Abstract
Increasing evidence has demonstrated that microRNAs (miRNAs) are involved in colon cancer initiation and progression, and may serve as diagnostic and prognostic biomarkers for colon cancer. Here, we investigated the levels of miR-9-1, miR-203-3p, miR-221-3p, miR-342-3p, miR-491-5p and miR-503-5p in 90 pairs of colon cancer and adjacent normal tissues, and explored the relationship between their expression and clinical outcome of colon cancer. Five miRNAs (miR-203-3p, miR-221-3p, miR-342-3p, miR-491-5p and miR-503-5p) were dysregulated in colon cancer tissue (P < 0.05). The levels of miR-503-5p in larger tumors (≥ 6 cm) were higher than those in smaller ones (< 6 cm) (P = 0.031), while the levels of miR-203-3p and miR-491-5p in patients aged 70 years and older were higher than those in patients aged younger than 70 years (P = 0.019 and 0.049, respectively). The high levels of miR-221-3p (HR = 2.416, 95% CI 1.314-4.445, P = 0.005), miR-342-3p (HR = 1.807, 95% CI 1.003-3.253, P = 0.049) and miR-491-5p (HR = 1.868, 95% CI 1.032-3.384, P = 0.039) were significantly associated with worse survival time. Moreover, combination analysis of miR-221-3p, miR-342-3p and miR-491-5p expression revealed that patients with 3 highly expressed miRNAs had lower survival rates compared with those with zero-to-two highly expressed miRNAs (HR = 2.100, 95% CI 1.157-3.813, P = 0.015), especially those with TNM stages I and II (HR = 4.204,95% CI 1.762-10.030, P = 0.001). Our results suggest that the three-miRNA signature may help doctors better predict prognosis and guide treatment decisions for colon cancer.
Keywords: miR-221-3p, miR-342-3p, miR-491-5p, prognosis, colon cancer
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide, accounting for 10% of all new cancer cases and over 1.23 million deaths per year [1]. China is one of countries with high incidence of CRC, in which CRC ranks third among all cancer sites in cancer incidence and fifth in cancer mortality [2]. In addition, CRC incidence and mortality rates are continuing to rise in China. For example, in Shanghai, CRC incidence rate increases annually by 4.2% since 1973 [3]. The prognosis of early-stageCRC is usually favorable, but nearly two-thirds of CRC cases have spread to adjacent or distant organs when diagnosed [4]. In recent years, significant progress has been made in the molecular pathogenesis and clinical treatment of CRC, but advanced CRC is still extremely difficult to treat. There is no substantial improvement in the prognosis of the majority of CRC patients, especially those in the later stages of the disease. CRC is rapidly becoming a main public health problem due to its high incidenceand mortality rates in China.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNA molecules that regulate gene expression at the post-transcriptional level. To date, more than 2500 miRNAs have been identified in human (miRBase database 20.0) [5]. It is assumed that a given miRNA can target hundreds of target genes and a gene can be targeted by multiple miRNAs [6]. miRNAs are involved in almost every biological process, such as cell proliferation, apoptosis and tumorigenesis. Dysregulation of miRNAs contributes to diverse diseases, including cancer [7]. miRNAs act as oncogenes and tumor suppressors in cancer initiation, progression and metastasis [8-12], and affect response to treatment and prognosis of cancer patients [11-18]. For example, Zhang et al. [13] identified six-miRNA signature in 178 patients with stage II colon cancer that can be used to predict the risk of the recurrence in patients and determine whether or not patients can benefit from chemotherapy. Chen et al. [12] found that miR-214 was downregulated in CRC with liver metastasis, leading to the upregulation of FGFR1 and the promotion of liver metastasis. Furthermore, FGFR1 is one of hot targets for anticancer drug development [19]. As a crucial factor in tumor occurrence and development, miRNA is likely to be new breakthroughs in cancer diagnosis and therapy.
Gene expression analysis can help improve the current understanding of miRNA function in cancer and may be used to develop predictive markers for patient classification. In this study, we investigated the levels of six miRNAs (miR-9-1, miR-203-3p, miR-221-3p, miR-342-3p, miR-491-5p and miR-503-5p) in 90 pairs of colon cancer and adjacent normal tissues, and evaluated the relationship between miRNA expression and clinical features and prognosis of patients with colon cancer.
Materials and methods
Patients and samples
A total of 90 pairs of colon cancer and paracancerous tissues were selected from the Biobank of National Engineer Center for Biochip at Shanghai. Patients were pathologically diagnosed with confirmed primary colon cancer during July 2006 and May 2007. Furthermore, all patients did not receive anti-cancer treatment, including chemotherapy and radiotherapy, before surgery. Ninety matched adjacent normal tissues were collected at sites more than 5 cm away from the tumor margin. The end point of the study was overall survival. All patients gave written informed consent according to the protocol approved by the Committees of National Engineer Center for Biochip at Shanghai and Tongren Hospital.
RNA isolation
Total RNA was isolated from the FFPE tissues by using the RecoverAllTM Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The concentration and quality of the isolated RNA were assessed on a NanoDrop ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE). All RNA samples were stored at -80°C until subsequent use.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Reverse transcription was performed with the miScript Reverse Transcription Kit (Qiagen, Hilden, Germany) in a final volume of 20μl containing 6μl of total RNA, 4 μl of 5× miScript RT buffer and 1 μl of miScript Reverse Transcriptase Mix. The reaction mixtures were incubated at 37°C for 60 min, at 95°C for 5 min, and then held at 4°C. Quantitative RT-PCR was carried out using the miScript SYBR Green PCR kit (Qiagen, Hilden, Germany) on an ABI 7900 Real-time PCR system (Applied Biosystems, CA, USA). Each qRT-PCR reaction was performed in a final volume of 20 μl containing 1× QuantiTect SYBR Green PCR Master mix (Qiagen, Hilden, Germany), 2 μl of the cDNA, 0.5 mM of each primer. The reaction mixtures were incubated at 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. All reactions were carried out in triplicate. The expression levels of miRNAs were normalized to the endogenous control RNU6B, and were calculated with the formula 2-ΔΔCt.
Statistical analyses
Comparison of miRNA levels between colon cancer and adjacent normal tissues was using a Student’s t test. The nonparametric test Mann-Whitney-Wilcoxon was used to assess the relationship between miRNA levels and other characteristics. The associations between miRNA levels and continuous variables were tested with the Spearman rank correlation. The expression levels of miRNA were divided into 2 groups (high and low expression groups) according to the median levels, respectively.The chi-square test was used to assess the association of miRNA expression with clinicopathologic characteristics. Survival analysis was performed using the Kaplan-Meier method, and differences between the groups were tested using the log rank test. Hazard ratio (HR) and 95% confidence intervals (CIs) were calculated using the Cox proportional hazard model. All analyses were two-sided, and a P value of less than 0.05 was considered statistically significant. All statistical analyses were performed using the SPSS software package (version 19.0, SPSS Inc., Chicago, IL, USA).
Results
Expression levels of miRNAs in colon cancer
There was no difference in miR-9-1 expression between cancer and normal tissues. Five miRNAs (miR-203-3p, miR-221-3p, miR-342-3p, miR-491-5p and miR-503-5p) were upregulated in colon cancer tissues compared with paracancerous tissues (P < 0.05) (Figure 1). In addition, there was positive correlation between miR-503-5p level and tumor size (P = 0.017). The levels of miR-503-5p in larger tumors (≥ 6 cm) were higher than those in smaller ones (< 6 cm) (P = 0.031) (Figure S1). Although miR-203-3p and miR-491-5p expression were not correlated with age, the levels of miR-203-3p and miR-491-5p in patients aged 70 years and older were higher than those in patients aged younger than 70 years (P = 0.019 and 0.049, respectively) (Figure S2).
Figure 1.

The expression levels of miR-9-1 (A), miR-203-3p (B), miR-221-3p (C), miR-342-3p (D), miR-491-5p (E) and miR-503-5p (F) in 90 pairs of colon cancer and adjacent normal tissues.
Associations of miRNA expression with clinicopathologic characteristics
Ninety patients were divided into two groups based on the expression levels of miRNAs (i.e., low and high expression groups). The expression level of miR-203-3p was significantly associated with histology grade (P = 0.016) (Table 1). No other difference was observed between miRNAs expression and clinicopathologic characteristics.
Table 1.
Association between miRNAs expression and the clinicopathological features
| Characteristics | miR-9-1 | miR-203-3p | miR-221-3p | ||||||
|
|
|
|
|||||||
| Low | High | P value | Low | High | P value | Low | High | P value | |
|
| |||||||||
| Age (years) | |||||||||
| ≥ 70 | 21 (46.7) | 25 (58.1) | 0.296 | 18 (40.9) | 28 (63.6) | 0.054 | 19 (43.2) | 27 (61.4) | 0.135 |
| < 70 | 24 (53.3) | 18 (41.9) | 26 (59.1) | 16 (36.4) | 25 (56.8) | 17 (38.6) | |||
| Sex | |||||||||
| Male | 23 (51.1) | 24 (53.3) | 1.000 | 27 (60.0) | 20 (44.4) | 0.205 | 25 (55.6) | 22 (48.9) | 0.673 |
| Female | 22 (48.9) | 21 (46.7) | 18 (40.0) | 25 (55.6) | 20 (44.4) | 23 (51.1) | |||
| Tumor size (cm) | |||||||||
| ≥ 6 | 18 (40.9) | 16 (36.4) | 0.827 | 14 (31.8) | 20 (45.5) | 0.280 | 14 (31.8) | 20 (45.5) | 0.274 |
| < 6 | 26 (59.1) | 28 (63.6) | 29 (67.4) | 25 (55.6) | 30 (68.2) | 24 (54.5) | |||
| Histology grade | |||||||||
| well | 1 (2.2) | 4 (8.9) | 0.371 | 0 (0) | 5 (11.1) | 0.016 | 3 (6.7) | 2 (4.4) | 0.781 |
| moderate | 26 (57.8) | 23 (51.1) | 22 (48.9) | 27 (60.0) | 23 (51.1) | 26 (57.8) | |||
| poor | 18 (40.0) | 18 (40.0) | 23 (51.1) | 13 (28.9) | 19 (42.2) | 17 (37.8) | |||
| LNM | |||||||||
| no | 19 (42.2) | 15 (33.3) | 0.515 | 15 (33.3) | 19 (42.2) | 0.515 | 13 (28.9) | 21 (46.7) | 0.127 |
| yes | 26 (57.8) | 30 (66.7) | 30 (66.7) | 26 (57.8) | 32 (71.1) | 24 (53.3) | |||
| TNM stage | |||||||||
| I | 6 (13.3) | 2 (4.5) | 0.317 | 3 (6.8) | 5 (11.4) | 0.566 | 5 (11.1) | 3 (6.8) | 0.317 |
| II | 20 (44.4) | 27 (61.4) | 27 (60.0) | 20 (45.5) | 27 (60.0) | 20 (45.5) | |||
| III | 18 (40.0) | 14 (31.8) | 14 (31.1) | 18 (40.9) | 12 (26.7) | 20 (45.5) | |||
| IV | 1 (2.2) | 1 (2.3) | 1 (2.2) | 1 (2.3) | 1 (2.2) | 1 (2.3) | |||
|
| |||||||||
| Characteristics | miR-342-3p | miR-491-5p | miR-503-5p | ||||||
|
|
|
|
|||||||
| Low | High | P value | Low | High | P value | Low | High | P value | |
|
| |||||||||
| Age (years) | |||||||||
| ≥ 70 | 20 (45.5) | 26 (59.1) | 0.286 | 19 (43.2) | 27 (61.4) | 0.135 | 19 (44.2) | 27 (60.0) | 0.200 |
| < 70 | 24 (54.5) | 18 (40.9) | 25 (56.8) | 17 (38.6) | 24 (55.8) | 18 (40.0) | |||
| Sex | |||||||||
| Male | 21 (46.7) | 26 (57.8) | 0.399 | 22 (48.9) | 25 (55.6) | 0.673 | 23 (51.1) | 24 (53.3) | 1.000 |
| Female | 24 (53.3) | 19 (42.2) | 23 (51.1) | 20 (44.4) | 22 (48.9) | 21 (46.7) | |||
| Tumor size (cm) | |||||||||
| ≥ 6 | 16 (36.4) | 18 (40.9) | 0.829 | 13 (30.2) | 21 (46.7) | 0.280 | 13 (30.2) | 21 (46.7) | 0.130 |
| < 6 | 27 (62.8) | 27 (62.8) | 30 (69.8) | 24 (53.3) | 20 (69.8) | 24 (53.3) | |||
| Histology grade | |||||||||
| well | 2 (4.4) | 3 (6.7) | 0.896 | 3 (6.7) | 2 (4.4) | 0.896 | 2 (6.7) | 2 (4.4) | 0.847 |
| moderate | 25 (55.6) | 24 (53.3) | 24 (53.3) | 25 (55.6) | 25 (55.6) | 24 (53.3) | |||
| poor | 18 (40.0) | 18 (40.0) | 18 (40.0) | 18 (40.0) | 17 (37.8) | 19 (42.2) | |||
| LNM | |||||||||
| no | 16 (35.6) | 18 (40.0) | 0.828 | 16 (35.6) | 18 (40.0) | 0.828 | 16 (35.6) | 18 (40.0) | 0.828 |
| yes | 29 (64.4) | 27 (60.0) | 29 (64.4) | 27 (60.0) | 29 (64.4) | 27 (60.0) | |||
| TNM stage | |||||||||
| I | 5 (11.1) | 3 (6.8) | 0.888 | 6 (13.3) | 2 (4.5) | 0.545 | 5 (11.1) | 3 (6.8) | 0.888 |
| II | 24 (53.3) | 23 (52.2) | 23 (51.1) | 24 (54.5) | 24 (53.3) | 23 (52.2) | |||
| III | 15 (33.3) | 17 (38.6) | 15 (33.3) | 17 (38.6) | 15 (33.3) | 17 (38.6) | |||
| IV | 1 (2.2) | 1 (2.3) | 1 (2.2) | 1 (2.3) | 1 (2.2) | 1 (2.3) | |||
Survival analyses
During follow-up (median: 27 months; range 3 to 85 months), 46 (51.1%) of 90 patients died. The median survival time was 65.5 months. The 3-, 5- and 7-year survival rates were 61.1%, 48.5% and 48.5%, respectively. Survival analyses showed that high levels of miR-221-3p (HR = 2.416, 95% CI 1.314-4.445, P = 0.005), miR-342-3p (HR = 1.807, 95% CI 1.003-3.253, P = 0.049) and miR-491-5p (HR = 1.868, 95% CI 1.032-3.384, P = 0.039) were significantly associated with worse survival time (Table 2) (Figure 2). However, only miR-221-3p expression remain significantly after adjusted for TNM and LNM (HR = 2.043, 95% CI 1.095-3.812, P = 0.025). Moreover, combination analysis of miR-221-3p, miR-342-3p and miR-491-5p expression revealed that patients with zero-to-two highly expressed miRNAs had a 60 percent 7-year overall survival rate, whereas patients with 3 highly expressed miRNAs had a 28 percent 7-year overall survival rate (adjusted HR = 2.100, 95% CI 1.157-3.813, P = 0.015) (Table 3).
Table 2.
Univariate and multivariate Cox regression analysis of overall survival in patients with colon cancer
| Features | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| age (years), ≥ 70 vs. < 70 | 1.755 (0.941-3.273) | 0.077 | ||
| sex, male v female | 1.203 (0.672-2.156) | 0.534 | ||
| tumor size (cm), ≥ 6 vs. < 6 | 0.761 (0.423-1.370) | 0.363 | ||
| tumor differentiation, poor vs well, moderate | 0.640 (0.358-1.144) | 0.132 | ||
| TNM stage, III+IV vs. I+II | 2.925 (1.628-5.258) | < 0.001 | 4.684 (1.292-16.981) | 0.019 |
| LNM, yes vs. no | 2.990 (1.664-5.374) | < 0.001 | 2.170 (0.473-9.960) | 0.319 |
| miR-9-1, high vs. low | 1.481 (0.826-2.653) | 0.187 | ||
| miR-203-3p high vs. low | 1.384 (0.774-2.475) | 0.273 | ||
| miR-221-3p, high vs. low | 2.416 (1.314-4.445) | 0.005 | 2.043 (1.095-3.812) | 0.025 |
| miR-342-3p, high vs. low | 1.807 (1.003-3.253) | 0.049 | 1.625 (0.895-2.950) | 0.110 |
| miR-491-5p, high vs. low | 1.868 (1.032-3.384) | 0.039 | 1.605 (0.878-2.934) | 0.125 |
Figure 2.
The expression levels of miR-221-3p, miR-342-3p and miR-491-5p in association with outcome in 90 patients. Kaplan-Meier curves of the overall survival for patients depending on miR-221-3p, miR-342-3p and miR-491-5p expression. A, miR-221-3p. B, miR-342-3p. C, miR-491-5p. D & E, combinatory effect of miR-221-3p, miR-342-3p and miR-491-5p expression.
Table 3.
Combined Effects of miR-221-3p, miR-342-3p and miR-491-5p expression on survival time of patients with colon cancer
| No. of highly expressed genes | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| 0 | 1 | 1 | ||
| 1 | 0.377 (0.086-1.660) | 0.197 | 0.363 (0.082-1.602) | 0.181 |
| 2 | 1.385 (0.559-3.433) | 0.482 | 1.186 (0.478-2.942) | 0.714 |
| 3 | 2.267 (1.163-4.420) | 0.016 | 1.907 (0.964-3.772) | 0.064 |
| 0-2 | 1 | 1 | ||
| 3 | 2.396 (1.339-4.288) | 0.003 | 2.100 (1.157-3.813) | 0.015 |
We further performed stratification analyses on clinicopathologic characteristics. Significant associations between miRNA expression and overall survival were found in patients with TNM stages I and II. The high levels of miR-221-3p (HR = 2.796, 95% CI 1.157-6.760, P = 0.022), miR-342-3p (HR = 2.929, 95% CI 1.180-7.266, P = 0.020) and miR-491-5p (HR = 2.836, 95% CI 1.143-7.038, P = 0.025) were inversely associated with overall survival (Figure 3). In addition, patients with 3 highly expressed miRNAs displayed a poorer survival compared with those with zero-to-two highly expressed miRNAs (HR = 4.204, 95% CI 1.762-10.030, P = 0.001).
Figure 3.
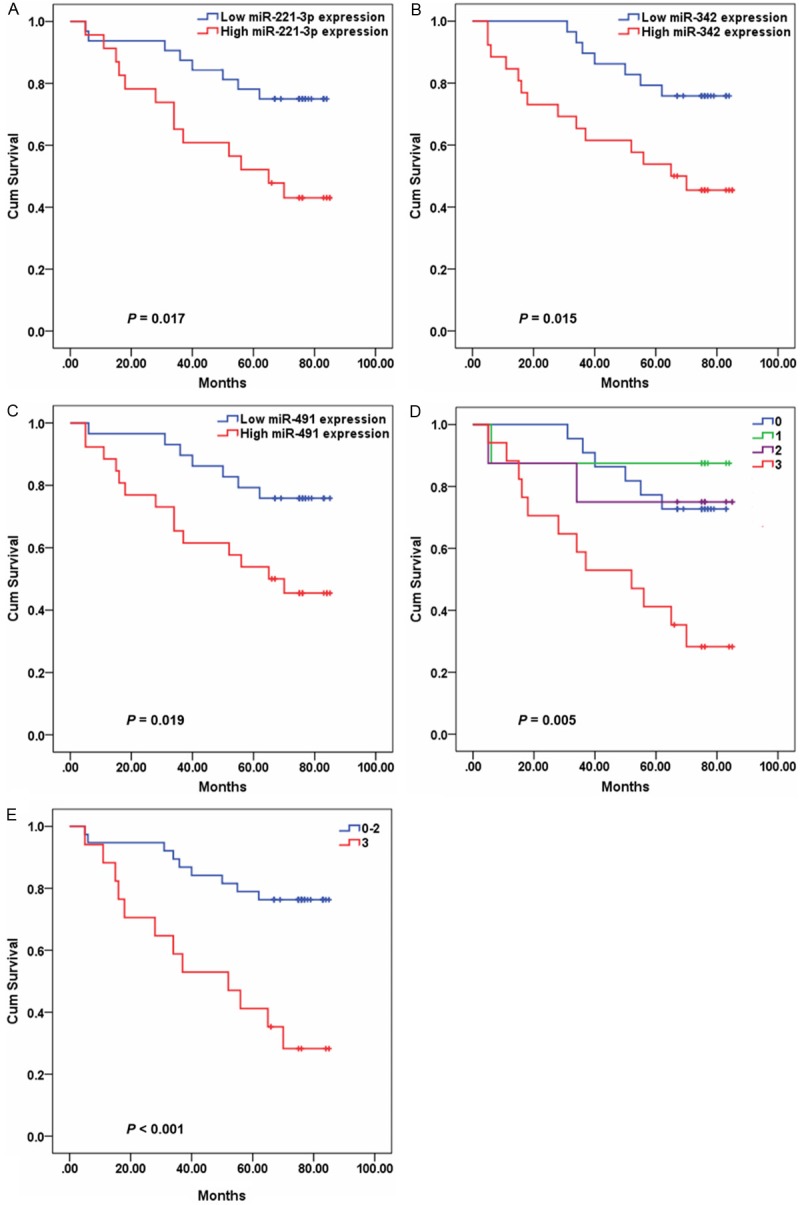
The expression levels of miR-221-3p, miR-342-3p and miR-491-5p in association with clinical outcome in 55 patients with TNM stages I and II. Kaplan-Meier curves of the overall survival for patients depending on miR-221-3p, miR-342-3p and miR-491-5p expression. A, miR-221-3p. B, miR-342-3p. C, miR-491-5p. D & E, combinatory effect of miR-221-3p, miR-342-3p and miR-491-5p expression.
Discussion
It is widely accepted that CRC is a group of diverse heterogeneous diseases arising through various molecular pathways. This heterogeneity determines tumor prognosis and response to therapy and brings great challenges not only in studying the molecular basis of the disease, but also in clinical patient management. To date, there are some molecular markers, such as microsatellite instability, the mutation status of BRAF, KRAS and PICK3CA, that are in use for treatment decisions and patient stratification. However, patient groups classified by these markers differ remarkably in their response to treatment. There is a strong need to identify novel biomarkers that better define CRC patients whose cancer is aggressive and lethal. In this study, we found that 5 miRNAs were overexpressed in human colon cancer tissues, and the expression levels of miR-221-3p, miR-342-3p and miR-491-5p affected the prognosis of patients with colon cancer.
Dysregulation of miR-221 has been reported in various types of cancer such as CRC [20,21], prostate [22] and pancreatic cancers [10]. Overexpression of miR-221 promotes many types of cancer cell proliferation, migration and invasion [10,20,21,23,24], but inhibits prostate cancer cell growth and invasion and induces apoptosis [22]. Accumulating evidence has indicated that dysregulation of miR-221 plays an important role in the development and progression of CRC. miR-221 demonstrated an oncogenic function in CRC [20,21,25]. Overexpression of miR-221 promoted CRC cell invasion and metastasis by targeting CDKN1C and RECK [20,21,25]. Anti-miR-221 improves the sensitivity of CRC cells to radiation in vivo by upregulation of PTEN, indicating that miR-221 is an important regulator of the radiation response [17]. In this study, we also demonstrated that miR-221-3p was upregulated in colon cancer, and had potential as a prognostic biomarker. However, in contradiction to the above mentioned results, Yuan et al. [26] found that the passenger strand of miR-221 was down-regulated in CRC and exhibit tumor suppressor-like properties.
miR-491 functions as a tumor suppressor gene in vitro, and is downregulated in several cancers, including oral squamous cell carcinoma (OSCC), pancreatic cancer, glioblastoma, breast cancer and hepatocellular carcinoma (HCC) [27-31]. Zhou et al. [31] found that miR-491 suppressed the metastasis of HCC by inhibiting the expression of matrix metalloproteinase and epithelial to mesenchymal transition. A new study revealed that arsenic trioxide restored the expression of miR-491 via demethylation, resulting in inhibition of the migration/invasion potential of liver cancer cells [18]. In colon cancer cell line DLD-1, miR-491 inhibits Bcl-XL expression and induces apoptosis [9]. Furthermore, miR-491-5p, one mature form of miR-491, suppresses the growth of OSCC, pancreatic cancer and glioblastoma by targeting GIT1, TP53, EGFR, CDK6 and Bcl-XL genes [27-29]. However, in the present study, we found that the level of miR-495-5p was increased in colon cancer, especially in patients aged 70 years and older. High miR-491-5p expression correlated with poor overall survival of patients with colon cancer, which was not in agreement with previous report that OSCC patients with low miR-491-5p expression had poor prognosis [27]. Furthermore, we also found that there was no difference in miR-491-5p expression between colon cancer and adjacent normal tissues in patients aged younger than 70 years (data not shown). These result indicated that there may exist different mechanismof miR-495-5p in older patients with colon cancer. It has been demonstrated that miRNAs act as oncogenes or tumor suppressors in a context dependent manner [32], even in the same type of cancer [33]. Moreover, Li et al. [29] found that miR-491-5p and miR-491-3p targeted different genes and may have different roles in glioblastoma. Further studies are necessary to shed light on the complex role of miR-491-5p in colon cancer.
Downregulation of miR-342 is not only observed in leukemia [34], lung adenocarcinoma [35], CRC [8,36] and breast cancer [37,38], but also affects the sensitivity of cancer cell to anti-cancer drugs [34,39]. In CRC, overexpression of miR-342 leads to a dramatic reduction of DNMT1 expression and thus reactivates ADAM23, Hint1, RASSF1A and RECK, which inhibit cell proliferation and invasion [8]. Furthermore, miR-342 expression has been reported to be associated with the prognosis of breast cancer [37]. However, contrary to the above mentioned results, we found that the level of miR-342-3p was weak but significantly increased in colon cancer, and was inversely associated with the prognosis of patients with colon cancer, which was similar to miR-491-5p. Grady et al. [36] reported that hypermethylation of miR-342 was found in 86% of CRC and in 67% of adenomas, while a recent study reported than only 32.6% of CRC were hypermethylated [40]. These results indicate that inter-tumor heterogenicity exist in CRC and miR-342-3p may play distinct role in CRC cells with different driver genes.
In conclusion, we showed for the first time in clinical specimens that patients with high miR-221-3p, miR-342-3p and miR-491-5p expression had higher risk of death, especially those with TNM stages I and II. Since most patients were 70 years or older in our study, further comprehensive studies are needed to validate these results and to establish them as markers for use in the diagnosis and prognosis of patients with colon cancer.
Acknowledgements
This work was supported by the ‘New 100 Talents Project’ of Shanghai Municipal Public Health Bureau, China (grant No. XBR2013124), and the Fund for International Scientific Cooperation of Shanghai Committee of Science and Technology, China (grant No. 13440701500).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.He J, Chen W. 2012 Chinese Cancer Registry Annual Report. Military Medical Science Press. 2012:27–31. [Google Scholar]
- 3.Wan DS. Epidemiologic trend of and strategies for colorectal cancer. Chin J Cancer. 2009;28:897–902. doi: 10.5732/cjc.008.10833. [DOI] [PubMed] [Google Scholar]
- 4.Hegde SR, Sun W, Lynch JP. Systemic and targeted therapy for advanced colon cancer. Expert Rev Gastroenterol Hepatol. 2008;2:135–149. doi: 10.1586/17474124.2.1.135. [DOI] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu R, Pan Z, Kang T, Huang W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis. 2011;32:1033–1042. doi: 10.1093/carcin/bgr081. [DOI] [PubMed] [Google Scholar]
- 9.Nakano H, Miyazawa T, Kinoshita K, Yamada Y, Yoshida T. Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal cancer cells. Int J Cancer. 2010;127:1072–1080. doi: 10.1002/ijc.25143. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, Wang FH, Chiao LJ, Pelicano H, Huang P, Li YH, Xu RH. Identification of miR-214 as a negative regulator of colorectal cancer liver metastasis via regulation of FGFR1 expression. Hepatology. 2014 doi: 10.1002/hep.27118. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, Zhao HW, Chen W, He YL, Wang HY, Xie D, Luo JH. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Duan B, Jiang L, Lin M, Sheng H, Huang J, Gao H. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res. 2013;6:71–77. [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41:1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 17.Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World J Gastroenterol. 2013;19:9307–9317. doi: 10.3748/wjg.v19.i48.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Jiang F, Mu J, Ye X, Si L, Ning S, Li Z, Li Y. Arsenic trioxide attenuates the invasion potential of human liver cancer cells through the demethylation-activated microRNA-491. Toxicol Lett. 2014;227:75–83. doi: 10.1016/j.toxlet.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, Cortes J, Iafrate AJ, Nuciforo P, Tabernero J. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25:552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun K, Wang W, Zeng JJ, Wu CT, Lei ST, Li GX. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;32:375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Luo M. MicroRNA-221 promotes colorectal cancer cell invasion and metastasis by targeting RECK. FEBS Lett. 2014;588:99–104. doi: 10.1016/j.febslet.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Strobel P, Gessler M, Riedmiller H, Spahn M. Survival in Patients with High-Risk Prostate Cancer Is Predicted by miR-221, Which Regulates Proliferation, Apoptosis, and Invasion of Prostate Cancer Cells by Inhibiting IRF2 and SOCS3. Cancer Res. 2014;74:2591–2603. doi: 10.1158/0008-5472.CAN-13-1606. [DOI] [PubMed] [Google Scholar]
- 23.Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang L, Yang A, Jia L. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47:268–73. doi: 10.5483/BMBRep.2014.47.5.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Martino MT, Gulla A, Cantafio ME, Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F, Cannataro M, Neri A, Giordano A, Tagliaferri P, Tassone P. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget. 2013;4:242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Sun K, Wu CT, Lei ST, Zeng JJ, Wu YJ, Li GX. [Effect of miR-221-specific inhibitor on the proliferation and apoptosis of human colorectal carcinoma cells] . Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:674–677. [PubMed] [Google Scholar]
- 26.Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, Wu M. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145:853–864. e859. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, Juang JL, Wang HC, Cheng AJ, Lu YC, Wang LH. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751–764. doi: 10.1158/0008-5472.CAN-13-1297. [DOI] [PubMed] [Google Scholar]
- 28.Guo R, Wang Y, Shi WY, Liu B, Hou SQ, Liu L. MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules. 2012;17:14733–14747. doi: 10.3390/molecules171214733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Liu Y, Granberg KJ, Wang Q, Moore LM, Ji P, Gumin J, Sulman EP, Calin GA, Haapasalo H, Nykter M, Shmulevich I, Fuller GN, Lang FF, Zhang W. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2014 doi: 10.1038/onc.2014.98. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leivonen SK, Sahlberg KK, Makela R, Due EU, Kallioniemi O, Borresen-Dale AL, Perala M. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104. doi: 10.1016/j.molonc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Li Y, Ye J, Jiang R, Yan H, Yang X, Liu Q, Zhang J. MicroRNA-491 is involved in metastasis of hepatocellular carcinoma by inhibitions of matrix metalloproteinase and epithelial to mesenchymal transition. Liver Int. 2013;33:1271–1280. doi: 10.1111/liv.12190. [DOI] [PubMed] [Google Scholar]
- 32.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Si Q, Xiao S, Xie Q, Lin J, Wang C, Chen L, Chen Q, Wang L. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30:353. doi: 10.1007/s12032-012-0353-2. [DOI] [PubMed] [Google Scholar]
- 34.De Marchis ML, Ballarino M, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU. 1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23:856–862. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- 35.Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23:1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 36.Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, Kroh EM, Allen A, Fritz BR, Markowitz SD, Tewari M. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- 37.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 38.Van der Auwera I, Limame R, van Dam P, Vermeulen PB, Dirix LY, Van Laere SJ. Integrated miRNA and mRNA expression profiling of the inflammatory breast cancer subtype. Br J Cancer. 2010;103:532–541. doi: 10.1038/sj.bjc.6605787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crippa E, Lusa L, De Cecco L, Marchesi E, Calin GA, Radice P, Manoukian S, Peissel B, Daidone MG, Gariboldi M, Pierotti MA. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS One. 2014;9:e87039. doi: 10.1371/journal.pone.0087039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva TD, Vidigal VM, Felipe AV, JM DEL, Neto RA, Saad SS, Forones NM. DNA methylationas an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6:1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



