;346:2760–2768. doi: 10.1016/j.carres.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard C, et al. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 26.Merighi M, et al. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 27.Ryjenkov DA, et al. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 28.Jain S, Ohman DE. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keiski CL, et al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure. 2010;18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitney JC, et al. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci USA. 2011;108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz J, et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, et al. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Linker A, Jones RS. A new polysaccharide resembling alginic acid isolated from pseudomonads. J Biol Chem. 1966;241:3845–3851. [PubMed] [Google Scholar]
- 35.Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pindar DF, Bucke C. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem J. 1975;152:617–622. doi: 10.1042/bj1520617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henry RL, et al. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol. 1992;12:158–161. doi: 10.1002/ppul.1950120306. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, et al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. J Am Med Assoc. 2005;293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 40.Franklin MJ, et al. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haug A, Larsen B. Biosynthesis of alginate. II Polymannuronic acid C-5-epimerase from Azotobacter vinelandii. Carbohydr Res. 1971;17:297–308. doi: 10.1016/s0008-6215(00)82537-9. [DOI] [PubMed] [Google Scholar]
- 42.Sherbrock-Cox V, et al. The purification and chemical characterisation of the alginate present in extracellular material produced by mucoid strains of Pseudomonas aeruginosa. Carbohydr Res. 1984;135:147–154. doi: 10.1016/0008-6215(84)85012-0. [DOI] [PubMed] [Google Scholar]
- 43.Larsen B, Haug A. Biosynthesis of alginate. 1 Composition and structure of alginate produced by Azotobacter vinelandii. Carbohydr Res. 1971;17:287–296. doi: 10.1016/s0008-6215(00)82536-7. [DOI] [PubMed] [Google Scholar]
- 44.Pier GB, et al. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skjak-Braek G, et al. Effect of acetylation on some solution and gelling properties of alginates. Carbohydr Res. 1989;185:131–138. [Google Scholar]
- 46.Nivens DE, et al. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oglesby LL, et al. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remminghorst U, Rehm BH. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol. 2009;12:512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Jain S, et al. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol. 2003;47:1123–1133. doi: 10.1046/j.1365-2958.2003.03361.x. [DOI] [PubMed] [Google Scholar]
- 51.Franklin MJ, Ohman DE. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993;175:5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin MJ, Ohman DE. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J Bacteriol. 1996;178:2186–2195. doi: 10.1128/jb.178.8.2186-2195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franklin MJ, Ohman DE. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3000–3007. doi: 10.1128/JB.184.11.3000-3007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franklin MJ, et al. Evidence that the algI/algJ gene cassette, required for O acetylation of Pseudomonas aeruginosa alginate, evolved by lateral gene transfer. J Bacteriol. 2004;186:4759–4773. doi: 10.1128/JB.186.14.4759-4773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robles-Price A, et al. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 2004;186:7369–7377. doi: 10.1128/JB.186.21.7369-7377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weadge JT, et al. Expression, purification, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgX. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2010;66:588–591. doi: 10.1107/S1744309110011851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jain S, Ohman DE. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect Immun. 2005;73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfram F, et al. Expression, purification, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgL. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2012;68:584–587. doi: 10.1107/S1744309112012808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Edqvist PJ, et al. Tetratricopeptide repeats in the type III secretion chaperone, LcrH: their role in substrate binding and secretion. Mol Microbiol. 2006;59:31–44. doi: 10.1111/j.1365-2958.2005.04923.x. [DOI] [PubMed] [Google Scholar]
- 61.Kim KH, et al. Crystal structure of beta-barrel assembly machinery BamCD protein complex. J Biol Chem. 2011;286:39116–39121. doi: 10.1074/jbc.M111.298166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hay ID, et al. Identification of a periplasmic AlgK–AlgX–MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol. 2012;93:215–227. doi: 10.1007/s00253-011-3430-0. [DOI] [PubMed] [Google Scholar]
- 63.Wood LF, Ohman DE. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J Bacteriol. 2006;188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 65.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 66.Hestrin S, et al. Synthesis of cellulose by resting cells of Acetobacter xylinum. Nature. 1947;159:64. doi: 10.1038/159064a0. [DOI] [PubMed] [Google Scholar]
- 67.Zogaj X, et al. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 68.Bassis CM, Visick KL. The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J Bacteriol. 2010;192:1269–1278. doi: 10.1128/JB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saxena IM, et al. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong HC, et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazur O, Zimmer J. Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ. J Biol Chem. 2011;286:17601–17606. doi: 10.1074/jbc.M111.227660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Quere B, Ghigo JM. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol Microbiol. 2009;72:724–740. doi: 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- 73.Hu SQ, et al. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci USA. 2010;107:17957–17961. doi: 10.1073/pnas.1000601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saxena IM, et al. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spiers AJ, et al. Biofilm formation at the air–liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol. 2003;50:15–27. doi: 10.1046/j.1365-2958.2003.03670.x. [DOI] [PubMed] [Google Scholar]
- 76.Ude S, et al. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol. 2006;8:1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, et al. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaplan JB, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lillard JW, Jr, et al. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 80.Parise G, et al. Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol. 2007;189:750–760. doi: 10.1128/JB.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Itoh Y, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin, poly-β-1,6-N-acetyl-D-glucosamine (PGA) J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ryan RP, et al. When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 2012;20:235–242. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 83.Little DJ, et al. The structure and metal dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-D-glucosamine. J Biol Chem. 2012;287:31126–31137. doi: 10.1074/jbc.M112.390005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tracy BS, et al. Acceptor specificity of the Pasteurella hyaluronan and chondroitin synthases and production of chimeric glycosaminoglycans. J Biol Chem. 2007;282:337–344. doi: 10.1074/jbc.M607569200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weigel PH, DeAngelis PL. Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem. 2007;282:36777–36781. doi: 10.1074/jbc.R700036200. [DOI] [PubMed] [Google Scholar]
- 86.Vasseur P, et al. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 87.Lee VT, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whitney JC, et al. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem. 2012;287:23582–23593. doi: 10.1074/jbc.M112.375378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Franklin MJ, et al. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stasinopoulos SJ, et al. Detection of two loci involved in (1→3)-β-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology. 1999;9:31–41. doi: 10.1093/glycob/9.1.31. [DOI] [PubMed] [Google Scholar]
- 91.Matias VR, et al. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramelot TA, et al. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 93.Yum S, et al. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol. 2009;387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 94.Ciesla WP, Jr , Bobak DA. Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J Biol Chem. 1998;273:16021–16026. doi: 10.1074/jbc.273.26.16021. [DOI] [PubMed] [Google Scholar]
- 95.Gille S, Pauly M. O-Acetylation of plant cell wall polysaccharides. Front Plant Sci. 2012;3:12. doi: 10.3389/fpls.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 97.Morgan JL, et al. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steiner S, et al. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein–protein interaction. EMBO J. 2012 doi: 10.1038/emboj.2012.315. In press. http://dx.doi.org/10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed]